Abstract
Due to the high level of heterogeneity and mutations inherent in human cancers, single agent therapies, or combination regimens which target the same pathway, are likely to fail. Emphasis must be placed upon the inhibition of pathways that are responsible for intrinsic and/or adaptive resistance to therapy. An active field of investigation is the development and testing of DNA repair inhibitors that promote the action of, and prevent resistance to, commonly used chemotherapy and radiotherapy. We used a novel protocol to evaluate the effectiveness of BRCA2 inhibition as a means to sensitize tumor cells to the DNA damaging drug cisplatin. Tumor cell metabolism (acidification and respiration) was monitored in real-time for a period of 72 hr to delineate treatment effectiveness on a minute by minute basis. In combination, we performed an assessment of metastatic frequency using a chicken embryo chorioallantoic membrane (CAM) model of extravasation and invasion. This protocol addresses some of the weaknesses of commonly used in vitro and in vivo methods to evaluate novel cancer therapy regimens. It can be used in addition to common methods such as cell proliferation assays, cell death assays, and in vivo murine xenograft studies, to more closely discriminate amongst candidate targets and agents, and select only the most promising candidates for further development.
Keywords: Medicine, Issue 96, chicken embryo chorio-allantoic membrane model, real-time metabolic monitoring, anti-cancer drug testing, pre-clinical development, DNA repair
Introduction
Acquired resistance to targeted and/or cytotoxic cancer treatment is an important clinical problem that can lead to treatment failure, relapse, and increased patient mortality1. Given the high level of heterogeneity in most tumors, it is a mathematical certainty that a tumor of sufficiently high cell number will contain a subset of cells resistant to single or combined therapies targeting molecular pathways on which those cells depend for survival2,3. Such tumor cells can be positively selected for during treatment, leading to disease recurrence. Development of novel therapies that simultaneously target different cancer cell survival mechanisms, either before or after treatment-mediated selection, is thus clinically important.
A high level of genome instability and mutation in tumor genomes is a fundamental characteristic that distinguishes cancer cells from non-tumor host cells4. Consequently, a useful strategy to increase the effectiveness of DNA-damaging chemotherapy and to prevent the development of resistance is to actively inhibit DNA repair in tumor cells5. This is an active field of investigation and a variety of novel DNA repair targets are being explored in a pre-clinical setting. A number of small molecule or antisense-based inhibitors of these targets have been developed and are undergoing testing6-8. The objective is to identify the most promising pre-clinical candidates and evaluate their safety and efficacy in clinical trials.
The high cost of clinical trials and the risk of failure (for a variety of reasons including sub-optimal pre-clinical evaluation) are formidable obstacles to progress in development of new therapies9. The use of appropriate and rigorous pre-clinical models to adequately evaluate new therapeutic targets and candidate drugs may decrease the high failure rate of clinical trials10.
Some commonly-used pre-clinical methods to evaluate the effectiveness of novel anti-cancer regimens are: a) measurement of capacity to reduce tumor cell proliferation in vitro (cell proliferation assays), b) therapy-induced reduction in capacity of tumor cells to form tissue culture colonies (colony formation assays), c) therapy-induced reduction in tumor cell metabolic activity in vitro (redox-dye conversion) , d) therapy-induced induction of in vitro tumor cell death (apoptotic, necrotic, autophagic, associated with mitosis and others)11, and e) in vivo therapy-induced reduction of growth or ablation of human and mouse xenografts12-14.
A major weakness of the listed in vitro methods is that none of them provide continuous real-time evaluation of the effect of candidate therapies. Rather, they provide information only at selected, widely-separated time points during the course of treatment. Such measurements have diminished capacity to accurately reflect the magnitude and timing of tumor cell responses. In vivo mouse xenograft models are also limited by high cost, length of time to complete, and risk of sub-optimal dosing and treatment timing (scheduling). In addition, there is evidence that rodent xenograft models are limited predictors of clinical efficacy in humans, compared to in vitro assessment of responses of primary human tumor cells and established human tumor cell lines to candidate therapeutic interventions15,16.
We devised a novel combination protocol to evaluate new drug combinations pre-clinically, in a manner that addresses the weaknesses of the more common procedures listed above. In place of proliferation, colony formation, or redox-dye conversion assays, we utilized a metabolism measurement unit to analyze cell adhesion, respiration, and acidification in real time during the entire treatment period17. Simultaneously, we investigated the effects of treatment combination in vivo by using a chicken embryo chorioallantoic membrane (CAM) model of invasion and metastasis18,19. We used these methods to evaluate the ability of an antisense oligonucleotide (ASO) targeting BRCA2 to potentiate the effectiveness of the commonly used chemotherapeutic drug cisplatin.
Protocol
NOTE: The following protocol was designed for use with adherent cells. Modifications are required to apply the method to non-adherent cells grown in suspension. The CAM experiments described in the protocol are designed for use with cells that express a fluorescent marker (e.g., GFP, RFP, etc.). Nine day old chicken embryos are required on day 2 of the experimental protocol for the CAM experiments.
1. Preparation and Transfection of Tumor Cells with Antisense Oligonucleotides (ASOs)
Aspirate medium from T75 flask(s) containing cells of interest, wash with 1XPBS, and add 2 ml of 0.25% trypsin/EDTA. Add 10 ml of complete growth medium to each flask and transfer the cells in solution to a 50 ml tube. Count the cells and adjust the volume so that the final concentration is 1.0x105 cells per ml.
Label two groups of T25 flasks: one group will contain eight flasks, labeled 1-8, and will be used for the real-time metabolism measurementexperiment; and the second group will also contain 8 flasks and will be used for the chicken embryo CAMexperiment. Add 2 ml of cells in solution to each flask and place in the incubator (37 °C, 5% CO2 O/N).
- The next morning (Day 0, Time 0), label three 5 ml tubes as follows: control (non-targeting ASO or siRNA), 'target of interest' (i.e., targeting ASO or siRNA), and Lipofectamine 2000 (or a similar transfection reagent). Dilute the stock ASO solutions to the appropriate concentration with serum free medium in the appropriate tube.
- Add the required amount of transfection reagent to the appropriate tube and incubate in serum-free medium for 5 min. Add the transfection reagent solution to the ASO tubes and incubate for 20 min.
- Add 250 µl of control ASO transfection reagent solution to each of flasks 1-4 in each group, and 250 µl of 'target of interest' ASO transfection reagent solution to each of flasks 5-8 in each group. Incubate for 4 hr (37 ᵒC, 5% CO2).
- During incubation, carry out the steps described in section 2.1. After the incubation, add 3 ml of complete medium to each flask in the CAM experiment group and return to the incubator O/N (37 ᵒC, 5% CO2). Process the metabolismexperiment group according to the description in section 2.2. Process the CAM experiment set according to the description in section 4.
2. Biosensor Chip Preparation and Cell Seeding
- In a sterile environment, carefully wash six biosensor chips with 400 µl of PBS. Sterilize the chips with 400 µl of 70% ethanol for 20 min. Wash three times with 400 µl of PBS.
- Place the chips inside sterile tissue culture dishes to prevent contamination. Place three chips in a dish labeled 'control', and three in a dish labeled 'target of interest'.
- Label two 50 ml tubes as 'control' and 'target of interest'. Wash cells with PBS and add an appropriate volume of 0.25% Trypsin/EDTA. Wait until the monolayer detaches and collect the cells from each flask. Deposit the solution into the appropriate tube.
- Count the cells and adjust the volume so that the final concentration is 6.0x105 cells/ml. Centrifuge and resuspend the cells if the initial concentration is too low.
Add 250 µl of the cell solution to each biosensor chip in the appropriate group (for a total of 1.25x105 cells per chip). Perform this task one group at a time to minimize the risk of confusing the chips. Place the sterile dishes containing the chips inside a humidity chamber to prevent evaporation, and place the humidity chamber in the incubator O/N (37 ᵒC, 5% CO2).
The next morning (Day 1), carefully aspirate the medium from each chip and replace it with 250 µl of growth medium supplemented with 0.2% fetal bovine serum (FBS) and 1x penicillin and streptomycin (P/S). Incubate for 4 hr (37 ᵒC, 5% CO2).
3. Metabolism Measurement System Preparation and Solution Preparation
- During the incubation, prepare the requisite drug solutions and the metabolism measurement system for the upcoming assay. NOTE: The metabolism measurement system contains a total of six modules. Each module accommodates one chip per experiment.
- Subdivide the assay into the following experimental groups: one chip from each of the 'control' and 'target of interest' groups to receive vehicle for the duration of the assay; two chips from each group to receive drug treatment for a defined period of time during the assay, followed by vehicle during the washout period.
- For drug sensitization studies, modify the length, timing, and concentration of drug treatment to suit the particular drug in question. NOTE: For cisplatin, the following steps were determined empirically with A549 cells, and may vary depending on cell line. Perform all stages with the default flow rate of 4 µl/min. 6 hr of growth medium + 0.2% FBS and 1x P/S 24 hr of 6 µM cisplatin in medium + 0.2% FBS 1x P/S 24 hr of growth medium + 0.2% FBS and 1x P/S 18 hr of growth medium + 0.2% FBS and 1x P/S 4 hr of 0.1% Triton-X NOTE: Deviations from these steps will require calculation to determine the required volume of medium and drug solutions based on the desired flow rate and duration of experiment. The following sections are based on the steps described above.
Label and fill fourteen 50 ml tubes with 50 ml of growth medium + 0.2% FBS and 1x P/S. Place six in one metabolism measurement system tube rack, six in another, and the remaining two in a third tube rack that also contains tubes with cisplatin. Seal the first two racks with an air permeable membrane to prevent evaporation.
- Prepare a solution of 6 µM cisplatin (of the appropriate volume) in medium + 0.2% FBS with 1x P/S. Dispense the required volume of cisplatin solution into four labeled 50 ml tubes.
- Place the tubes in the remaining four slots in the third metabolism measurement tube rack. Ensure that the tubes are placed in the appropriate slots that correspond to the desired biomodules. Seal the tubes with an air permeable membrane.
Prepare and label six 50 ml tubes and fill them with 20 ml each of 0.1% Triton-X solution in medium. The Triton-X solution will lyse any remaining cells and provide a 'zero' value for each chip. Place the tubes in a fourth metabolism measurement unit tube rack and seal with an air permable membrane to prevent excessive evaporation.
Load the chips into the biomodules and initiate the experiment.
4. CAM Experiment Drug Treatment and Injection
NOTE: The steps and procedure for preparing chicken embryos for a CAM experiment are described elsewhere19
Begin processing the CAM experiment 48 hr post-transfection (Day 2). Sub-divide the 'control' and 'target of interest' groups into two flasks for vehicle, and two for cisplatin. Aspirate the medium from the flasks and replenish with either 3 ml of fresh medium, or 6 µM cisplatin. Incubate for 6 hr (37 ᵒC, 5% CO2).
- Aspirate the medium, then wash and trypsinize the cells; collect the trypsinized cells in solution into four appropriately labeled 15 ml tubes.
- Centrifuge and wash the cells three times with PBS. Re-suspend the cell pellet in 5 ml of PBS and count the cells.
- Adjust the volume so that the final cell concentration is 1.0x106 cells/ml. Transfer the cell solution into labeled 5 ml tubes for injection, and place on ice. Invert the tubes gently prior to injection to ensure that the cells are well mixed.
- Set aside a total of 24 chicken embryos (9 days old) for injection (6 per group). Perform the injection using a glass needle attached to a flexible tube connected to a 1 ml syringe. The individual performing the injection should be blinded to the treatment group.
- Using a stereo microscope, inject 1.0x105 cells in a volume of 100 µl into the venous circulation of the CAM. Use a slow, pulsating technique to inject the full volume of cells. Gently dab the injection site with a soft wipe to stem the flow of blood and absorb any spillage. Label each embryo container appropriately following each injection.
Confirm the presence and distribution of fluorescent cells in the vasculature of the CAM following injection by visualizing them using a fluorescent microscope. Take note of any embryos in which the number of tumor cells appears low, or the distribution is poor.
Place the chicken embryos in a humidity chamber and store them in an incubator (37 °C, 60% humidity). Wait 7-9 days before enumerating metastatic foci.
5. Metastatic Focus Enumeration
During the 7-9 day incubation period, monitor the embryos and discard any dead ones. Ensure that the humidity chamber remains moist at all times during the incubation period.
Using a fluorescent stereo microscope at 20X magnification, scan the entire upper surface of the embryo CAM in a regular, grid-like pattern.
Count the number of metastatic foci in each field of view, then tally the number for a final total per embryo. Doing so for the six embryos in each group will provide an average number of metastatic foci per treatment.
Representative Results
Results and figures adapted with permission from published work22.
BRCA2 inhibition induces a decrease in respiration in cisplatin treated tumor cells
Human A549 lung cancer cells treated with BRCA2-targeting ASO and cisplatin exhibited an early and irreversible decrease in respiration, compared to cells which received control ASO and cisplatin alone; after 24 hr of cisplatin treatment the difference in respiration between BRCA2 ASO and control ASO treated cells was 39% (Figure 1A). Importantly, this decrease in respiration started before a detectable drop in cell adhesion, suggesting that this occurred independent of changes in cell number. Respiration began to decrease approximately 10 hr before the first observable drop in adhesion, suggesting that a drop in respiration may be a formative event that precedes cell death (Figure 1B). No difference in acidification was observed during the 72 hr experiment time-frame, suggesting that BRCA2 ASO and cisplatin treatment has little to no effect on glucose metabolism (Figure 1C).
BRCA2 inhibition combined with cisplatin treatment decreases the frequency of metastasis
Human A549 lung cancer cells were treated with control ASO or BRCA2 ASO in the presence or absence of cisplatin, and injected into the venous circulation of the CAM (Figure 2A). Nine days following injection, cells treated with BRCA2 ASO and cisplatin exhibited a 77% lower frequency of metastatic foci compared to cells treated with control ASO and cisplatin, or BRCA2 ASO alone (Figure 2B). This suggests that BRCA2 ASO and cisplatin treatment has the potential to decrease or prevent metastatic burden in patients with solid tumors.
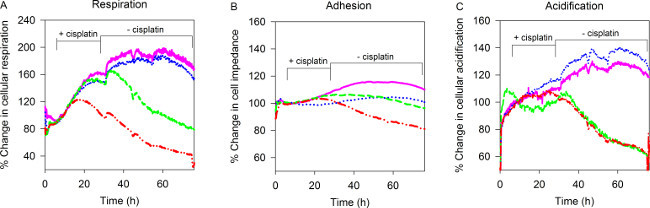 Figure 1. Real-time monitoring of tumor cell adherence, respiration, and acidification. A549 cells were transfected with BRCA2 ASO, plated onto biosensor chips, and treated with cisplatin for 24hrs using the Bionas Discovery System. Measurements of (A) respiration, (B) adherence, and (C) acidification were performed every 4 min over a period of 72 hr. Pink = control ASO, blue = BRCA2 ASO, green = control ASO + cisplatin, red = BRCA2 ASO + cisplatin. Figure adapted from published work with permission22. Please click here to view a larger version of this figure.
Figure 1. Real-time monitoring of tumor cell adherence, respiration, and acidification. A549 cells were transfected with BRCA2 ASO, plated onto biosensor chips, and treated with cisplatin for 24hrs using the Bionas Discovery System. Measurements of (A) respiration, (B) adherence, and (C) acidification were performed every 4 min over a period of 72 hr. Pink = control ASO, blue = BRCA2 ASO, green = control ASO + cisplatin, red = BRCA2 ASO + cisplatin. Figure adapted from published work with permission22. Please click here to view a larger version of this figure.
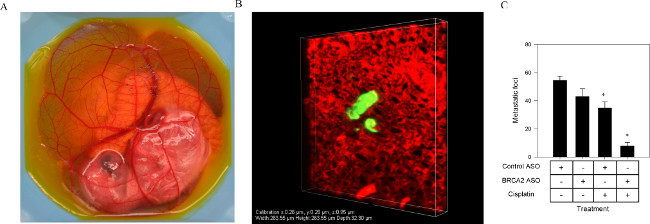 Figure 2. Enumerating metastatic frequency using a chicken embryo CAM model. A549 cells transfected with BRCA2 ASO were treated with cisplatin for 6 hr, and then injected into the venous circulation of a 9 day old chicken embryo (A). Metastatic foci (B) (visualized using a confocal microscope with a 40x objective) were counted nine days following injection (C). Figure adapted from published work with permission22. Please click here to view a larger version of this figure.
Figure 2. Enumerating metastatic frequency using a chicken embryo CAM model. A549 cells transfected with BRCA2 ASO were treated with cisplatin for 6 hr, and then injected into the venous circulation of a 9 day old chicken embryo (A). Metastatic foci (B) (visualized using a confocal microscope with a 40x objective) were counted nine days following injection (C). Figure adapted from published work with permission22. Please click here to view a larger version of this figure.
Discussion
Due to the inherent cost and risk associated with clinical trials, there is a need to develop better and more rigorous pre-clinical testing methodology to adequately evaluate novel anti-cancer treatment regimens. Current commonly-used techniques all exhibit weaknesses that may limit their capacity to discriminate between potentially promising therapeutic targets/agents, and those that may be less effective. We devised a protocol to evaluate new anti-cancer approaches that addresses many of the shortcomings of traditional methods.
A particularly important feature of the described protocol is the ability to measure cellular responses in real time, and to track changes in metabolism and adherence minute by minute. This allows for the identification of peak drug effects, optimal dosing and a more detailed understanding of the interaction between the components of drug combination treatment. Simultaneously, it provides insight into discrete metabolic alterations in cancer cells, which is information that is not provided by cell counting, apoptosis, or dye-conversion assays. A limitation of this assay is the inability to accurately determine the type of cell death that is occurring during drug treatment. Changes in viability will be reflected by changes in adherence to the biosensor chip, but this does not provide information regarding the manner of cell death. It is also possible to use fluorescence microscopy and live cell imaging techniques to determine similar metabolic data, and this can be used in addition to the experiments described in this protocol20,21.
Furthermore, use of the chicken embryo CAM model to study metastatic frequency following treatment is an effective way to determine whether a new target, drug, or drug combination decreases the frequency of cancer cells which extravasate and/or invade surrounding tissue. This is a particularly relevant question from a clinical standpoint, because metastatic disease causes a high proportion of mortality amongst cancer patients. A potential limiting factor is that actual drug treatment does not occur in the CAM. Instead it occurs ex vivo and the pre-treated cells are then injected into the CAM. Therefore, this assay does not provide information regarding drug uptake or distribution in vivo. In addition, the intravenous CAM injections are technically challenging and require significant practice/experience.
We used this protocol to evaluate the ability of a BRCA2-targeting ASO to sensitize tumor cells to the DNA damaging drug cisplatin. The results of our experiments identified BRCA2 as a promising target for therapeutic attack, and also provided rationale for further pre-clinical development of the BRCA2 ASO as a candidate drug. We envision this protocol being used to identify novel targets for combination treatment, particularly in the burgeoning field of DNA repair inhibition. We feel this protocol will be best used in addition to the more common techniques in cancer biology, in an effort to more stringently evaluate new anti-cancer approaches.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was made possible by grants to JK from the Ontario Centres of Excellence and the Ontario Research Fund.
We would like to thank Siddika Pardhan and Dr. Peter Ferguson for technical assistance during filming.
References
- Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer. 2012;12:587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- Bozic I, et al. Evolutionary dynamics of cancer in response to targeted combination therapy. Elife. 2013;2:e00747. doi: 10.7554/eLife.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz LA, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Rytelewski M, et al. Inhibition of BRCA2 and Thymidylate Synthase Creates Multidrug Sensitive Tumor Cells via the Induction of Combined 'Complementary Lethality'. Mol Ther Nucleic Acids. 2013;2:e78. doi: 10.1038/mtna.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- Shaheen M, Allen C, Nickoloff JA, Hromas R. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood. 2011;117:6074–6082. doi: 10.1182/blood-2011-01-313734. [DOI] [PubMed] [Google Scholar]
- Green S, Benedetti J, Smith A, Crowley J. Clinical trials in oncology. Vol. 28. CRC press; 2012. [Google Scholar]
- Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:531–533. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggeri BA, Camp F, Miknyoczki S. Animal models of disease: pre-clinical animal models of cancer and their applications and utility in drug discovery. Biochem Pharmacol. 2014;87:150–161. doi: 10.1016/j.bcp.2013.06.020. [DOI] [PubMed] [Google Scholar]
- Kwon MC, Berns A. Mouse models for lung cancer. Mol Oncol. 2013;7:165–177. doi: 10.1016/j.molonc.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M, Ordonez L, Clarke AR. What are the best routes to effectively model human colorectal cancer. Mol Oncol. 2013;7:178–189. doi: 10.1016/j.molonc.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alborzinia H, et al. Real-time monitoring of cisplatin-induced cell death. PLoS One. 2011;6:e19714. doi: 10.1371/journal.pone.0019714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic D, et al. KISS1R induces invasiveness of estrogen receptor-negative human mammary epithelial and breast cancer cells. Endocrinology. 2013;154:1999–2014. doi: 10.1210/en.2012-2164. [DOI] [PubMed] [Google Scholar]
- Leong HS, Chambers AF, Lewis JD. Assessing cancer cell migration and metastatic growth in vivo in the chick embryo using fluorescence intravital imaging. Methods Mol Biol. 2012;872:1–14. doi: 10.1007/978-1-61779-797-2_1. [DOI] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantama M, Hung YP, Yellen G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J Am Chem Soc. 2011;133:10034–10037. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytelewski M, et al. BRCA2 inhibition enhances cisplatin-mediated alterations in tumor cell proliferation, metabolism, and metastasis. Mol. Onc. 2014;8(8):1429–1440. doi: 10.1016/j.molonc.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]


