Abstract
Approximately 30% of patients with chronic myelomonocytic leukemia (CMML) have karyotypic abnormalities and this low frequency has made using cytogenetic data for the prognostication of CMML patients challenging. Recently, a three-tiered cytogenetic risk stratification system for CMML patients has been proposed by the Spanish study group. Here we assessed the prognostic impact of cytogenetic abnormalities on overall survival (OS) and leukemia-free survival (LFS) in 417 CMML patients from our institution. Overall, the Spanish cytogenetic risk effectively stratified patients into different risk groups, with a median OS of 33 months in the low-, 24 months in intermediate- and 14 months in the high-risk groups. Within the proposed high risk group; however, marked differences in OS were observed. Patients with isolated trisomy 8 showed a median OS of 22 months, similar to the intermediate-risk group (p=0.132), but significantly better than other patients in the high-risk group (p=0.018). Furthermore, patients with more than 3 chromosomal abnormalities showed a significantly shorter OS compared with patients with 3 abnormalities (8 vs. 15 months, p=0.004), suggesting possible a separate risk category. If we simply moved trisomy 8 to the intermediate risk category, the modified cytogenetic grouping would provide a better separation of OS and LFS; and its prognostic impact was independent of other risk parameters. Our study results strongly advocate for the incorporation of cytogenetic information in the risk model for CMML.
Keywords: Chronic myelomonocytic leukemia (CMML), Cytogenetics risk classification, Overall survival, trisomy 8, complex karyotype
INTRODUCTION
Chronic myelomonocytic leukemia (CMML) is a myelodysplastic/myeloproliferative neoplasm (MDS/MPN) characterized by persistent monocytosis. The clinical course of CMML patients is variable, with a reported life expectancy ranging from a few months to several years. Clinical parameters, such as older age, low hemoglobin level, low platelet count, high white blood cell (WBC) count, presence of circulating blasts, presence of circulating immature myeloid cells (IMC), high percentage of bone marrow blasts, and high serum levels of lactate dehydrogenase (LDH) or beta2-microglobulin have been reported to be associated with shorter survivals [1–11]. Recently, cytogenetic data [5–6] and gene mutations [7, 11–12] have been recognized as important factors in determining prognosis in CMML patients.
Cytogenetic abnormalities are detected in about 30% of CMML patients [3, 5, 13–16]. Due to this low frequency, cytogenetic risk stratification in CMML patients is challenging and criteria are not well-established. Proposed risk models, including a recent study that integrated gene mutation data into a prognostic score for CMML [7, 11], have not incorporated karyotype information. In 2011, Such and colleagues studied a cohort of 414 CMML patients from the Spanish MDS Registry [5]. This study showed that patients with an abnormal karyotype have a worse overall survival (OS) compared with the patients with a normal karyotype. The authors further divided karyotypical abnormalities into three risk groups: low-risk including a normal karyotype or loss of Y chromosome; high-risk including trisomy 8, abnormalities of chromosome 7, or a complex karyotype with 3 or more cytogenetic abnormalities; and an intermediate-risk including all other abnormalities. The median survival for patients in a low-, intermediate-, and high-risk cytogenetic groups was 37, 18, and 11 months, respectively. This Spanish cytogenetic risk stratification shares great similarities with the Revised International Prognostic Scoring System (IPSS-R) for myelodysplastic syndromes (MDS) [17], but differs in the risk assignment of trisomy 8 and definition of a complex karyotype. In the IPSS-R, +8 was assigned to the intermediate risk, and a complex karyotype was further grouped into “poor risk group” (3 abnormalities) and “very poor risk group” (>3 abnormalities). Subsequently, a prognostic scoring system that includes cytogenetic data for patients with CMML was proposed by the same Spanish study group [6].
In this study, we reviewed conventional cytogenetic data for 417 CMML patients who were diagnosed and treated at our institution and examined the application of the Spanish cytogenetic risk stratification system in our patients. In addition, with this large patient cohort, we also examined the clinical significance of some rare chromosomal alterations in CMML patients. With the information we obtained, we examined the significance of cytogenetic risk grouping in the recently proposed Mayo prognostic model for CMML patients [11]
MATERIALS AND METHODS
Patients
We searched the archives at The University of Texas M. D. Anderson Cancer Center (MDACC) between January 1, 2003 through June 30, 2013 for cases of CMML as defined in the 2008 WHO Classification [18]. For cases in which the initial diagnosis of CMML was made at other hospitals, the pathologic materials were reviewed in our department to confirm the diagnosis. Complete blood count (CBC) data were obtained at the time of diagnosis. Blasts were counted on peripheral blood (PB) based on 200 cells as well as bone marrow (BM) smears based on 500 cells. The blast count included myeloblasts, monoblasts and promonocytes. Circulating IMC included any of the following cells in PB: myeloblasts, promyelocytes, myelocytes, and metamyelocytes. Based on the blast count, CMML cases were further subclassified as CMML-1 (<5% in the PB and <10% in the BM) and CMML-2 (5% to 19% in the PB, or 10%–19% in the BM). Patients without essential clinical and cytogenetic information or patients with blasts ≥20% in PB or BM that fulfilled the criteria for acute myeloid leukemia (AML) were excluded from this study. The study was approved by the institutional review board of MDACC.
Patients received risk-adapted therapies, including supportive care, hydroxyurea, hypomethylating agents, low dose/single dose chemotherapy and standard induction chemotherapy. Thirty-seven patients received hematopoietic stem cell transplant (HSCT).
Conventional cytogenetic analysis
Conventional chromosomal analysis was performed on G-banded metaphase cells prepared from unstimulated 24- and 48-hour BM aspirate cultures at the time of diagnosis using standard techniques. The median number of metaphases analyzed was 20 (range, 10 to 50). The karyotype was documented according to the International System for Human Cytogenetic Nomenclature (ISCN 2013) [19].
Composition of Cytogenetic Subgroups
Abnormal karyotypes were grouped into three risk categories using the scoring system proposed by Spanish study group. In addition, we also examined the significance of cytogenetic subgroups within these three large groups. Single karyotypical alterations occurring in at least five patients were assessed separately, whereas all other single abnormalities occurring in less than five patients were grouped into “other single abnormalities”. Patients with two cytogenetic abnormalities were further divided into three subgroups: trisomy 8 with one additional clonal aberration; monosomy 7/or del(7q) with one additional clonal aberration; and any other two abnormalities. Complex karyotypes were subdivided into two subgroups, those with exactly three abnormalities, and those with greater than three abnormalities.
Statistical analysis
The unpaired t-test was used for numerical comparisons between groups. Chi-square and Fisher’s exact tests were applied for categorical variables. Interval to AML transformation was calculated from the time of diagnosis of CMML to the time of transformation into AML. Overall survival (OS) was estimated by the Kaplan–Meier method from the date of diagnosis of CMML until death from any cause (censored at last follow-up for alive patients, or censored at the time of HSCT for patients who received HSCT). In the analysis of prognostic factors, variables included age, gender, hemoglobin level, white blood cell count (WBC), absolute neutrophil count (ANC), absolute monocyte count (AMC), absolute lymphocyte count (ALC), platelet count, PB blasts, BM blasts, circulating IMC, BM cellularity, and cytogenetic risk groups. Univariate analysis was performed to determine the association between single variables and OS or leukemia-free survival. Multivariate analysis was performed by the Cox proportional regression model with OS or time to AML transformation as the end point. Analyses were performed exclusively for subgroups with a minimum of five patients. P values ≤0.05 were considered significant.
RESULTS
Demographics and Clinical Findings
The demographic and clinical features are shown in Table 1. The median age of the whole patient cohort at time of diagnosis was 69 years (range, 27–92 years). The male to female ratio was 2.3 to 1. Based on the percentage of blasts in PB and BM, 331 (79.3%) patients had CMML-1, and 86 (20.6%) patients had CMML-2.
Table 1.
Demographics and clinical characteristics of CMML patients with or without chromosomal abnormalities
| Total N (%) |
Normal karyotype | Abnormal karyotype | P* | |
|---|---|---|---|---|
| Number of Patients | 417 | 292 (70%) | 125 (30%) | |
| Age (years) | ||||
| Median (range) | 69 (27–92) | 69 (30–92) | 69 (27–87) | 0.1771 |
| <70 (No. of patients) | 211 | 146 | 65 | 0.7486 |
| ≥ 70 (No. of patients) | 206 | 146 | 60 | |
| Gender | 0.998 | |||
| Male | 292 | 204 | 88 | |
| Female | 125 | 88 | 37 | |
| WHO classification | 0.1678 | |||
| CMML-1 | 331 | 237 | 94 | |
| CMML-2 | 86 | 55 | 31 | |
| WBC counts (x109/L) | ||||
| Median (range) | 16 (2.6–209) | 17.4 (2.6 – 209) | 15.4 (3.3 – 116) | 0.1956 |
| <13 (No. of patients) | 187 | 117 | 70 | 0.455 |
| ≥ 13 (No. of patients) | 230 | 175 | 55 | |
| Hemoglobin (g/dL) | ||||
| Median (range) | 10.7 (3.8–16.7) | 10.9 (5.9–16.7) | 10.2 (3.8 – 15.8) | 0.0013 |
| ≥ 10 (No. of patients) | 265 | 192 | 73 | 0.1529 |
| <10 (No. of patients) | 152 | 100 | 52 | |
| Platelet (x109/L) | ||||
| Median (range) | 102 (5–820) | 108 (5 – 809) | 90 (6 – 820) | 0.0089 |
| ≥ 100 (No. of patients) | 217 | 162 | 55 | 0.0316 |
| <100 (No. of patients) | 200 | 130 | 70 | |
| PB blasts (%) | ||||
| Median (range) | 0 (0–15) | 0 (0–15) | 0 (0–13) | 0.0746 |
| < 5 (No. of patients) | 391 | 278 | 113 | 0.0630 |
| ≥ 5 (No. of patients) | 26 | 14 | 12 | |
| PB Immature Myeloid Cells | ||||
| Presence | 226 | 149 | 77 | 0.0536 |
| Absence | 191 | 143 | 48 | |
| PB Monocyte (x109/L) | ||||
| Median (range) | 3.3 (1.03–71.9) | 3.23 (1.03–64.4) | 3.36 (1.03–71.9) | 0.7783 |
| ≥ 3 (No. of patients) | 217 | 145 | 72 | 0.1642 |
| <3 (No. of patients) | 200 | 147 | 53 | |
| BM cellularity (%) | ||||
| Median (range) | 90 (30–100) | 90 (30–100) | 80 (30–100) | 0.0876 |
| BM blasts (%) | ||||
| Median (range) | 4 (0–9) | 4 (0–19) | 5 (0–18) | 0.5103 |
| < 10 (No. of patients) | 333 | 233 | 100 | 0.9618 |
| ≥ 10 (No. of patients) | 84 | 59 | 25 | |
| Survival (months) | ||||
| Overall Survival (median) | 27 | 33 | 19 | <0.0001 |
| AML transformation | ||||
| Accumulated (%) | 25.9 | 23.6 | 31.2 | 0.1109 |
| 2-year (%) | 18.2 | 13.7 | 28.8 | 0.0005 |
Comparisons between patients with or without cytogenetic abnormalities.
AML: Acute myeloid leukemia; BM: bone marrow; CMML: chronic myelomonocytic leukemia; PB: peripheral blood; WBC: white blood cell count.
Clonal karyotypic abnormalities were detected in 125 (30%) patients. The clinicopathological features of patients with or without cytogenetic abnormalities are shown in Table 1. In brief, patients with or without cytogenetic abnormalities had a comparable age at diagnosis, similar gender distribution, comparable WBC and absolute monocyte counts, and similar percentages of BM blasts. However, patients with an abnormal karyotype had a significantly lower hemoglobin level and platelet count, borderline higher PB blasts and immature myeloid cells, and a significantly shorter OS. Although accumulated AML transformation probability was not different, patients with an abnormal karyotype had a higher rate of AML transformation at the 2-year follow-up interval.
Cytogenetic abnormalities
The cytogenetic abnormalities for all 125 patients with abnormal karyotypes are summarized in Table 2.
Table 2.
Association between Cytogenetics Subgroups with Overall Survival and AML Transformation
| Cytogenetics Categories | Case# | Age* (Years) | Alive /Dead | Overall Survival | AML Transformation | ||||
|---|---|---|---|---|---|---|---|---|---|
| Time* (Mon) | Hazard Ratio (95% CI) | P+ | Rate | Hazard Ratio (95% CI) | P+ | ||||
| Normal karyotype | 292 | 69 | 120/172 | 33 | 1 | 69 (23.6%) | 1 | ||
| Single abnormality | 80 | 69 | 34/46 | 23 | |||||
| +8 | 17 | 69 | 6/11 | 22 | 1.55 (0.75 – 3.18) | 0.235 | 5 (29.4%) | 1.73 (1.03 – 6.34) | 0.046 |
| −7 or del(7q) | 14 | 71 | 4/10 | 14 | 3.52 (3.89 – 13.16) | <0.001 | 6 (36%) | 3.18 (1.72 – 14.64) | 0.008 |
| −Y | 13 | 77 | 7/6 | 30 | 1.14 (0.48 – 2.79) | 0.755 | 0 | undefined | |
| Del(20q) | 11 | 72 | 4/7 | 24 | 1.44 (0.92 – 4.25) | 0.076 | 2 (18%) | 1.34 (0.28 – 7.62) | 0.645 |
| −X or del(Xq) | 5 | 63 | 3/2 | 25 | 1.16 (0.26 – 4.21) | 0.849 | 0 | undefined | |
| Other single | 20 | 68 | 10/10 | 22 | 1.25 (0.67 – 2.33) | 0.486 | 6 (30%) | 1.17 (0.49 – 2.83) | 0.734 |
| 2 abnormalities | 20 | 67 | 3/17 | 16 | |||||
| −7 or del(7q) plus 1 | 6 | 63 | 1/5 | 14 | 3.46 (2.09 – 16.84) | 0.038 | 4 (67%) | 4.29 (3.29 – 19.84) | 0.002 |
| +8 plus 1 | 5 | 74 | 0/5 | 14 | 2.83 (1.458 – 7.48) | 0.015 | 3 (60%) | 4.42 (2.72 – 18.99) | 0.006 |
| Other two | 9 | 77 | 3/6 | 18 | 1.99 (0.97 – 5.77) | 0.064 | 4 (44%) | 2.16 (1.69 – 8.77) | 0.035 |
| 3 abnormalities | 9 | 69 | 3/6 | 15 | 2.91 (1.74 – 16.29) | 0.006 | 5 (56%) | 5.13 (3.13 – 26.67) | <0.001 |
| >3 abnormalities | 16 | 68 | 3/13 | 8 | 9.44 (4.94 – 18.06) | <0.001 | 3 (19%) | 3.68 (1.13 – 11.98) | 0.031 |
Data presented as median.
Compared to normal karyotype, significant while p<0.05
AML: acute myeloid leukemia; Mon: months.
Single abnormalities
Eighty (64%) patients had single (isolated) abnormalities. Abnormalities occurring in at least 5 patients included: trisomy 8 (n=17), −7 or del(7q) (n=14), −Y (n=13), del(20q) (n=11), −X or del(Xq) (n=5) (Table 2). Abnormalities occurring in 2 patients included: trisomy 21, del(9q), del(13q), and t(11;19)(q23;p13.1). In addition, 12 cytogenetic abnormalities were observed only in single patients.
Two abnormalities
Two chromosomal abnormalities were detected in 20 (16%) patients. The most common combination was −7 and trisomy 21 (n=3). Monosomy 7/del(7q) plus one additional abnormality were observed in 6 patients (4.8%), and trisomy 8 plus one additional abnormality were observed in 5 patients (4.0%) (Table 2).
Complex abnormalities
A complex karyotype was observed in 25(20%) patients and the median number of abnormalities was 6 (range, 3–15). Nine patients had exactly 3 abnormalities and 16 patients had greater than 3 abnormalities. Chromosome 5 abnormalities, either monosomy 5 or del(5q), were observed exclusively in the context of a complex karyotype, especially in patients with >3 abnormalities (11 of 16 patients).
Prognostic Value of Cytogenetic Subgroups
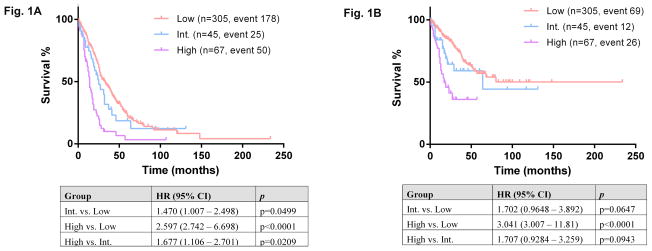
Using the Spanish cytogenetic risk stratification system, 305 (73.1%) of our patients were assigned to the low-risk group, 45 (10.8%) patients intermediate-risk group, and 67 (16.1%) high-risk group. The median OS was 33 months for patients in low-risk group (reference group, HR=1), 24 months for patients in intermediate-risk group (HR=1.47, p=0.0499), and 14 months for patients in high-risk group (HR=2.597, p<0.0001) (Figure 1A). Risk of AML transformation was 22.6% for low-risk (reference HR=1), 28.2% for intermediate-risk (HR=1.702, p=0.0647), and 39.4% for high-risk patients (HR=3.041, p=<0.0001) (Figure 1B)
Figure 1.
Overall survival (A) and leukemia-free survival (B) among three risk groups: low, intermediate, and high, based on Spanish cytogenetic risk stratification. Int: intermediate.
The prognostic value of each cytogenetic subgroup was analyzed and these data are summarized in Table 2. Patients with a normal karyotype were used as the reference group.
Low-risk group
Patients with isolated –Y showed a very similar OS to the patients with a normal karyotype (median OS: 30 versus 33 months, p=0.755). None of the patients with –Y experienced AML transformation.
Intermediate-risk group
Within the intermediate-risk group, patients with two abnormalities showed a trend toward an inferior OS (p=0.064) and had a higher probability of AML transformation (p=0.035), compared with patients with a normal karyotype. Of the single karyotypical abnormalities, patients with del(20q) showed a trend toward an inferior OS (p=0.076) but not for LFS (p=0.645). The remaining patients with single abnormalities in the intermediate-risk group, including –X/del(Xq), showed no significant difference in OS or LFS compared with patients with a normal karyotype.
High-risk group
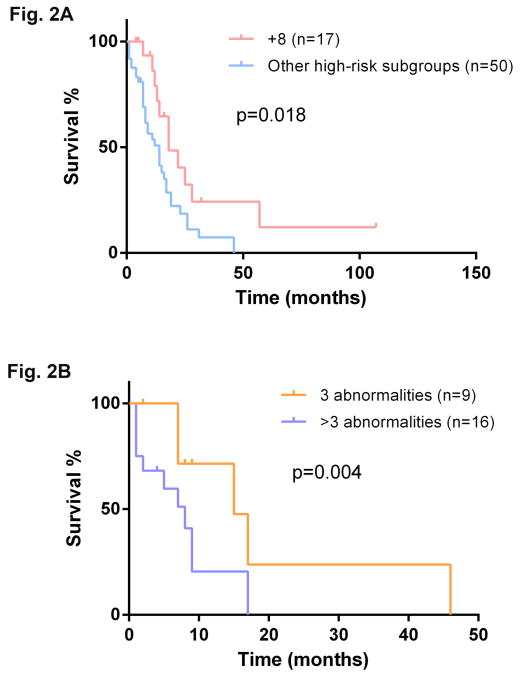
trisomy 8 showed a higher risk of AML transformation (p=0.046), but a comparable OS to patients with a normal karyotype (p=0.235). Compared with other patients in the high-risk group, patients with trisomy 8 had a significant better OS (22 versus 14 months, p= 0.018) (Figure 2A), though the LFS was not significantly different (p= 0.243). On the other hand, the OS of patients with trisomy 8 was more similar to that of patients in the intermediate-risk group (median OS: 22 versus 24 months, p=0.132). In addition, patients with >3 abnormalities showed a significant shorter OS compared with patients who had exactly 3 abnormalities (8 versus 15 months, p=0.004) (Figure 2B)
Figure 2.
A: Comparison of overall survival between patients with isolated trisomy 8 and patients with other high-risk cytogenetic abnormalities based on Spanish cytogenetic risk stratification.
B: Comparison of overall survival between patients with exact 3 abnormalities and patients with >3 abnormalities
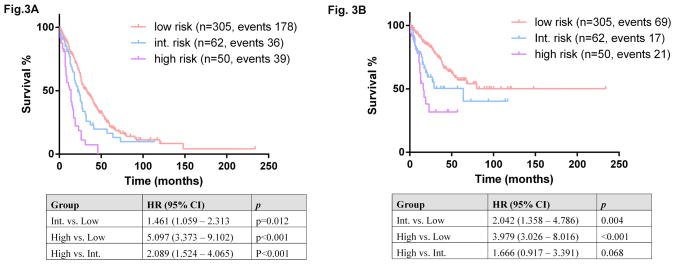
Based on these data, we modified the Spanish cytogenetic risk stratification system by moving patients with isolated trisomy 8 from the high-risk group to the intermediate-risk group, which resulted in a better separation of OS among the three risk groups (Figure 3A). This modification also resulted in a better separation of LFT among three risk groups (Figure 3B).
Figure 3.
Overall survival (A) and leukemia-free survival (B) among three risk groups: low, intermediate, and high, based on the modified Spanish cytogenetic risk stratification. Int: intermediate.
Independent Prognostic Value of Modified Cytogenetic Risk Groups
In the univariate analysis, factors adversely influencing OS and LFS included a higher WBC count (p<0.001 and p=0.007), lower hemoglobin level (p<0.001 and p<0.001), lower platelet count (p<0.001 and p<0.001), higher ANC (p=0.013 and p=0.03), higher ALC (p=0.002 and p=0.004), higher AMC (p=0.005 and p=0.002), the presence of circulating IMC (p<0.001 and p=0.001), the presence of circulating blasts (p=0.001 and p<0.001) and intermediate- or high- risk cytogenetics (p<0.001 and p<0.001). A higher BM cellularity was significant for OS (p=0.039) but not for LFS whereas a higher BM blast count were significant for LFS (p=0.007) but insignificant for OS. In multivariate analysis, the parameters remained to be significant for survivals are listed in Table 3. A lower hemoglobin level (<10 g/dl), higher AMC (≥3x10^9/L), the presence of circulating IMC, and intermediate and high cytogenetic risk groups were negative factors for OS as well as for LFS (see Table 3). A higher WBC count (≥13 x01^9/L) and a lower platelet count (<100 x10^9/L) negatively impacted OS but not LFS; whereas BM blasts adversely affected LFS but not OS.
Table 3.
Multivariate Cox Proportional Hazard Regression Analysis for Overall Survival and Leukemia-free Survival.
| Overall survival | Leukemia-free Survival | |||
|---|---|---|---|---|
| Variables | Hazard ratio (95% CI) | p | Hazard ratio (95% CI) | P |
|
Hemoglobin level <10 vs. ≥ 10 g/dl |
1.599 (1.214 – 2.106) | 0.001 | 1.760 (1.151 – 2.692) | 0.009 |
|
White blood cell count ≥13 vs. <13 x10^9/L |
1.707 (1.184 – 2.461) | 0.004 | 1.141 (0.858 – 1.979) | 0.224 |
|
Platelet count <100 vs. ≥ 100 x10^9/L |
1.232 (1.124 – 1.656) | 0.039 | 0.946 (0.783 – 1.355) | 0.201 |
|
Absolute monocyte count ≥3 vs. <3x10^9/L |
1.5 (1.148 – 1.966) | 0.023 | 1.612 (1.024 – 2.754) | 0.045 |
|
Circulating Immature Myeloid Cells Presence vs. Absence |
1.405 (1.165 – 1.854) | 0.016 | 1.758 (1.211 – 2.807) | 0.018 |
|
Bone marrow blasts ≥10 vs. <10% |
1.023 (0.829 – 1.374) | 0.124 | 2.377 (1.537 – 3.676) | <0.001 |
| Cytogenetic risk categories | ||||
| - Intermediate vs. Low | 1.493 (1.093 – 2.554) | 0.043 | 2.693 (1.593 – 4.554) | 0.008 |
| - High vs. Low | 3.360 (2.493 – 5.285) | 0.001 | 3.802 (2.268 – 6.804) | <0.001 |
We also tested our modified cytogenetic risk stratification system in the Mayo prognostic model [11], which was based on four risk factors: hemoglobin <10 g/dL, platelets <100 x10^9/L, the presence of circulating IMC, and AMC >10x10^9/L. Our modified cytogenetic risk stratification remained to be significant for OS in this model: the intermediate risk group had a HR 1.429, p=0.046; high risk group had a HR 3.408, p<0.001. The risk factors of the Mayo prognostic model, when co-analyzed with our cytogenetic risk groups, all except AMC (HR=1.125, p=0.246) remained to be significant. Notably, if using an AMC >3 x10^9/L, all 5 factors would be significant in this model.
DISCUSSION
In this patient cohort of 417 CMML patients from a single institution, we show that clonal cytogenetic abnormalities are present in less than one third of patients. In the cytogenetically abnormal subgroup, nearly two thirds of patients have isolated chromosomal abnormalities, either numerical or structural; and 20% of patients have a complex karyotype. The frequency and types of abnormal karyotypes are similar to those reported earlier in other series [3, 5, 13–16].
In this study, we did not identify any cytogenetic abnormalities that are unique for CMML patients. The most common single abnormalities, like trisomy 8, monsomy7/del(7q), loss of Y, and del(20q), are also commonly observed in patients with MDS [20]. Unlike MDS, however, sole abnormalities of chromosome 5 were not observed in this cohort of CMML patients. It is unclear why isolated chromosome 5 abnormalities are rare in CMML; one speculation is that loss of chromosome 5 genomic material may not impart a growth advantage to monocytes.
Trisomy 8 is one of the most common cytogenetic abnormalities detected in myeloid neoplasms; however, the prognostic role of trisomy 8 seems variable in different subtypes of myeloid neoplasms. Trisomy 8 as an isolated abnormality has been widely accepted to impart an intermediate risk in AML and MDS patients [17, 18], whereas it is considered to be a high-risk finding in patients with primary myelofibrosis [21]. In the cohort we presented here, patients with trisomy 8 showed very heterogeneous clinical course and outcome, similar to what have been reported for MDS patients [22]. The median OS for patients with sole trisomy 8 was 22 months (range, 4 – 107 months), very similar to that of patients in the intermediate-risk group but better than patients in the high-risk group. Reassigning trisomy 8 from the high-risk to the intermediate-risk group in this study resulted in a better separation between different risk groups both in OS and LFS. Based on these data, we suggest that isolated trisomy 8 likely belongs to the intermediate-risk group. It is not entirely clear why patients with isolated trisomy 8 have a variable clinical course. Cryptic cytogenetic abnormalities, such as del(7p) and del(12p), which can be detected by array comparative genomic hybridization in patients with trisomy 8 [23], or concomitant gene mutations are possible explanations [7].
In the prognostic system for CMML patents proposed by Such and colleagues, a complex karyotype was defined as three or more structural or numerical abnormalities [5]. By contrast, in the IPSS-R [17] prognostic system for MDS patients, a complex karyotype is further subgrouped into “3 abnormalities” and “more than 3 abnormalities”; with the latter representing the highest risk in MDS. The prognosis of MDS patients deteriorates with the increasing number of chromosomal abnormalities and complexity, reflexing the genetic instability of the neoplastic clone [13]. By analogy, we used this approach for the CMML patient cohort in this study. We showed that patients with >3 abnormalities had a significantly inferior survival compared with patients with 3 abnormalities. We suggest that this modification of the Spanish CMML cytogenetic prognostic system would further stratify CMML patients within the high-risk group. Risk of AML transformation was lower in patients with >3 abnormalities, but this lower AML risk could be partially explained by a very short survival observed in patients with >3 abnormalities. These patients likely died of bone marrow failure or CMML related complications before transformation to AML can develop.
Of the very infrequent isolated abnormalities observed in less than 5 patients, one rare abnormality t(11;19)(q23;p13.1) involving MLL gene was identified in two patients. Both patients developed AML rapidly (in 4 and 9 months), and died shortly (9 and 13 months after CMML diagnosis, respectively). A similar aggressive clinical course was observed in two patients with trisomy 21. However, the small number of patients with individual single chromosomal abnormalities precludes a meaningful analysis of their potential association with survival or risk of AML.
We showed that if we modified the Spanish cytogenetic risk grouping simply by moving trisomy 8 to the intermediate risk group, it resulted in a better separation of OS and LFS among three risk groups. Both the intermediate and high risk groups independently and negatively impacted OS and LFS. This modified cytogenetic grouping remained to be prognostically relevant when incorporated in the recently proposed Mayo clinic Model for CMML [11]. Interesting, an AMC cut off of 3x10^9/L rather than 10x10^9/L in the Mayo clinic model seemed to be a better strata in our patients cohort. Notably, only about 11% of our patients had an AMC >10x10^9/L, likely contributing to the difference we observed.
Recently, ASXL1 mutational status have been found to be associated with an inferior outcome in patients with CMML and incorporated in the model proposed by Itzykson et al [7]. It has been shown to greatly improve the Mayo prognostic model to delineate risk categories [11]. Our study is retrospective, and we did not have ASXL1 mutation status in majority of the patients. Incorporating ASXL1 mutation status along with cytogenetic risk grouping with the known adverse parameters may provide a more robust risk scoring system.
In summary, cytogenetic data can play an important and independent role in the assessment of the prognosis of CMML patients. The cytogenetic prognostic system suggested by Spanish study group in 2011 was an excellent beginning, but we suggest here some revisions that we believe would better stratify patients into prognostically different groups. In particular, isolated trisomy 8 in CMML patients is better considered an intermediate-risk finding. In addition, we suggest that CMML patients with >3 abnormalities have an extremely short survival and therefore deserve a separate risk category. The reclassification of isolated trisomy 8 and the division of cytogenetic abnormalities into 3 versus >3 is analogous to the approach of the IPSS-R used for MDS patients.
Acknowledgments
We would like to extend our appreciation to our colleagues, for the helpful discussions and support throughout this study.
References
- 1.Worsley A, Oscier DG, Stevens J, et al. Prognostic features of chronic myelomonocytic leukaemia: a modified Bournemouth score gives the best prediction of survival. Br J Haematol. 1988;68:17–21. doi: 10.1111/j.1365-2141.1988.tb04173.x. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez-Medina I, Bueno J, Torrequebrada A, et al. Two groups of chronic myelomonocytic leukaemia: myelodysplastic and myeloproliferative. Prognostic implications in a series of a single center. Leuk Res. 2002;26:821–824. doi: 10.1016/s0145-2126(02)00021-8. [DOI] [PubMed] [Google Scholar]
- 3.Onida F, Kantarjian HM, Smith TL, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99:840–849. doi: 10.1182/blood.v99.3.840. [DOI] [PubMed] [Google Scholar]
- 4.Germing U, Kundgen A, Gattermann N. Risk assessment in chronic myelomonocytic leukemia (CMML) Leuk Lymphoma. 2004;45:1311–1318. doi: 10.1080/1042819042000207271. [DOI] [PubMed] [Google Scholar]
- 5.Such E, Cervera J, Costa D, et al. Cytogenetic risk stratification in chronic myelomonocytic leukemia. Haematologica. 2011;96:375–383. doi: 10.3324/haematol.2010.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Such E, Germing U, Malcovati L, et al. Development and validation of a prognostic scoring system for patients with chronic myelomonocytic leukemia. Blood. 2013;121:3005–3015. doi: 10.1182/blood-2012-08-452938. [DOI] [PubMed] [Google Scholar]
- 7.Itzykson R, Kosmider O, Renneville A, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31:2428–2436. doi: 10.1200/JCO.2012.47.3314. [DOI] [PubMed] [Google Scholar]
- 8.Beran M, Wen S, Shen Y, et al. Prognostic factors and risk assessment in chronic myelomonocytic leukemia: validation study of the M.D. Anderson Prognostic Scoring System. Leuk Lymphoma. 2007;48:1150–1160. doi: 10.1080/10428190701216386. [DOI] [PubMed] [Google Scholar]
- 9.Parikh SA, Tefferi A. Chronic myelomonocytic leukemia: 2013 update on diagnosis, risk stratification, and management. Am J Hematol. 2013;88:967–974. doi: 10.1002/ajh.23574. [DOI] [PubMed] [Google Scholar]
- 10.Patnaik MM, Padron E, Laborde RR, et al. Mayo prognostic model for WHO-defined chronic myelomonocytic leukemia: ASXL1 and spliceosome component mutations and outcomes. Leukemia. 2013;27:1504–1510. doi: 10.1038/leu.2013.88. [DOI] [PubMed] [Google Scholar]
- 11.Patnaik MM, Itzykson R, Lasho TL, et al. ASXL1 and SETBP1 mutations and their prognostic contribution in chronic myelomonocytic leukemia: a two center study of 466 patients. Leukemia. 2014 doi: 10.1038/leu.2014.125. in press. [DOI] [PubMed] [Google Scholar]
- 12.Kohlmann A, Grossmann V, Klein HU, et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72. 8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010;28:3858–3865. doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- 13.Haase D, Germing U, Schanz J, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 2007;110:4385–4395. doi: 10.1182/blood-2007-03-082404. [DOI] [PubMed] [Google Scholar]
- 14.Sole F, Espinet B, Sanz GF, et al. Incidence, characterization and prognostic significance of chromosomal abnormalities in 640 patients with primary myelodysplastic syndromes. Br J Haematol. 2000;108:346–356. doi: 10.1046/j.1365-2141.2000.01868.x. [DOI] [PubMed] [Google Scholar]
- 15.Sole F, Luno E, Sanzo C, et al. Identification of novel cytogenetic markers with prognostic significance in a series of 968 patients with primary myelodysplastic syndromes. Haematologica. 2005;90:1168–1178. [PubMed] [Google Scholar]
- 16.Bacher U, Haferlach T, Kern W, et al. Conventional cytogenetics of myeloproliferative diseases other than CML contribute valid information. Ann Hematol. 2005;84:250–257. doi: 10.1007/s00277-004-0977-1. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swerdlow SH, Campo C, Harris NL, et al. International Agency for Research on Cancer, editor. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: 2008. [Google Scholar]
- 19.Shaffer LG, McGowan-Jordan J, Schmid M. An international system for human cytogenetic nomenclature. S. Karger; Basel: 2013. [Google Scholar]
- 20.Schanz J, Tuchler H, Sole F, et al. New comprehensive cytogenetic scoring system for primary myelodysplastic syndromes (MDS) and oligoblastic acute myeloid leukemia after MDS derived from an international database merge. J Clin Oncol. 2012;30:820–829. doi: 10.1200/JCO.2011.35.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussein K, Pardanani AD, Van Dyke DL, et al. International Prognostic Scoring System-independent cytogenetic risk categorization in primary myelofibrosis. Blood. 2010;115:496–499. doi: 10.1182/blood-2009-08-240135. [DOI] [PubMed] [Google Scholar]
- 22.Pozdnyakova O, Miron PM, Tang G, et al. Cytogenetic abnormalities in a series of 1,029 patients with primary myelodysplastic syndromes: a report from the US with a focus on some undefined single chromosomal abnormalities. Cancer. 2008;113:3331–3340. doi: 10.1002/cncr.23977. [DOI] [PubMed] [Google Scholar]
- 23.Paulsson K, Heidenblad M, Strombeck B, et al. High-resolution genome-wide array-based comparative genome hybridization reveals cryptic chromosome changes in AML and MDS cases with trisomy 8 as the sole cytogenetic aberration. Leukemia. 2006;20:840–846. doi: 10.1038/sj.leu.2404145. [DOI] [PubMed] [Google Scholar]