Summary
Human cytomegalovirus (HCMV) infection rewires host cell metabolism, up-regulating flux from glucose into acetyl-CoA to feed fatty acid metabolism, with saturated very long-chain fatty acids (VLFCA) required for production of infectious virion progeny. The human genome encodes seven elongase enzymes (ELOVL) that extend long chain fatty acids into VLCFA. Here we identify ELOVL7 as pivotal for HCMV infection. HCMV induces ELOVL7 by more than 150-fold. This induction is dependent on mTOR and SREBP-1. ELOVL7 knockdown or mTOR inhibition impairs HCMV-induced fatty acid elongation, HCMV particle release, and infectivity per particle. ELOVL7 overexpression enhances HCMV replication. During HCMV infection, mTOR activity is maintained by the viral protein pUL38. Expression of pUL38 is sufficient to induce ELOVL7, and pUL38-deficient virus is partially defective in ELOVL7 induction and fatty acid elongation. Thus, through its ability to modulate mTOR and SREBP-1, HCMV induces ELOVL7 to synthesize the saturated VLCFA required for efficient virus replication.
Introduction
Human cytomegalovirus (HCMV), a β-herpesvirus, is a widely dispersed enveloped virus that establishes a life-long persistent infection in greater than 60% of the world population (Mocarski et al., 2007). HCMV is a major cause of birth defects, an opportunistic infection in HIV-1/AIDS patients, and a life-threatening post-transplant complication in allograft recipients (Britt, 2008). In addition, it has been associated with glioblastoma and other cancers (Cobbs, 2013; Cobbs et al., 2002), cardiovascular disease (Streblow et al., 2008) and immune senescence (Moss, 2010).
To meet their needs viruses modulate specific cellular metabolic networks, but little is known about how most viruses exploit cellular metabolic and lipid environments. HCMV provides a model system for studying how viruses rewire cellular physiology since a global change in metabolism of infected cells occurs during replication (Chambers et al., 2010; Munger et al., 2008; Rabinowitz et al., 2011; Vastag et al., 2011). HCMV increases glucose uptake (Munger et al., 2006; Yu et al., 2011), glycolytic and TCA fluxes (Munger et al., 2008), as well as lipid metabolism (Koyuncu et al., 2013; Sanchez and Dong, 2010; Yu et al., 2012). HCMV, like all enveloped viruses, depends on its host cells to provide the lipids required to build its envelope. Previously we reported that infected cells use carbons derived from glucose to synthesize very long-chain fatty acids (VLCFA), which are required for efficient viral replication (Koyuncu et al., 2013).
Fatty acid (FA) synthesis is initiated by FA synthase (FAS) forming long-chain FAs up to 16 carbons in length (C16) by connecting carbons, two at time, using malonyl-CoA as a substrate. This process is controlled by the synthesis of malonyl-CoA from acetyl-CoA by acetyl-CoA carboxylase 1 (ACC-1). The product of FAS is the 16-carbon FA palmitate (C16:0, the number following the colon represents the number of double bonds in the FA). Palmitate can be processed further to make a longer chain or desaturated to introduce a double bond among the carbons in the tail. In human cells, longer FAs are made by one or more of the seven elongases (ELOVL1-7), again by using 2 carbon units from malonyl-CoA.
ELOVLs are important for various biological processes, such as proper development (Harkewicz et al., 2012; Li et al., 2007). They may also play a role in disease processes; for example, ELOVL7 has been implicated in the growth of prostate tumor cells (Tamura et al., 2009). ELOVLs show substrate preference depending on the chain length and degree of saturation of the FA chain to be elongated. In general, saturated FAs can be elongated by ELOVL1, 3, 4 and 7 (Ohno et al., 2010; Tamura et al., 2009), monounsaturated FAs by ELOVL1, 3, 5, 6 and 7 (Kitazawa et al., 2009; Ofman et al., 2010; Ohno et al., 2010), and polyunsaturated FAs by ELOVL 2, 4, and 5 (Harkewicz et al., 2012; Leonard et al., 2002; Ohno et al., 2010). ELOVL1, 4, 5, 6 are ubiquitously expressed whereas the others are more tissue tropic (Ohno et al., 2010). The expression of ELOVLs is controlled by transcription factors such as sterol regulatory element-binding proteins (SREBPs) but it is likely that cells also use additional mechanisms to control their expression and activity (Jakobsson et al., 2006; Moon et al., 2001).
The requirement for FA elongation is a poorly understood aspect of HCMV biology. Here we examine the role of ELOVLs in HCMV replication, identifying a pivotal role specifically for ELOVL7. Metabolic tracers were used to establish that HCMV infection induces the ELOVL7-dependent synthesis of lipids with VLCFA tails that are subsequently used to build the virion envelope. ELOVL7 was found to be essential for the efficient release of viral particles and to the infectivity of cell-free progeny. Additionally, we demonstrate that HCMV, in part via pUL38, strongly induces the expression and activity of ELOVL7 by manipulating the mammalian target of rapamycin kinase (mTOR) and SREBP1. Our results illustrate that HCMV commandeers multiple cellular mechanisms (e.g. kinase and transcriptional activity) to ensure ELOVL7 produces saturated VLCFAs that are required to build a properly functioning viral envelope.
Results
Glucose-derived two-carbon units contribute to the virion envelope via elongation
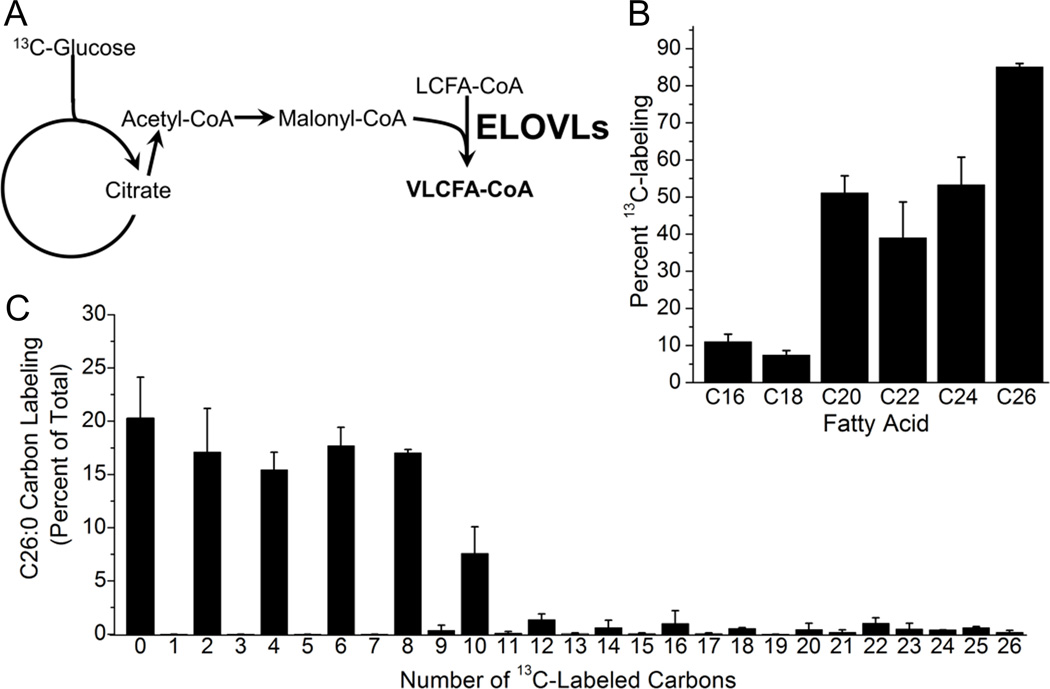
Enveloped viruses, such as HMCV, either (i) derive the lipids to build their envelope from those present in the host cell prior to infection and/or (ii) induce the cell to synthesize the required lipids during the viral replication cycle. HCMV infection of fibroblast cells results in up-regulated FA elongation flux, resulting in a several-fold increase in VLCFAs in the host cell after infection with HCMV at a high multiplicity of infection (3 infectious units (IU) per cell) (Koyuncu et al., 2013). We hypothesized that these newly synthesized saturated VLCFAs are used to build virion lipids. This hypothesis was directly tested using uniformly labeled 13C-glucose as a metabolic tracer. Flow of carbons derived from glucose into FA synthesis and elongation results in labeling of the FA hydrocarbon tails (Figure 1A). The experiments were performed under serum-free conditions to eliminate contaminating sources of FAs from fetal bovine serum, with lipids extracted from the biological samples saponified to release the FA tails, enabling the analysis of these tails by LC-MS. Most C16:0 and C18:0 FA tails in the viral envelope were unlabeled (Figure 1B). In contrast, longer chains were predominantly labeled with more than 80% of the measured C26:0 containing at least one 13C-labeled two-carbon unit. Evaluation also of unsaturated FA revealed that virion FA labeling was greatest with increasing chain length and a lower degree of saturation (Figures S1A). The labeling of FAs from virions was compared to FA isolated from uninfected and infected fibroblasts. Consistent with our previous work, viral infection increased 13C-incorparation into host cell FAs, particularly the VLCFAs (Koyuncu et al., 2013 and Figure S1A). Overall the extent of labeling in the virions was similar to infected cells, with most extensive labeling of saturated VLCFAs (Figure S1A). The observation that saturated FAs are enriched in the viral envelope in comparison to unsaturated FAs (Figure S1B) further underscores their importance to HCMV replication.
Figure 1.
HCMV induces fatty acid elongation to produce VLCFAs that are incorporated into the virion envelope.
(A) Schematic of pathway from glucose to fatty acid elongation. The straight line down from glucose indicates glycolysis and the circle indicates the TCA cycle. Fatty acid elongases (ELOVLs) use malonyl-CoA as a substrate to elongate shorter fatty acids.
(B) Incorporation of labeled carbons from 13C-glucose into saturated FAs in purified virions. Virions were isolated from cells grown in serum-free DMEM containing all glucose U-13C-labeled. For each FA species the data is reported as the percentage that contains at least one 13C-labeled two-carbon unit.
(C) The labeling pattern of C26:0 from virions grown as in (b).
All data are represented as mean ± SEM of three independent experiments. See also Figure S1.
Next, we evaluated the isotope labeling pattern of the virion VLCFAs. FAS generates one C16:0 LCFA using 2-carbon units from one acetyl-CoA and seven malonyl-CoA molecules. The resulting C16 FA can be elongated by ELOVLs using 2-carbon units derived from malonyl-CoA. In the case of C26:0, de novo synthesis would result in 12–26 labeled carbons depending on the extent to which the acetyl-CoA/malonyl-CoA pools are labeled, while the incorporation of 10 or fewer 13C-carbons would demonstrate that the VLCFAs are made by elongation of preexisting FAs. In virions the C26:0 contained 10 or fewer 13C-labeled carbons demonstrating that most of the VLCFAs are made by ELOVLs extending shorter-chained FAs that existed in the cell prior to infection (Figure 1C). The C26:0 labeling pattern in virion was similar to that of infected cells (Koyuncu et al., 2013 and Figure S1C). Thus, induction of malonyl-CoA synthesis from glucose by HCMV contributes to the virion envelope via saturated FA elongation.
To further define the relationship between HCMV replication and FA elongation the 13C-labeling of FAs in cells infected at various multiplicities was examined. Fibroblast cells infected at multiplicity as low as 0.5 IU per cell showed an increased labeling of saturated FAs, with the overall labeling of VLCFAs dependent on the amount of input virus (Figure S1D). Similarly, the cellular concentration of saturated VLCFA, especially those of 26 carbons or longer, was dependent on the multiplicity of infection (Figure S1E). These observations reinforce the conclusion that HCMV induces FA elongation. Collectively, these data demonstrate that during viral replication FA elongation is upregulated to build lipids containing saturated VLCFA tails to be used for the viral envelope.
Elongation catalyzed by ELOVL7 is required for efficient HCMV replication
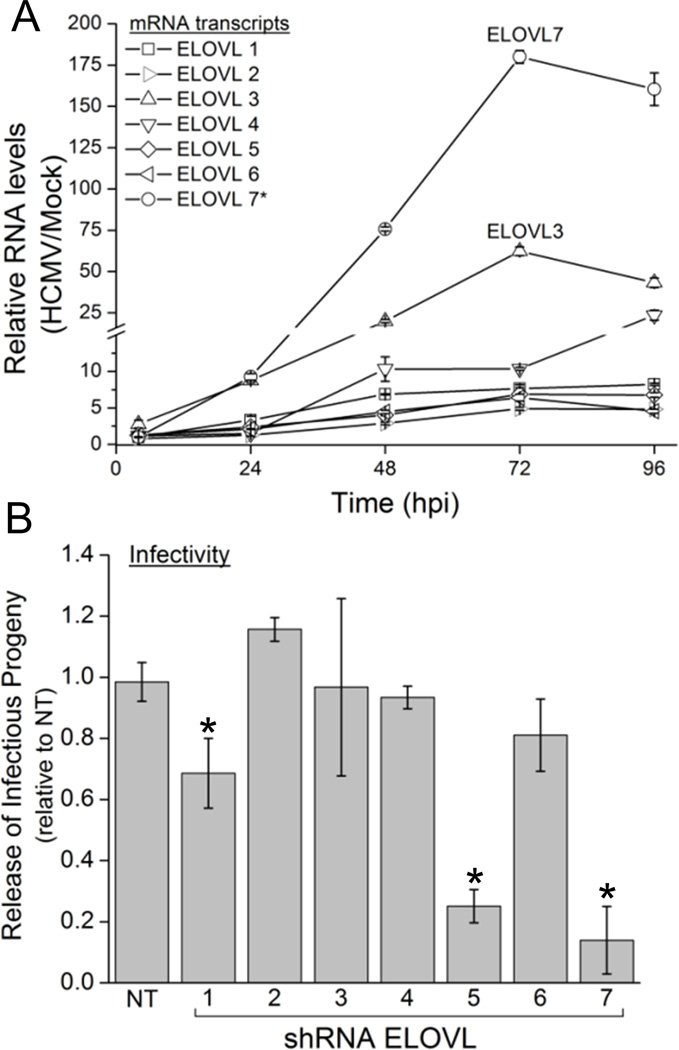
We were curious which of the seven known ELOVLs contribute to the observed FA elongation during HCMV infection. We first determined how the expression of each ELOVL was altered by HCMV. The mechanisms controlling ELOVL activity have not been thoroughly studied, but their activity is regulated at least in part at the level of gene expression (Jakobsson et al., 2006; Wang et al., 2006); thus the RNA levels of each ELOVL was tested following infection at a multiplicity of 3 IU/cell. By 48 hours post infection (hpi), the expression of all seven ELOVLs was at least 2-fold increased (Figure 2A), confirming findings of a previously performed screen (Koyuncu et al., 2013). The expression of some ELOVLs continued to increase as replication progressed. Of note, ELOVL7 levels were below a level that could be reliably detected in uninfected cells; however, its RNA was observed at low levels in two out of three experiments in the 4 hpi samples. Remarkably, ELOVL7 RNA levels increased over 150-fold from the level observed at 4 hpi; thus, compared to uninfected cells, HCMV infected cells express ≥150-fold greater amounts of ELOVL7 mRNA.
Figure 2.
Fatty acid elongase 7 is strongly induced by HCMV and required for efficient production of infectious progeny.
(A) ELOVL1-7 mRNA levels during the course of HCMV replication. MRC-5 fibroblasts infected at 3 IU/cell were compared to mock infected cells for ELOVL1-6. The (*) denotes that, because the ELOVL7 transcript was not detectable in uninfected cells, the ELOVL7 the data is reported as fold change from 4 hpi.
(B) HCMV replication in MRC-5 fibroblasts with shRNA knockdown of ELOVL1-7 (compared to non-targeting (NT) sequence control). Knockdown cells were infected at 0.5 IU/cell and at 96 hpi the medium was assayed for infectious progeny. (* p<0.05, t-test)
All data are represented as mean ± SEM of at least three independent experiments. See also Figure S2.
To test which, if any, of the seven ELOVLs is required for viral replication an shRNA knockdown strategy was employed. Three shRNA clones targeting each ELOVL were used to generate a stable knockdown in MRC-5 fibroblasts. Viral replication was decreased by all clones tested for ELOVL5 and 7, and by one clone of ELOVL1 (Figure S2A). To increase the knockdown efficiency in infected cells and to counteract the induction of ELOVLs by HCMV we pooled the three clones per ELOVL and generated knockdown cells that received each lentivirus at a multiplicity of at least 3 IU/cell. Using this pooling method a greater than 60% knockdown of mRNA at 96 hpi was achieved for all ELOVLs; most ELOVL mRNA levels including, ELOVL7, were decreased by 80–90% (Figure S2B). Under these conditions, the decrease in ELOVL5 and 7 resulted in a marked reduction in the production of extracellular infectious progeny (Figures 2B and S2C). Knockdown of ELOVL1 resulted in a modest reduction (Figure 2B), but this is likely due to the knockdown affecting the health of the host cell since these fibroblasts had a severe growth defect (Figure S2D). Thus, the shRNA knockdown experiments demonstrate that ELOVL5 and 7 are required for efficient HCMV replication.
Since ELOVL7 is reported to elongate saturated VLCFAs whereas ELOVL5 is thought to produce PUFAs (Leonard et al., 2002; Tamura et al., 2009), and given the dramatic increase in ELOVL7 RNA levels caused by HCMV infection, we hypothesized that ELOVL7 activity is essential for the observed enrichment in saturated VLCFAs in infected cells and virions. Thus, we focused on saturated FAs and ELOVL7.
ELOVL7 is essential for virus-induced VLCFA synthesis
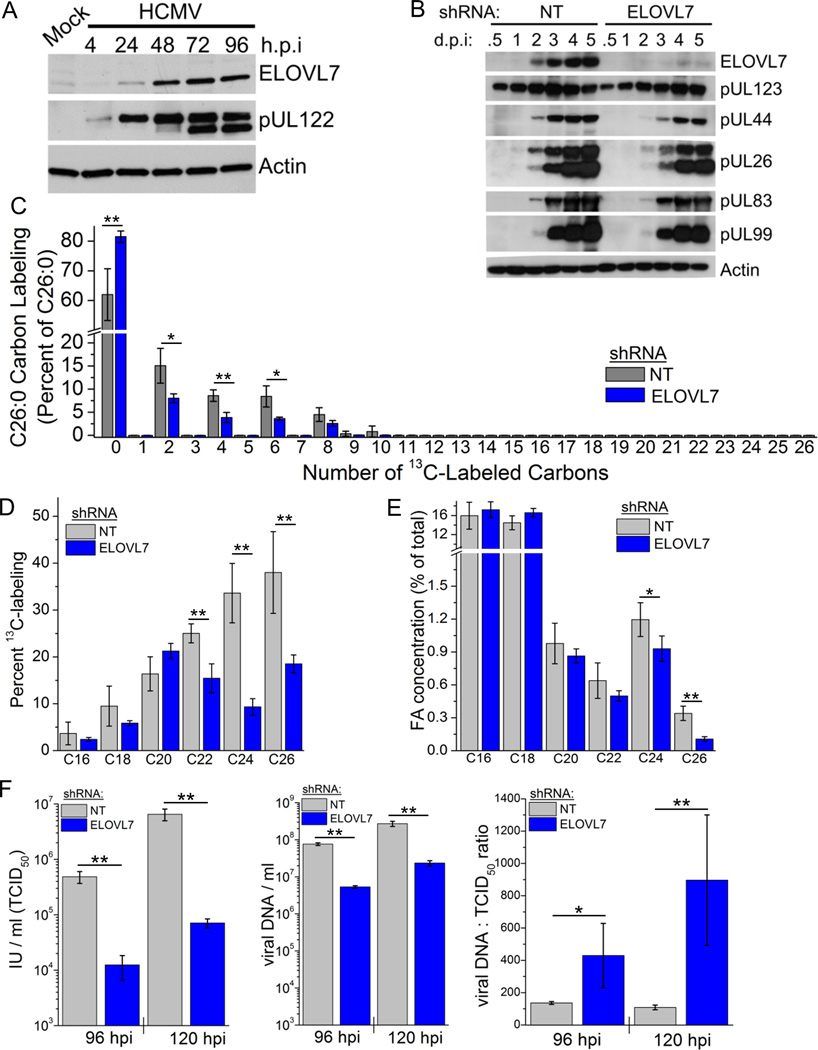
As expected from the RNA results, ELOVL7 protein was detected at a low level in uninfected fibroblasts but strongly increased during the course of infection (Figure 3A). ELOVL7 knockdown cells were used to evaluate if the loss of this enzyme altered the kinetics of viral replication. The replication of HCMV can be divided into three phases: immediate-early (IE; viral gene expression that occurs following viral entry), early (genes expressed subsequently to IE genes and prior to viral DNA replication), and late (those that require viral DNA replication). In these experiments, ELOVL7 protein knockdown was maintained for 120 h following infection (Figure 3B). Although the early proteins assayed (pUL44 and pUL26) accumulated with a slight delay, all viral proteins, including late viral proteins (pUL99 and pUL83) accumulated to normal levels in ELOVL7-deficient cells. Viral DNA synthesis was lower but not significantly altered in knockdown cells as predicted since the expression of the late viral proteins is chiefly dependent on DNA replication (Figure S2E). Altogether, these data suggest that the decrease in the release of infectious progeny by the knockdown cells is due to a defect downstream of late protein synthesis.
Figure 3.
ELOVL7 synthesizes fatty acids required for virion infectivity.
(A) ELOVL7 protein level during HCMV replication (3 IU/cell). An immediate-early protein (pUL122) is shown as a marker of viral replication.
(B) Kinetics of the HCMV replication cycle in ELOVL7 knockdown cells (compared to NT control cells). Immediate-early protein 1 (pUL123; IE1), early proteins pUL44 and pUL26, leaky-late protein pUL83, and late pUL99 protein were examined.
(C) C26:0 labeling pattern in knockdown cells grown in DMEM without fetal bovine serum with all glucose U-13C-labeled. Cells were harvested at 72 hpi following infection at (1 IU/cell, 72 hpi).
(D) Saturated FA labeling in cells grown under labeling conditions described in part (C).
(E) Concentration of saturated FAs in cells grown in serum-free DMEM (72 hpi, 1 IU/cell).
(F) The release of infectious progeny, DNA-containing particles, and particle-to-infectious progeny ratio (1 IU/cell). Left panel: Infectious progeny as measured by TCID50 (left panel). Total (infectious and non-infectious) cell-free viral particles were determined by the number of genomes (viral DNA) per ml of medium collected from the cells (middle panel). The particle to IU ratio was also compared (right panel).
All data are represented as mean ± SEM of three independent experiments. (* p<0.05, ** p<0.01, t-test)
The ability of ELOVL7 knockdown cells to produce saturated FAs during HCMV infection was examined using 13C-glucose. Knockdown of ELOVL7 substantially reduced the labeling of C26:0 and other VLCFAs (Figure 3C, D). This reduced production resulted in a selective depletion of VLCFA tails, especially of C26:0, the VLCFA that is most dramatically increased during viral replication (Figures 3E and S2G). The increasing extent of depletion with greater FA length is consistent with the synthesis of the longer VLCFA chains requiring more elongation steps (e.g., five for C26:0 versus two for C20:0) and/or with ELOVL7 being particularly important for the later steps. Thus, during HCMV infection, ELOVL7 is required to produce saturated VLCFA tails.
To evaluate the consequence of deficiency of such VLCFAs on viral replication, we quantitated the number of infectious particles released by control and ELOVL7 knockdown cells by TCID50. Infectious particles released into the medium at 96 and 120 hpi were decreased by a factor of ~20 in the knockdown cells (Figure 3F, left panel). A partial recovery of infectivity was restored by adding one product of ELOVL7, C26:0, to knockdown cells demonstrating that the loss of saturated VLCFA is responsible for the decrease in infectivity (Figure S2F); a more complete recovery of infectivity may likely require the addition of other VLCFAs, each at a proper concentration, or might require subcellular localization of the VLCFAs which is not achieved via exogenous feeding. Interestingly, adding C26:0 to control cells caused a minor loss, ~2-fold, of infectivity suggesting that an optimal level (and/or localization) of saturated VLCFAs is required for viral replication (Figure S2F).
To examine whether this decreased activity reflected a failure to release particles or defective infectivity of the particles that are released, we measured the particle number by determining the amount of encapsidated viral DNA released by cells. The particle number was substantially reduced by ELOLV7 knockdown, but not as much as the number of infectious progeny (Figure 3F, middle panel). The particle to IU ratio released by control cells was ~100 particles per virion at both time points assayed; in contrast the ratio was ~400 and ~900 particles per virion in knockdown cells at 96 and 120 hpi, respectively (Figure 3F, right panel). Thus, ELOVL7 is required both for particle release and for infectivity of the released particles.
Overexpression of ELOVL7 enhances production of infectious HCMV progeny
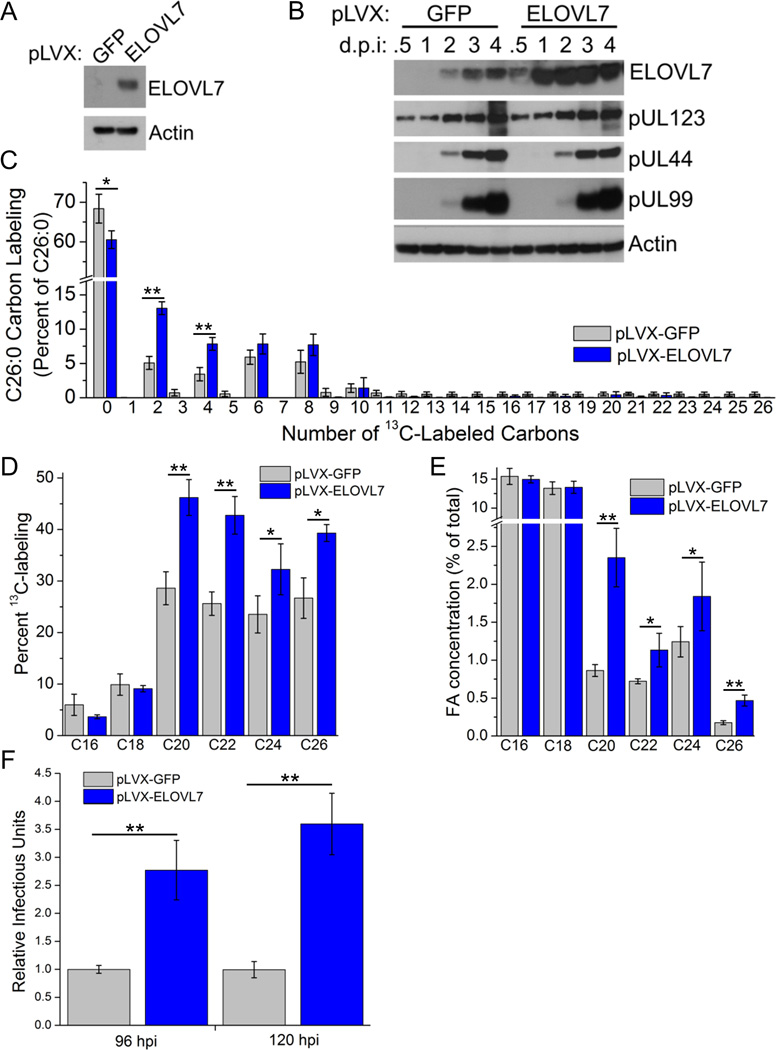
We have demonstrated the ELOVL7 is necessary for viral replication. To further monitor the ability of ELOVL7 to optimize HMCV infectivity, cells stably overexpressing ELOVL7 were generated by lentiviral transduction (Figure 4A). Viral protein expression in ELOVL7 overexpressing cells was indistinguishable from that in controls cells stably expressing GFP suggesting that increased levels of ELOVL7 do not affect viral protein expression and replication kinetics (Figure 4B). The overall incorporation of 13C from labeled glucose into saturated VLCFAs was enhanced by the overexpression of ELOVL7, demonstrating increased elongation activity in these cells (Figure 4C, D), which resulted in higher concentrations of the associated saturated VLCFA products (Figure 4E). A significant concentration increase was seen for fatty acids of 20 carbons and longer. Thus, ELOVL7 is sufficient to produce significant quantities of these FAs, although the ELOVL7 knockdown data suggest that it is necessary primarily for only those of 24 or more carbons (Figure 2D, E), perhaps because other elongases can also produce C20 or in the context of overexpression ELOVL7 releases C20 after elongating C16–C18. Importantly, ELOVL7 overexpression enhanced the ability of host cells to support viral replication as indicated by a greater than 2-fold increase in the release of infectious progeny compared to controls cells expressing GFP (Figure 4F). These experiments support the view that the ability of host cells to generate lipids with saturated VLCFAs is a critical factor that limits HCMV replication in fibroblasts. Overall we conclude that ELOVL7 plays an important role in HCMV replication by providing the VLCFAs necessary to build infectious virions.
Figure 4.
HCMV replication is enhanced by overexpression of ELOVL7.
(A) ELOVL7 protein levels in uninfected MRC-5 fibroblast cells following ELOVL7 overexpression using pLVX-lentiviral stable expression (GFP-expressing cells were used as a control).
(B) Kinetics of the HCMV replication cycle (1 IU/cell).
(C) C26:0 labeling pattern (1 IU/cell, 72 hpi).
(D) Saturated FA labeling (1 IU/cell, 72 hpi).
(E) Concentration of saturated FAs (72 hpi, 1 IU/cell).
(F) Production of cell-free infectious virus (1 IU/cell).
All data are represented as mean ± SEM of three independent experiments. (* p<0.05, ** p<0.01, t-test)
Viral protein pUL38 activates mammalian target of rapamycin (mTOR) and thereby induces ELOVL7
Our results establish that HCMV strongly induces the expression of ELOVL7 to generate VLCFAs required to form virions. To probe the mechanism underlying the manipulation of ELOVL7 by HCMV, we examined the role of mTOR in the ELOVL7 expression. mTOR complex 1 (mTORC1) kinase activity is required to maintain active translation (Mamane et al., 2006), and the kinase also controls key aspects of lipid metabolism via several mechanisms, including SREBP-1 activation (Laplante and Sabatini, 2009; Lewis et al., 2011). HCMV induces stress response signaling that would normally block mTOR activity. However, HCMV encodes a viral protein, pUL38, which binds and antagonizes the tuberous sclerosis protein complex, a suppressor of mTOR (Moorman et al., 2008). Through this and additional mechanisms, the kinase is maintained in an active state following infection (Clippinger and Alwine, 2012; Kudchodkar et al., 2004, 2006; Moorman et al., 2008; Moorman and Shenk, 2010).
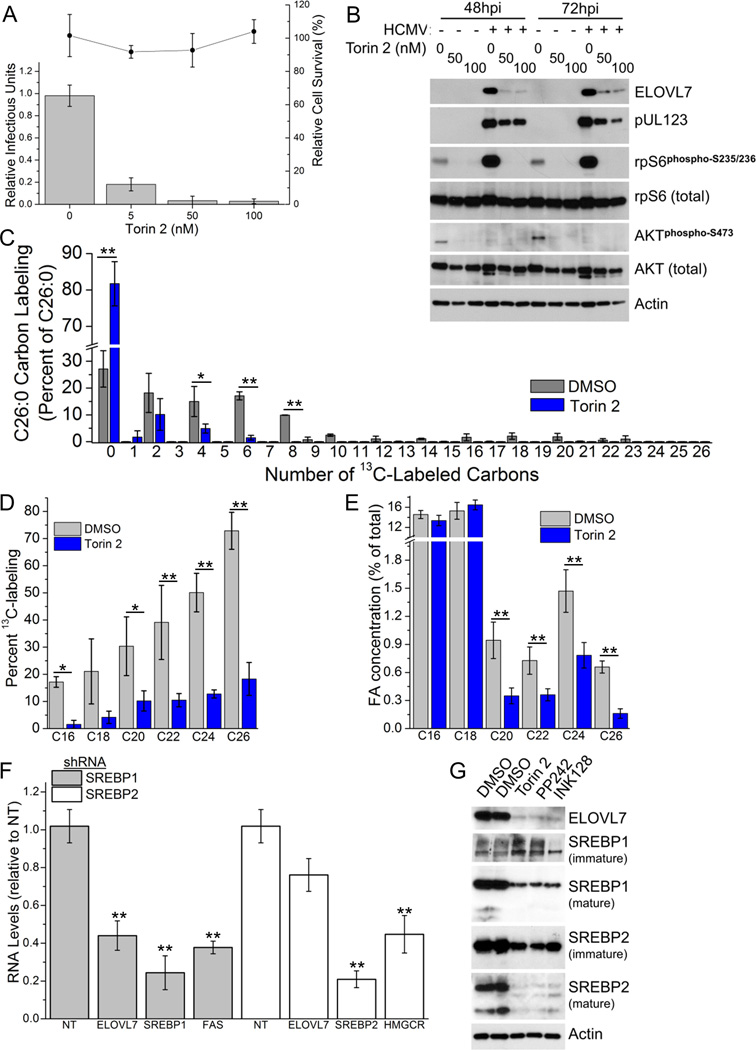
To determine the importance of mTOR activity in HCMV replication and ELOVL7 induction, cells were treated with torin 2, a selective and potent inhibitor of mTORC1 and mTORC2 (Liu et al., 2011a). Torin 2 blocked HCMV replication without any discernable cytotoxic effects (Figure 5A). The ability of torin 2 to inhibit mTOR activity was evaluated by examining the phosphorylation of two known targets of mTOR: ribosomal protein S6 and AKT. Phosphorylation of both targets was completely blocked by 50 nM torin 2 (Figure 5B). Inhibition of mTOR resulted in a slight reduction in the accumulation of an HCMV immediate-early protein (pUL123, IE1) at 48 and 72 hpi, similar to the results previously reported for torin 1 treatment, a related mTOR inhibitor (Moorman and Shenk, 2010). Treatment with torin 2 markedly reduced the HCMV-induced up-regulation of ELOVL7 transcript (Figure S3C) and protein (Figure 5B). Similar results were observed with two additional—but structurally distinct—mTOR inhibitors (Figure S3A–B). Although drug treatment decreased ELOVL7 expression, a restoration of viral replication was not observed in cells expressing ELOVL7 cells likely due to the polymorphic function of mTOR during HCMV infection (Figure S3D). Moreover, torin 2 treatment resulted in a major reduction in FA elongation when compared to DMSO-treated controls cells, as evident by less labeling of C26:0 (Figure 5C), and other long and very long chain saturated FAs (Figure 5D). Unlike the ELOVL7 knockdown experiments (Figure 3), torin 2 treatment altered the labeling of shorter-chained FAs suggesting that mTOR inhibition is also altering the activity of other enzymes involved in FA metabolism. Consistent with a reduction of elongase activity observed under labeling conditions, torin 2 treatment decreased the concentrations of the VCLFA species important for viral replication (Figure 5E). These data demonstrate that, in addition to its other roles (e.g. in protein translation), mTOR is important for lipid metabolism during HCMV infection.
Figure 5.
HCMV-induced mTOR and SREBP1 activity is required for lipid remodeling. In all panels, cells were treated with torin 2 to inhibit mTOR or DMSO as a control.
(A) Torin 2 blocked HCMV infectivity (bars) at various concentrations without altering cell survival of uninfected cells (circles).
(B) ELOVL7 and pUL123 protein levels. AKT and rpS6 are shown as controls to verify mTOR inhibition.
(C) C26:0 labeling pattern under 0.1 µM torin 2 treatment (3 IU/cell, 72 hpi).
(D) Saturated FA labeling under 0.1 µM torin 2 treatment (3 IU/cell, 72 hpi).
(E) Concentration of saturated FAs under 0.1 µM torin 2 treatment (3 IU/cell, 72 hpi).
(F) mRNA levels of ELOVL7 in SREBP1 and SREBP2 knockdown cells. FAS and HMGCR levels were used to monitor the decrease in activity of SREBP1 and 2, respectively.
(G) Maturation of SREBPs in infected cells treated with various mTOR inhibitors (0.1 µM torin 2, 1 µM PP242, and 0.5 µM INK128).
All data are represented as mean ± SEM of three independent experiments (* p<0.05, ** p<0.01, t-test). See also Figure S3.
mTOR may control of the expression of ELOVL7 by influencing the maturation and activity of SREBPs. SREBPs are transcription factors well known to regulate the expression of genes involved in FA metabolism (Ye and DeBose-Boyd, 2011), and HCMV stimulates the maturation of SREBP1 and 2 (Spencer et al., 2011; Yu et al., 2012). To investigate this further we examined the levels of ELOVL7 in host cells in which SREBP1 or SREBP2 levels were reduced using shRNA. The level of each transcription factor was reduced by approximately 80% compared to non-targeting shRNA control cells (Figure 5F). RNA for ELOVL7 was reduced by 2-fold in cells with reduced SREBP1; this reduction was similar to the RNA level of a known gene regulated by SREBP1, FAS (Ye and DeBose-Boyd, 2011). In contrast, only a 20% reduction in ELOVL7 RNA level was observed with knockdown of SREBP2—compared to a 2-fold decrease the RNA level of a SREBP2 regulated gene, HMG-CoA reductase—demonstrating that SREBPs, particularly SREBP1, regulates the level of ELOVL7 in HCMV infected cells. Because the mTOR inhibitor torin 1 has previously been shown to impact SREBP2 maturation in HCMV infected cells (Spencer et al., 2011; Yu et al., 2012), we examined the level of mature SREBPs in cells treated with three different mTOR inhibitors (Figure 5G). As expected, maturation of SREBP2 was dramatically reduced by each inhibitor. Moreover, the level of mature SREBP1 was reduced upon mTOR inhibition, thereby establishing a pathway via mTOR and SREBP1 by which HCMV induces ELOVL7 and FA elongation.
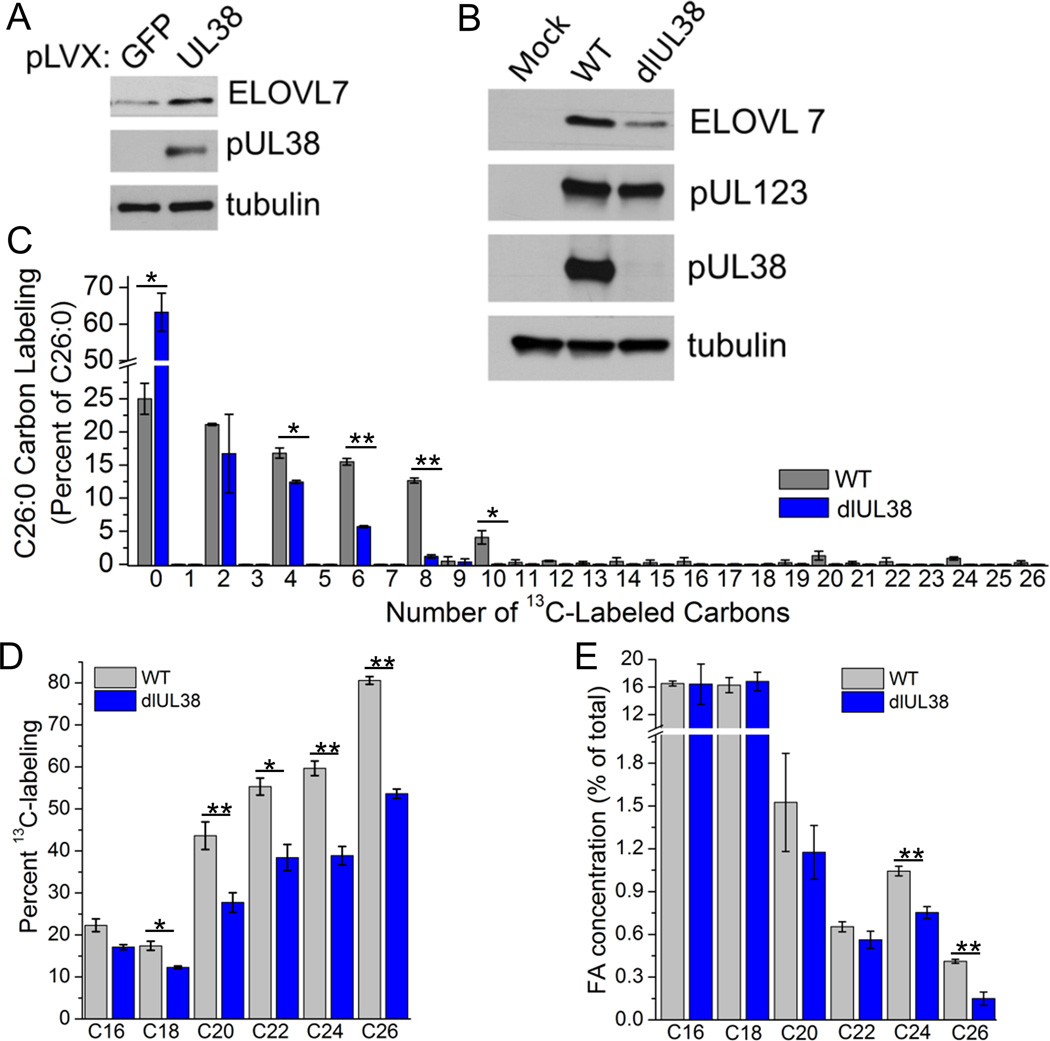
To evaluate more directly whether the HCMV-encoded, mTOR-activating pUL38 protein contributes to ELOVL7 expression we generated uninfected cells that stably expressed the protein. pUL38 alone increased the level of ELOVL7 protein compared to uninfected cells but not to the level observed in infected fibroblasts; however this observation demonstrates that it has the ability to control ELOVL7 expression independent of other virus-encoded factors (Figure 6A). Although pUL38 expression in uninfected cells induced ELOVL7, it was not sufficient to increase FA elongation, presumably due to the lack of up-regulation of upstream metabolic events (Figure S4A). To further probe the role of pUL38 in the expression of ELOVL7, cells were infected with a mutant virus, ADdlUL38, which lacks the UL38 gene (Terhune et al., 2007). Since cells infected with virus containing a UL38 deletion show dramatic cell death at 72 hpi (Terhune et al., 2007), we limited our analysis to 48 hpi. The mutant induced ELOVL7 expression, but not as strongly as wild-type virus (Figure 6B). Thus, although pUL38 is not the sole mechanism by which HCMV induces ELOVL7, pUL38 is necessary for its full expression.
Figure 6.
Viral pUL38 protein drives ELOVL7 expression.
(A) ELOVL7 protein level in uninfected cells expressing viral protein pUL38 (or GFP as a control) using pLVX-lentiviral stable expression.
(B) ELOVL7 protein expression in cells infected with WT (AD169) or ADdlUL38, a mutant virus lacking the UL38 gene (3 IU/cell, 48 hpi).
(C) C26:0 labeling pattern (3 IU/cell, 48 hpi).
(D) Saturated FA labeling (3 IU/cell, 48 hpi).
(E) Concentration of saturated FAs (3 IU/cell, 48 hpi).
All data are represented as mean ± SEM of three independent experiments (* p<0.05, ** p<0.01, t-test). See also Figure S4.
Consistent with a loss of ELOVL7 expression, we observed a substantial reduction in elongation of C26:0 and other saturated FA in ADdlUL38-infected cells at 48 hpi when compared to cells infected with WT virus (Figure 6C, D). The concentration of C26:0 was decreased by >2-fold in ADdlUL38-infected cells, while those of shorter lengths were less affected (Figure 6E). Similar to WT virus, ADdlUL38 replication was enhanced—but less than 2-fold—in ELOVL7 overexpressing cells at 120 hpi (Figure S4B compared to Figure 4F). Additionally, ELOVL7 particles released by ADdlUL38-infected cells had a similar disruption in the particle to IU ratio as those released by ELOVL7 knockdown cells infected with WT virus (Figures S4C and 3F). In sum, these data establish that pUL38, through its ability to control mTOR activity, aids in the synthesis of VLCFAs that are required to build infectious virions.
Discussion
FA metabolism is an important determinant for the replication of diverse viruses. Cells infected with another herpes virus, Kaposi’s sarcoma associated virus, exhibited elevated FA synthesis compared to uninfected endothelial cells (Delgado et al., 2012). Some RNA viruses, such as hepatitis C and rift valley fever viruses, remodel lipids during their assembly process (Moser et al., 2012; Nasheri et al., 2013; Petersen et al., 2014). In the case of dengue virus, FAS is relocalized to the location of viral replication so that C16:0 is produced at the intracellular site where the virus utilizes it (Heaton et al., 2010). Most previous studies have examined the role of FAS. The study described here highlights the importance of FA elongases, specifically ELOVL7, during viral replication.
HCMV instructs host cells to build lipids enriched with saturated VLCFAs that are required to construct a properly functioning virion. VLCFAs could contribute to the organization of lipid-embedded virion envelope proteins or to membrane dynamics during entry or egress of virions. Entry, assembly and egress of virions are dependent on multiple viral proteins and cellular processes (Alwine, 2012; Buchkovich et al., 2008; Gibson, 2008; Womack and Shenk, 2010). Our findings reveal that pUL38 contributes to one or more of these events by ensuring that ELOVL7 synthesizes the lipids containing saturated VLCFA that are incorporated into the virion, in addition to its role in controlling cell survival (Qian et al., 2011; Savaryn et al., 2013; Terhune et al., 2007).
Healthy primary fibroblasts have a low level of ELOVL7 expression (Janssen et al., 2009 and Figures 2A/3A); we showed that efficient HCMV replication in fibroblast cells requires ELOVL7 expression and activity. HMCV has broad cellular tropism and tissue distribution during acute infection (Mocarski et al., 2007) and one question raised by our results is: Are tissues that express ELOVL7 more susceptible to HCMV infection? In one case of fatal congenital infection, HCMV infected cells were found in tissues that also express ELOVL7, such as the pancreas, kidneys and liver (Bissinger et al., 2002; Ohno et al., 2010). However, the lungs were a key target for HCMV infection even though ELOVL7 expression in normal lung tissue was only faintly detected (Ohno et al., 2010). Based on our findings we would predict that ELOVL7 expression in the lungs would increase upon infection resulting in greater cellular susceptibility to viral replication.
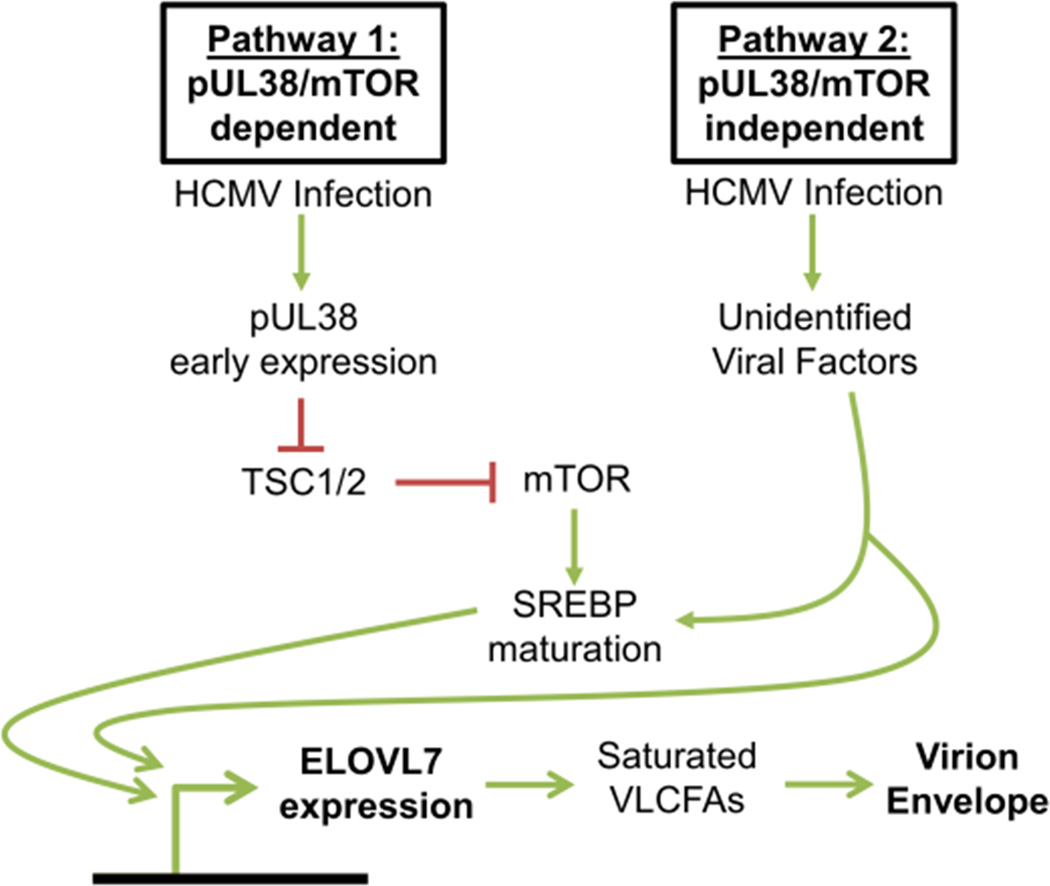
We propose a model for the steps of viral replication leading to the synthesis of the specialized lipids that are incorporated into virions (Figure 7). pUL38 is expressed with early kinetics allowing for mTOR signaling by preventing the block that would otherwise be introduced by the tuberous sclerosis protein complex (Moorman et al., 2008). Subsequently, mTOR stimulates the activity of SREBPs whose mature forms migrate from the ER to the nucleus to upregulate ELOVL7 gene expression. Additional downstream processes may also be influenced by mTOR, such translation of ELOVL7 mRNA. Although mTOR inhibition strongly blocks ELOVL7 protein induction, there is a modest induction in the presence of torin 2 (Figure 5B), and a pUL38-deficient virus can induce the elongase to a limited extent (Figure 6B). These observations argue that a second additional pathway—a pUL38 and/or mTOR independent pathway—also contributes to the elevated ELOVL7 accumulation induced by HCMV. The viral factor(s) responsible for this redundant pathway awaits identification but, since ELOVL7 levels are increased by 24 hpi (Figures 2A and 3A), it is likely that these additional viral factor(s) are expressed with immediate-early or early kinetics. It is unclear if the pUL38/mTOR independent pathway also requires SREBP maturation or relies on other cellular factors such as carbohydrate-response element binding protein (ChREBP), PKR-like endoplasmic reticulum (ER) kinase (PERK), or viperin that may contribute to the control of lipogenesis during HCMV replication (Seo and Cresswell, 2013; Yu et al., 2014; Yu et al., 2013). The observation that HCMV utilizes multiple mechanisms to stimulate ELOVL7 expression highlights the critical role of its activity to produce the VLCFAs that are required for viral replication.
Figure 7.
Schematic of mechanisms by which HCMV induces lipidome remodeling by ELOVL7.
ELOVL5 is also required to produce a maximal virus yield (Figure 2B). This elongase produces polyunsaturated FAs (PUFAs) (Leonard et al., 2002; Ohno et al., 2010). Our FA analyses reveal that although some PUFAs that may be produced by ELOVL5 were labeled (Figure S1A), they are only a minor constituent in the virion envelope (Figure S1B). Thus, ELOVL5 might not function to optimize lipid FA substituents in virus particles. Rather, it could counteract virus-induced cell stress. HCMV infection triggers various host cellular stresses (Alwine, 2008; Qian et al., 2011) and saturated VLCFAs, such as those elevated by HCMV, can induce an ER-stress response (Deguil et al., 2011; Karaskov et al., 2006). Indeed, exogenous C26:0 modestly inhibited the production of HCMV (Figure S2F), possibly through an elevated cell stress response. PUFAs, such as those produced by ELOVL5, aid in limiting the effect of cellular stresses (Pineau et al., 2009). Further studies are required to delineate the role of ELOVL5 during HCMV replication.
Overall our results demonstrate that HCMV infection dramatically induces the accumulation of ELOVL7 to synthesize lipids with saturated VLCFAs required for the production of infectious progeny. Furthermore, HCMV employs multiple mechanisms to ensure that ELOVL7 is expressed in fibroblast cells. Although the HCMV envelopment process remains largely a mystery, this work reveals the contribution of three host factors—mTOR, SREBP1, and ELOVL7—and one viral factor, pUL38, to the construction of a functional virion. Our HCMV results highlight the need to study the role of FA elongation and ELOVLs in the replication of other viruses to understand how enveloped viruses interact with host cells to ensure the generation of a lipid environment that favors viral progeny production.
Experimental Procedures
Cells, Virus and Experimental Setup
Human lung fibroblast cells, MRC-5, cultured in DMEM containing 10% FBS were used in this study. For experiments involving infection cells were maintained at confluence for three days and switched to DMEM without serum one day prior to infections. HCMV virus stocks of strain AD169 were made by clarifying the medium from MRC-5 cells via BAD, a bacterial artificial chromosomes (BAC) expressing the AD169 genome (Yu et al., 2002), followed by pelleting through 20% sorbitol and resuspending in serum-free DMEM. For viral envelope analysis, virions were prepared by further purification through two sequential sodium tartrate gradients (Liu et al., 2011b). Virus titers were determined using tissue culture infectious dose (TCID50). Relative infectious units (IU) were assessed using an anti-pUL123 (IE1) monoclonal antibody (clone 1B12) followed by visualization using an anti-mouse Alex-488 secondary antibody (Jackson ImmunoResearch) and quantified using an Operetta Instrument (Perkin-Elmer) as previously described (Grady et al., 2013; Terry et al., 2012). Virus lacking the UL38 gene was generated using the ADdlUL38 BAC (Terhune et al., 2007). Since the ADdlUL38 virus shows a limited cytopathic effect, the viral stocks for this mutant virus was determined by methanol fixing the cells on the TCID50 plates, then staining them using the anti-pUL123 antibody followed by a fluorescent secondary antibody to visualize infected cells. For 13C-labeling experiments, following infection the cells were grown in DMEM containing uniformly carbon labeled glucose (Cambridge Isotopes) at 4.5 g/L. Particle to TCID IU ratio was assayed by quantitating viral DNA using UL123-specific primers (Womack and Shenk, 2010).
MRC-5 cells expressing pUL38 and ELOVL7 from an EF1α promoter were generated by cloning the UL38 gene from BAD and ELOVL7 cDNA (Accession No. BC130310; Open Biosystems) into the pLVX-EF1α vector (Clontech). The cloned plasmids, along with psPAX2 and MD.2G plasmids (Addgene plasmids 12259 and 12260) were used to generate lentiviral particles for stable expression of the protein of interest in fibrobalsts. Following lentiviral treatment the cells were selected using 2 µg/ml puromycin. As a control, cells that expressed GFP from the pLVXEF1α plasmid were generated in parallel.
The following mTOR inhibitors were used in this study: torin 2 (50 and 100 nm; Tocris Biosciences), INK128 (500 nM; Cayman Chemicals), and PP242 (1 µM; Tocris Biosciences). For all experiments utilizing inhibitors the medium was replaced at 48 hpi to replenish the compound being examined. Cell viability was determined using AQueous One (Promega).
Fatty Acid Analysis
Lipid extraction and FA analysis were performed as previously described (Koyuncu et al., 2013). Briefly, cells and virions were lysed using pre-chilled 50% methanol containing 0.05 M HCl. Chloroform was added to extract lipids followed by saponification and isolation of the liberated FAs tails. The FAs were analyzed by liquid chromatography coupled high-resolution mass spectrometry (LC-MS) (Kamphorst et al., 2011; Koyuncu et al., 2013). Two LC-MS methods were employed. To analyze all FAs the samples were run on a Luna C8 reversed-phase column (Phenomemex) on a gradient from buffer A (97:3 water:methanol with 10 mM tributylamine and 15 mM acetic acid (pH 4.5)) to buffer B (100% methanol) during a 50 min run time. For low abundant FAs (C22–30), the samples were concentrated 5-fold and analyzed using a similar gradient during a 40 min run time that excluded the shorter chain, more abundant FAs (e.g. <C20). Both methods utilized a Thermo Exactive orbitrap mass spectrometer running in negative mode to identify and measure each FA species. MS data was analyzed using MAVEN: Metabolomic Analysis and Visualization Engine (Clasquin et al., 2012; Melamud et al., 2010) and MATLAB (The MathWorks). For labeling experiments, the results were corrected for naturally occurring carbon-13 (Munger et al., 2008).
Reduction of ELOVL Activity
Gene knockdown of ELOVLs was achieved by shRNA. Lenti-viral particles were generated in 293T cells following transfection of pLKO.1-puro clones targeting ELOVL1-7 or a non-targeting (NT) control (Sigma), pMD.2G, and psPAX2. Transduced MRC-5 cells were selected by 2 µg/ml puromycin for 4 days and allowed to grow for two days without puromycin prior to being used in an experiment. Table S1 contains information regarding shRNA clones.
Protein and mRNA Analysis
Proteins were examined by Western blot. Primary antibodies used are listed in the supplement (Table S2). All blots were blocked using 5% BSA in a Tris-buffered saline with 0.05% tween-20 (TBS-T), except rabbit anti-ELOVL7 blots which were blocked in 5% milk in TBS-T. For RNA isolation, cells were lysed using Trizol followed by purification using a Direct-zol kit (Zymo Research). mRNA was converted to cDNA via reverse-transcription using a oligo-dT primer and 1 or 2 µg of total RNA (Taqman, Life Technologies). Levels were measured by real-time qPCR using SYBR Green (Life Technologies). Table S3 lists the sequences for all primers used in this study.
Supplementary Material
Acknowledgments
We are grateful to Scott Terhune for providing the ADdlUL38 BAC and for comments from the members of the Shenk and Rabinowitz lab groups. This work was supported by NIH grants AI78063, CA82396, and GM71508 and an American Heart Association postdoctoral fellowship to JGP (12POST9190001).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
JGP, TS and JDR conceived and designed the experiments; JGP performed the experiments; JGP, TS and JDR interpreted the results and wrote the manuscript.
References
- Alwine JC. Modulation of host cell stress responses by human cytomegalovirus. Curr Top Microbiol Immunol. 2008;325:263–279. doi: 10.1007/978-3-540-77349-8_15. [DOI] [PubMed] [Google Scholar]
- Alwine JC. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog. 2012;8:e1002878. doi: 10.1371/journal.ppat.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissinger AL, Sinzger C, Kaiserling E, Jahn G. Human cytomegalovirus as a direct pathogen: correlation of multiorgan involvement and cell distribution with clinical and pathological findings in a case of congenital inclusion disease. Journal of medical virology. 2002;67:200–206. doi: 10.1002/jmv.2208. [DOI] [PubMed] [Google Scholar]
- Britt W. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol. 2008;325:417–470. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- Buchkovich NJ, Maguire TG, Yu Y, Paton AW, Paton JC, Alwine JC. Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. J Virol. 2008;82:31–39. doi: 10.1128/JVI.01881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JW, Maguire TG, Alwine JC. Glutamine metabolism is essential for human cytomegalovirus infection. J Virol. 2010;84:1867–1873. doi: 10.1128/JVI.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasquin MF, Melamud E, Rabinowitz JD. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis [et al] 2012;Chapter 14(Unit14 11) doi: 10.1002/0471250953.bi1411s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clippinger AJ, Alwine JC. Dynein mediates the localization and activation of mTOR in normal and human cytomegalovirus-infected cells. Genes Dev. 2012;26:2015–2026. doi: 10.1101/gad.196147.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbs CS. Cytomegalovirus and brain tumor: epidemiology, biology and therapeutic aspects. Current opinion in oncology. 2013;25:682–688. doi: 10.1097/CCO.0000000000000005. [DOI] [PubMed] [Google Scholar]
- Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, Nabors LB, Cobbs CG, Britt WJ. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–3350. [PubMed] [Google Scholar]
- Deguil J, Pineau L, Rowland Snyder EC, Dupont S, Beney L, Gil A, Frapper G, Ferreira T. Modulation of lipid-induced ER stress by fatty acid shape. Traffic. 2011;12:349–362. doi: 10.1111/j.1600-0854.2010.01150.x. [DOI] [PubMed] [Google Scholar]
- Delgado T, Sanchez EL, Camarda R, Lagunoff M. Global metabolic profiling of infection by an oncogenic virus: KSHV induces and requires lipogenesis for survival of latent infection. PLoS Pathog. 2012;8:e1002866. doi: 10.1371/journal.ppat.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W. Structure and formation of the cytomegalovirus virion. Curr Top Microbiol Immunol. 2008;325:187–204. doi: 10.1007/978-3-540-77349-8_11. [DOI] [PubMed] [Google Scholar]
- Grady SL, Purdy JG, Rabinowitz JD, Shenk T. Argininosuccinate synthetase 1 depletion produces a metabolic state conducive to herpes simplex virus 1 infection. Proc Natl Acad Sci U S A. 2013;110:E5006–E5015. doi: 10.1073/pnas.1321305110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkewicz R, Du H, Tong Z, Alkuraya H, Bedell M, Sun W, Wang X, Hsu YH, Esteve-Rudd J, Hughes G, et al. Essential role of ELOVL4 protein in very long chain fatty acid synthesis and retinal function. J Biol Chem. 2012;287:11469–11480. doi: 10.1074/jbc.M111.256073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Progress in lipid research. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Janssen RJ, Distelmaier F, Smeets R, Wijnhoven T, Ostergaard E, Jaspers NG, Raams A, Kemp S, Rodenburg RJ, Willems PH, et al. Contiguous gene deletion of ELOVL7, ERCC8 and NDUFAF2 in a patient with a fatal multisystem disorder. Human molecular genetics. 2009;18:3365–3374. doi: 10.1093/hmg/ddp276. [DOI] [PubMed] [Google Scholar]
- Kamphorst JJ, Fan J, Lu W, White E, Rabinowitz JD. Liquid chromatography-high resolution mass spectrometry analysis of fatty acid metabolism. Anal Chem. 2011;83:9114–9122. doi: 10.1021/ac202220b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaskov E, Scott C, Zhang L, Teodoro T, Ravazzola M, Volchuk A. Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology. 2006;147:3398–3407. doi: 10.1210/en.2005-1494. [DOI] [PubMed] [Google Scholar]
- Kitazawa H, Miyamoto Y, Shimamura K, Nagumo A, Tokita S. Development of a high-density assay for long-chain fatty acyl-CoA elongases. Lipids. 2009;44:765–773. doi: 10.1007/s11745-009-3320-8. [DOI] [PubMed] [Google Scholar]
- Koyuncu E, Purdy JG, Rabinowitz JD, Shenk T. Saturated very long chain fatty acids are required for the production of infectious human cytomegalovirus progeny. PLoS Pathog. 2013;9:e1003333. doi: 10.1371/journal.ppat.1003333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol. 2004;78:11030–11039. doi: 10.1128/JVI.78.20.11030-11039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection alters the substrate specificities and rapamycin sensitivities of raptor- and rictor-containing complexes. Proc Natl Acad Sci U S A. 2006;103:14182–14187. doi: 10.1073/pnas.0605825103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AE, Kelder B, Bobik EG, Chuang LT, Lewis CJ, Kopchick JJ, Mukerji P, Huang YS. Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids. 2002;37:733–740. doi: 10.1007/s11745-002-0955-6. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Griffiths B, Santos CR, Pende M, Schulze A. Regulation of the SREBP transcription factors by mTORC1. Biochemical Society transactions. 2011;39:495–499. doi: 10.1042/BST0390495. [DOI] [PubMed] [Google Scholar]
- Li W, Sandhoff R, Kono M, Zerfas P, Hoffmann V, Ding BC, Proia RL, Deng CX. Depletion of ceramides with very long chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in ELOVL4 deficient mice. International journal of biological sciences. 2007;3:120–128. doi: 10.7150/ijbs.3.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang J, Kang SA, Thoreen CC, Hur W, Ahmed T, Sabatini DM, Gray NS. Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. Journal of medicinal chemistry. 2011a;54:1473–1480. doi: 10.1021/jm101520v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ST, Sharon-Friling R, Ivanova P, Milne SB, Myers DS, Rabinowitz JD, Brown HA, Shenk T. Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proc Natl Acad Sci U S A. 2011b;108:12869–12874. doi: 10.1073/pnas.1109796108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ES, Shenk T, Pass RF, editors. Cytomegaloviruses. Philadelphia, PA: Lippincott, Williams and Wilkins; 2007. [Google Scholar]
- Moon YA, Hammer RE, Horton JD. Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. J Lipid Res. 2009;50:412–423. doi: 10.1194/jlr.M800383-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YA, Shah NA, Mohapatra S, Warrington JA, Horton JD. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J Biol Chem. 2001;276:45358–45366. doi: 10.1074/jbc.M108413200. [DOI] [PubMed] [Google Scholar]
- Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe. 2008;3:253–262. doi: 10.1016/j.chom.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman NJ, Shenk T. Rapamycin-resistant mTORC1 kinase activity is required for herpesvirus replication. J Virol. 2010;84:5260–5269. doi: 10.1128/JVI.02733-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser TS, Schieffer D, Cherry S. AMP-activated kinase restricts Rift Valley fever virus infection by inhibiting fatty acid synthesis. PLoS Pathog. 2012;8:e1002661. doi: 10.1371/journal.ppat.1002661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss P. The emerging role of cytomegalovirus in driving immune senescence: a novel therapeutic opportunity for improving health in the elderly. Current opinion in immunology. 2010;22:529–534. doi: 10.1016/j.coi.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasheri N, Joyce M, Rouleau Y, Yang P, Yao S, Tyrrell DL, Pezacki JP. Modulation of fatty acid synthase enzyme activity and expression during hepatitis C virus replication. Chemistry & biology. 2013;20:570–582. doi: 10.1016/j.chembiol.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Ofman R, Dijkstra IM, van Roermund CW, Burger N, Turkenburg M, van Cruchten A, van Engen CE, Wanders RJ, Kemp S. The role of ELOVL1 in very long-chain fatty acid homeostasis and X-linked adrenoleukodystrophy. EMBO Mol Med. 2010;2:90–97. doi: 10.1002/emmm.201000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno Y, Suto S, Yamanaka M, Mizutani Y, Mitsutake S, Igarashi Y, Sassa T, Kihara A. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc Natl Acad Sci U S A. 2010;107:18439–18444. doi: 10.1073/pnas.1005572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Drake MJ, Bruce EA, Riblett AM, Didigu CA, Wilen CB, Malani N, Male F, Lee FH, Bushman FD, et al. The major cellular sterol regulatory pathway is required for Andes virus infection. PLoS Pathog. 2014;10:e1003911. doi: 10.1371/journal.ppat.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau L, Colas J, Dupont S, Beney L, Fleurat-Lessard P, Berjeaud JM, Berges T, Ferreira T. Lipid-induced ER stress: synergistic effects of sterols and saturated fatty acids. Traffic. 2009;10:673–690. doi: 10.1111/j.1600-0854.2009.00903.x. [DOI] [PubMed] [Google Scholar]
- Qian Z, Xuan B, Gualberto N, Yu D. The human cytomegalovirus protein pUL38 suppresses endoplasmic reticulum stress-mediated cell death independently of its ability to induce mTORC1 activation. J Virol. 2011;85:9103–9113. doi: 10.1128/JVI.00572-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, Purdy JG, Vastag L, Shenk T, Koyuncu E. Metabolomics in drug target discovery. Cold Spring Harb Symp Quant Biol. 2011;76:235–246. doi: 10.1101/sqb.2011.76.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, Dong JJ. Alteration of lipid metabolism in cells infected with human cytomegalovirus. Virology. 2010 doi: 10.1016/j.virol.2010.04.026. [DOI] [PubMed] [Google Scholar]
- Savaryn JP, Reitsma JM, Bigley TM, Halligan BD, Qian Z, Yu D, Terhune SS. Human cytomegalovirus pUL29/28 and pUL38 repression of p53-regulated p21CIP1 and caspase 1 promoters during infection. J Virol. 2013;87:2463–2474. doi: 10.1128/JVI.01926-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JY, Cresswell P. Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog. 2013;9:e1003497. doi: 10.1371/journal.ppat.1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CM, Schafer XL, Moorman NJ, Munger J. Human cytomegalovirus induces the activity and expression of acetyl-coenzyme A carboxylase, a fatty acid biosynthetic enzyme whose inhibition attenuates viral replication. J Virol. 2011;85:5814–5824. doi: 10.1128/JVI.02630-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streblow DN, Dumortier J, Moses AV, Orloff SL, Nelson JA. Mechanisms of cytomegalovirus-accelerated vascular disease: induction of paracrine factors that promote angiogenesis and wound healing. Current topics in microbiology and immunology. 2008;325:397–415. doi: 10.1007/978-3-540-77349-8_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Makino A, Hullin-Matsuda F, Kobayashi T, Furihata M, Chung S, Ashida S, Miki T, Fujioka T, Shuin T, et al. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res. 2009;69:8133–8140. doi: 10.1158/0008-5472.CAN-09-0775. [DOI] [PubMed] [Google Scholar]
- Terhune S, Torigoi E, Moorman N, Silva M, Qian Z, Shenk T, Yu D. Human cytomegalovirus UL38 protein blocks apoptosis. J Virol. 2007;81:3109–3123. doi: 10.1128/JVI.02124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ, Vastag L, Rabinowitz JD, Shenk T. Human kinome profiling identifies a requirement for AMP-activated protein kinase during human cytomegalovirus infection. Proc Natl Acad Sci U S A. 2012;109:3071–3076. doi: 10.1073/pnas.1200494109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastag L, Koyuncu E, Grady SL, Shenk TE, Rabinowitz JD. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7:e1002124. doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Botolin D, Xu J, Christian B, Mitchell E, Jayaprakasam B, Nair MG, Peters JM, Busik JV, Olson LK, et al. Regulation of hepatic fatty acid elongase and desaturase expression in diabetes and obesity. J Lipid Res. 2006;47:2028–2041. doi: 10.1194/jlr.M600177-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womack A, Shenk T. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. mBio. 2010;1 doi: 10.1128/mBio.00282-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, DeBose-Boyd RA. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Smith GA, Enquist LW, Shenk T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J Virol. 2002;76:2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. J Virol. 2011;85:1573–1580. doi: 10.1128/JVI.01967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection induces adipocyte-like lipogenesis through activation of sterol regulatory element binding protein 1. J Virol. 2012;86:2942–2949. doi: 10.1128/JVI.06467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maguire TG, Alwine JC. ChREBP, a glucose-responsive transcriptional factor, enhances glucose metabolism to support biosynthesis in human cytomegalovirus-infected cells. Proc Natl Acad Sci U S A. 2014;111:1951–1956. doi: 10.1073/pnas.1310779111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Pierciey FJ, Jr, Maguire TG, Alwine JC. PKR-like endoplasmic reticulum kinase is necessary for lipogenic activation during HCMV infection. PLoS Pathog. 2013;9:e1003266. doi: 10.1371/journal.ppat.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.