Abstract
There is a pressing need for new medicines (new molecular entities; NMEs) for rare diseases as few of the 6800 rare diseases (according to the NIH) have approved treatments. Drug discovery strategies for the 102 orphan NMEs approved by the US FDA between 1999 and 2012 were analyzed to learn from past success: 46 NMEs were first in class; 51 were followers; and five were imaging agents. First-in-class medicines were discovered with phenotypic assays (15), target-based approaches (12) and biologic strategies (18). Identification of genetic causes in areas with more basic and translational research such as cancer and in-born errors in metabolism contributed to success regardless of discovery strategy. In conclusion, greater knowledge increases the chance of success and empirical solutions can be effective when knowledge is incomplete.
A disease is considered rare in the USA if it is believed to affect fewer than 200,000 individuals. There are approximately 6800 such diseases, according to the NIH. While each disease is rare, when considered together they affect nearly 30 million people or almost one in ten. In the EU, a disease is considered rare if it has a prevalence of fewer than five in 10,000 people. Some rare diseases have less than a dozen known cases, whereas others are more common, such as multiple sclerosis, cystic fibrosis and Duchenne muscular dystrophy. Collectively, these disorders affect 6–7% of the population in the developed world [1].
There are relatively few treatment options for most patients with a rare disease. Many new medicines (new molecular entities; NMEs) are needed. This creates challenges and opportunities. The US FDA definition of a NME is an active ingredient that has never before been marketed in the USA in any form. The knowledge to inform the discovery of medicines for each disease can be minimal. While over 80% of the rare disease have a genetic origin [1], the relationship between the genetic defect and the disease phenotype is rarely clear. There is currently insufficient research and funding to provide the understanding and knowledge to adequately address these unmet medical needs.
New strategies are needed that will increase the treatment options for more rare diseases. This includes strategies to increase funding to rare disease drug discovery and strategies to increase the impact of those funds. Incentives to discover NMEs for rare diseases include the Orphan Drug Act (ODA) and pediatric priority review vouchers (PRVs). The ODA established in 1983 provides for granting special status to a drug or biological product (‘drug’) to treat a rare disease or condition. This status is referred to as orphan designation. Orphan designation qualifies the sponsor of the drug for various development incentives, including tax credits for qualified clinical testing. A marketing application for a prescription drug product that has received orphan designation is not subject to a prescription drug user fee unless the application includes an indication for other than the rare disease or condition for which the drug was designated. A PRV is a voucher issued to the sponsor of a “rare pediatric disease product application” that entitles the holder of such voucher to priority review (instead of a longer standard review) of a single New Drug Application (NDA) or Biological License Application (BLA) after the date of approval of the rare pediatric disease product application.
Strategies that provide for more NMEs and efficient use of available resources are also needed. Over-all productivity in drug discovery is decreasing. There has been a dramatic increase in research and development spending without the corresponding increase in NMEs. The current trend is to spend more to increase knowledge, however this has not increased the clinical success rate. The low productivity is unacceptable for rare disease drug discovery. Funds need to be used more efficiently to identify new, useful medical treatments for rare diseases. While an increase in productivity would be of great benefit to the all therapeutic areas, an increase is mandatory for rare diseases in order for current funding levels to further impact the large unmet medical need.
Increased success in rare disease drug discovery will require better diagnostics, an understanding of disease that provides good translational biomarkers, and clearer clinical development programs. The mechanisms underlying rare diseases are not well understood, patients are hard to identify and diagnose and no regulatory precedent for the disease may exist (among others), all of which makes designing and conducting drug development programs very difficult. While these factors must be taken into account when starting a drug discovery program they are not the focus of this analysis.
The question addressed in the analysis is: what are the drug discovery strategies that produce NMEs?
Addressing this question provides knowledge from past experience to help inform future success. Success is defined as the approval of a medicine for use in patients. This implies that the medicine was effective for the unmet medical need at a safe dose. To this end we analyzed the NMEs that were approved with orphan drug status by the FDA between 1999 and 2012.
For the purpose of this work drug discovery is defined as the identification of a NME candidate for testing in humans for efficacy and safety. The clinical testing is the development process. A drug is discovered when nominated for clinical trials; theoretically from this point it should fail or succeed based on its own merit. Its merit will be determined in the clinical development program and biopharmaceutics evaluations. One key to increasing overall drug discovery productivity is to increase the success rate of clinical candidates.
The practice of drug discovery and development is an iterative cycle of learning and confirming (Figure 1). The process to discover a NME is driven by an unmet medical need. It requires some level of understanding of the disease and the corresponding physiology that can be used to design and implement a drug discovery strategy. Ideally the understanding provides a translational biomarker that can be used to align preclinical drug discovery with clinical development. Examples of biomarkers include viral load for a virus infection or blood glucose levels for diabetes. Knowledge of the pathophysiology of the disease, genomic links and the identity of physiological regulators contribute to understanding ways to correct a disease phenotype; for example, a genetic mutation in α-galactosidase A leading to a strategy to discover a treatment for Fabry's disease. In practice this knowledge is used to establish assays that will identify drug leads. The leads are generally small molecules or biologics including recombinant proteins and monoclonal antibodies. The leads are then optimized to drug candidates that have Sufficient efficacy, safety and biopharmaceutics properties for testing in human clinical studies. The clinical candidates then undergo clinical development and when successful will be approved as a NME by regulatory agencies such as the FDA. Iterative learning and confirming happens at every stage of the process (Figure 1).
Figure 1. Drug discovery and development cycle.
The approval of a medicine to treat an unmet medical need including a rare disease involves an iterative cycle of testing and learning. This figure describes some of the important phases in the process. The process of discovery and development of a new medicine is initiated in response to an unmet medical need to treat a disease. Physiological, genetic and chemical knowledge provide an understanding of the disease. This knowledge will lead to the identification of translation biomarkers that are used to evaluate the effectiveness of a potential medicine. The available knowledge informs drug discovery strategies that are used as starting points for the practical process of discovering a new molecular entity. TDD is associated with modulating a specific gene product known as the target, PDD is a strategy driven by assays that measure phenotypes associated with the disease. Ideally these phenotypes will be the associated with the translational biomarkers. These two strategies generally are focused on small molecules and are medicinal chemistry intensive, in contrast to biologics that use recombinant proteins and antibodies as therapeutics. It should be noted that the knowledge to choose a strategy is generally incomplete; however, the more iterations that occur in the drug discovery/development cycle, the more complete the knowledge and the better chance that a molecule will make it to registration. The discovery strategies will result in a lead molecule, ideally with activity against the translational biomarker. The molecule will work by a MMOA that provides an optimal therapeutic index. These molecules will then be optimized for biopharmaceutics properties and safety to provide a drug candidate. At this point, the process of drug discovery is complete and the molecule should succeed or fail based on its own merit. Opportunities to improve efficiency in drug discovery will increase the probability that clinical candidates will make it to registration. The left hand of the circle is the development phase of drug discovery, which involves testing for safety and efficacy in humans leading to registration. Multiple iterations are generally required before a medicine with sufficient efficacy at a safe dose is discovered, tested in humans and registered. MMOA: Molecular mechanism of action; PDD: Phenotypic drug discovery; pt: Point; TDD: Target-based drug discovery.
The fewer iterations that are required to register an NME the more efficient the process. Rarely does the process occur in one iteration. For first-in-class medicines, where the available knowledge is incomplete, there will be more failures and requiring more iterations. This is less of an issue with followers.
Drug discovery has evolved different strategies based on the available knowledge. One approach starts with a hypothesis related to the disease. Historically this hypothesis was related to a phenotype such as clearing an infection by killing the infectious organism or blocking experimentally induced seizures in animals as a phenotype for epilepsy. More recently an increased understanding of the molecular basis of disease has led to the molecular approach to drug discovery, in which genetics and other molecular sciences provide the basis for a molecular hypothesis.
Consequently, current small-molecule drug discovery strategies are polarized into two major types: target-based drug discovery (TDD) and phenotypic drug discovery (PDD). The focus of TDD is a gene product, known as a target. Molecular and chemical knowledge are used to investigate specific molecular hypotheses. The focus of PDD is assays that measure a clinically meaningful phenotype in a physiologically relevant system such as an animal or cell; the assays do not require prior understanding of the molecular mechanism of action (MMOA). Drug discovery with biologics is primarily a target informed approach. Successful drug discovery uses all strategies but there is debate and preferences on which is used first and under what circumstances.
In a previous analysis we looked to learn some of the features that contributed to successful drug discovery in all disease areas. The discovery strategies and the MMOA for NMEs and new biologics that were approved by the FDA between 1999 and 2008 were analyzed [2]. The results showed that the contribution of phenotypic screening to the discovery of first-in-class small-molecule drugs exceeded that of target-based approaches – with 28 and 17 of these drugs coming from the two approaches, respectively – in an era in which the major focus was on target-based approaches.
A conclusion from that analysis was that lower productivity partly reflects TDD's lack of consideration of the molecular complexities of how the drugs work. Knowing the parts of an efficient machine – a watch, an automobile or a computer – is not enough to describe how it works. The parts must collaborate in precise ways to provide accurate time, reliable transportation or processed information. The phrase “molecular mechanism of action” was used to describe the complexities of how medicines interact with biology to provide an effective and safe response. These observations led to the proposal that the value of phenotypic assays is to identify first-in-class medicines and their respective MMOAs, since phenotypic assays do not require a priori identification of the target and MMOA. The empirical, phenotypic approach enables drug discovery to proceed with incomplete information.
Drug development for rare diseases often poses additional challenges in comparison to common diseases, including fewer patients available for inclusion in clinical trials and a poorly understood natural history of the disease. Given the diversity of rare diseases and the variety of therapeutic strategies (ranging from repurposing of established small-molecule drugs to pioneering gene therapies), there is also clearly not a ‘one-size-fits-all’ strategy for orphan drugs [3]. It is of interest to learn what strategies have been successful.
Analysis
The analysis includes categorization between first in class and followers, discovery strategy such as target-based, phenotypic, natural substance or biologic and how genetic information regarding the disease contributed to the discovery strategy. Also included are descriptions of the mechanism of action for representative medicines, in many cases the details are provided in the mechanism of action section (12.1) of the medicines label available on the FDA's website [4]. The list of approved medicines and their classification as orphans were found on the FDA website.
Numbers of NMEs
In the 14-year period from 1999 to 2012, the FDA approved a total of 102 NMEs (67 small-molecule medicines and five imaging agents) and new therapeutic biologics (30 drugs). Of these, 46 drugs were identified as first in class from information in the product labels on the FDA website, and primary research and review publications (see Supplementary Table 1).
Discovery approaches
We categorized the list of 102 agents as first-in-class drugs (46 drugs), follower drugs (51 drugs) and imaging agents (five agents; these were not further analyzed) (Supplementary Table 1). The method of discovery of each new drug was identified as either TDD, PDD, modification of a natural substance, biologic based or other. Overall, 30 NMEs were discovered using target-based approaches, 28 NMEs were discovered using phenotypic-based approaches, eight NMEs were based on modifications of natural substances and 29 of the agents were biologics.
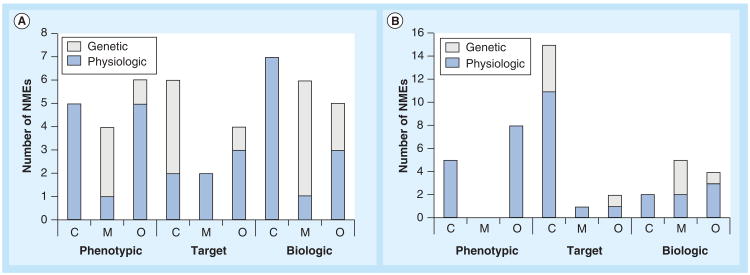
The discovery strategies for first in class and followers were fairly well balanced between phenotypic, target based and biologics. For first in class there were 15 that used PDD, 12 that used TDD and 18 for biologics. For followers the breakdown was 13 for PDD, 18 for TDD and 11 for biologics (Figure 2).
Figure 2. The distribution of new molecular entities with orphan drug designation approved by the US FDA between 1999 and 2012, according to drug discovery strategy.
(A) First in class; (B) followers. The graph illustrates the number of NMEs in each category. Also illustrated in the graph is the contribution of genetic information (white) and physiologic information (blue/dark) to the NMEs discovered with each strategy. Genetic information informed 26 out of the 102 NMEs approved between 1999 and 2012. Biologics were the most successful for first in class, followed by phenotypic. Target-based strategies were the least successful for first in class and the most successful for followers.
NME: New molecular entity.
The cumulative distribution of first-in-class NMEs by discovery strategy is shown in Figure 3A. The relative rate of first-in-class approvals for phenotypic and biologic discovery is fairly consistent through the period of the analysis. The rate of approvals for first-in-class target-based increased between the first years of the analysis (1999–2006) and the final years (2006–2012). The cumulative distribution for each category is shown in Supplementary Figure 1A for first in class and Supplementary Figure 1B for followers.
Figure 3. Cumulative distribution of approvals by discovery strategy.
(A) Comparison of phenotypic, target and biologic drug discovery strategies for first-in-class NMEs. (B) Comparison of the contribution of genetic and physiologic informed discovery to all NMEs.
NME: New molecular entity.
We can conclude that overall small-molecule drug discovery (TDD + PDD) provide more NMEs than biologics across all disease areas; 58 to 29 for overall, 27 to 18 for first in class, and 31 to 11 for followers. The drug discovery strategies for first in class distributed between phenotypic (33%), target-based (27%) and biologics (40%) (Figure 2A) is similar compared with what was observed for first-in-class medicines approved for all disease between 1999 and 2008 [2]. The success of PDD is still somewhat surprising since the majority of drug discovery resources are focused on TDD and biologics. More insights into the success of the different strategies is seen by analysis of genetic contributions in the context of the therapeutic areas.
Therapeutic area
Cancer indications comprised 44% (43 of 97) of the approved NMEs, followed by metabolic diseases 23% (22 of 97%) There were nine NMEs each for inflammation/immunomodulation, neurologic and vascular/cardiovascular. All other therapy areas had only five in total. In cancer there were slightly more followers than first in class (22 to 18) while for metabolic there was a 2:1 ratio of first in class to followers (Figure 4 & Supplementary Tables 1 & 2).
Figure 4. Drug discovery strategies by therapeutic area.
The therapeutic areas for NMEs approved by the US FDA between 1999 and 2012 with orphan designation. (A) First in class; (B) follower. The details are shown in Supplementary Tables 1 & 2. The therapeutic areas were classified as C, M and O. Others included infection and inflammation including arthritis, cardiovascular and vascular, neurologic and CNS, urinary, pain and respiratory. White bars are genetic informed approvals and blue/dark bars are physiologic informed approvals.
C: Cancer; M: Metabolic and endocrine; NME: New molecular entity; O: Other.
Further investigation into the details based on discovery strategy showed that targeted-based discovery was the most successful in cancer, both for first in class and followers. This was strongly influenced by the large number of kinase inhibitors approved for cancer indications. As described below, genetics had relatively the most significant contributions to cancer through identification of kinase gain-of-function mutations and diseases with in-born errors of metabolism that were treated with enzyme replacement therapy.
Genetics
The contribution of genetic knowledge to inform orphan drug discovery was also evaluated. Medicines whose discovery strategy was informed by a specific mutation were identified (Table 1). A drug discovery program was considered informed by genetics if Sufficient understanding of specific mutations were available to inform the choice of drug discovery assays. Essentially the association of Specific genetic mutations in a rare disease was used to inform the drug discovery strategy. Twenty-six of the 102 orphan medicines drug discovery were genetically informed, approximately 25%. For example, eight orphan drugs inhibit kinases where Specific mutations had been shown to have gain-of-function mutations that lead to cancers; five NMEs were for mutations in bcr-abl tyrosine kinase for CML, ruxolitinib inhibit mutant JAK2 kinase for myelofibrosis, crizotinib inhibits alk kinase for non-small-cell lung carcinoma (NSCLC) (Table 1). Mutations that cause enzyme deficiencies or loss of function leading to in-born errors in metabolism were treated with eight NMEs. Identification of the mutations, the gene products for the mutations and the underlying pathophysiology lead to the discovery strategy. For the remaining nine orphan drugs both genetics and a mechanistic understanding of the physiological system were used to inform the discovery strategy. Two of the indications (hereditary angioedema [HAE] and crypoyrin-associated periodic syndromes [CAPS]) had two approved medicines. Altogether, the genetics informed the discovery of medicines for 18 rare diseases (four for kinases involved in cancer, six for enzyme replacement therapy and eight from mechanism informed) in the 14-year period.
Table 1.
New molecular entities with orphan diseases designation approved by the US FDA between 1999 and 2012 that were informed by genetics.
| NME | Discovery strategy | Therapeutic area | Indication | Genetic mutation | Mechanism | Ref. |
|---|---|---|---|---|---|---|
| Ruxolitinib† | Target | Cancer | Myelofibrosis | JAK2V617F-positive MPN | K | [5] |
| Crizotinib† | Target | Cancer | NSCLC | Alk kinase | K | [6] |
| Vemurafenib† | Target | Cancer | Late-stage melanoma | BRAF | K | [7] |
| Imatinib mesylate† | Target | Cancer | CML | bcr-abl tyrosine kinase | K | [8] |
| Bosutinib monohydrate | Target | Cancer | CML | bcr-abl - imatinib resistant forms | K | [9] |
| Ponatinib | Target | Cancer | CML | bcr-abl tyrosine kinase T315I mutant | K | [10,11] |
| Nilotinib | Target | Cancer | CML | bcr-abl inactive form | K | [12] |
| Dasatinib | Target | Cancer | CML | bcr-abl | K | [13] |
| Velaglucerase alfa | Biologic | Genetic disease metabolic | Gaucher's disease | Enzyme replacement | E | [4] |
| Taliglucerase alfa | Biologic | Genetic disease metabolic | Gaucher's disease | Enzyme replacement | E | [4] |
| Alaglucerase alfa2 | Biologic | Genetic disease metabolic | Pompe's disease | Enzyme replacement | E | [4] |
| Alglucosidase alfa† | Biologic | Genetic disease metabolic | Pompe's disease | Enzyme deficiency | E | [4] |
| Sapropterin dihydrochloride† | Mod. nat. subst. | Genetic disease metabolic | Hyperphenylalaninemia | Phenylalanine hydroxylase deficiency augmented with synthetic cofactor BH4 | Mech informed | [14–17] |
| Idursulfase† | Biologic | Genetic disease metabolic | Hunter syndrome | Enzyme replacement | E | [18] |
| Galsulfase† | Biologic | Genetic disease metabolic | MPS VI | Enzyme replacement | E | [19] |
| Laronidase† | Biologic | Genetic disease metabolic | MPS I | Enzyme replacement | E | [20] |
| Agalsidase beta† | Biologic | Genetic disease metabolic | Fabry's disease | Enzyme replacement | E | [21] |
| Nitisinone† | Phenotype | Genetic disease metabolic | Type 1 tyrosinemia | 4-hydroxypyruvate dehydrogenase | Mech informed | [22–24] |
| Carglumic acid† | Phenotype | Genetic disease metabolic | Hyperammonemia due to NAGS deficiency | NAGS enzyme deficiency | Mech informed | [4] |
| Ivacaftor† | Phenotypic | Respiratory | Cystic fibrosis | Potentiating the open probability of the G551D-CFTR channel | Mech informed | [25,26] |
| Ecallantide† | Target | Vascular | HAE | Plasma kallikrein inhibitor | Mech informed | [4] |
| Icatibant | Target | Vascular | HAE | Bradykinen receptor antagonist | Mech informed | [4] |
| Canakinumab† | Biologic | Autoimmune | Cryopyrin-associated periodic syndromes | Biologic mAb/mutations in NLRP-3 | Mech informed | [4] |
| Rilonacept | Biologic | Autoimmune | Cryopyrin-associated periodic syndromes | Soluble decoy receptor that binds IL-1β mutations in NLRP-3 | Mech informed | |
| Eculizumab† | Biologic | Vascular | Paroxysmal nocturnal hemoglobinuria | mAb to complement C5 genetic mutation in patients with PNH | Mech informed | [27–29] |
| Miglustat† | Phenotype | Genetic disease metabolic | Gaucher's disease | Deficiency of glucocerebrosidase | Mech informed | [30,31] |
First in class.
CML: Chronic myelogenous leukemia; E: Enzyme replacement therapy; HEA: Hereditary angioedema; K: Kinase; mAb: Monoclonal antibody; Mech informed: Mechanism informed; Mod. nat. subst.: Modified natural substance; MPN: Paroxysmal nocturnal hemglobinuria; MPS: Mucopolysaccharidosis; NAGS: N-acetylglutamate syntha; NME: New molecular entity; NSCLC: Non-small-cell lung carcinoma.
How did knowledge of the Specific genetic mutants inform drug discovery? The kinase mutations were gain-of-function mutations that could be addressed with inhibitors and modern drug discovery molecular optimization and screening approaches including structure-based design. The enzyme replacement therapy replaces the defective gene product. Both of these approaches had the benefit of an understanding of the molecular mechanisms of disease and the corresponding technical expertise and resources to facilitate the discoveries.
The contribution of genetics to the other drug discovery successes provides insights as to how genetic information can be integrated with biological and chemical knowledge to identify NMEs.
HAE is a rare genetic disorder caused by mutations to C1-esterase-inhibitor (C1-INH) located on chromosome 11q. HAE is characterized by low levels of C1-INH activity and low levels of C4. C1-INH functions to regulate the activation of the complement and intrinsic coagulation (contact system pathway) and is a major endogenous inhibitor of plasma kallkrein. The kallikrein–kinin system is a complex proteolytic cascade involved in the initiation of both inflammatory and coagulation pathways. One critical aspect of this pathway is the conversion of high-molecular-weight (HMW) kininogen to bradykinin by the protease plasma kallikrein. In HAE, normal regulation of plasma kallkrein activity and the classical complement cascade is not present. During attacks, unregulated activity of plasma kallikrein results in excessive bradykinin generation. Bradykinin is a vasodilator which is thought by some to be responsible for the characteristic HAE symptoms of localized swelling, inflammation, and pain. Ecallantide binds to plasma kallikrein and blocks its binding site, inhibiting the conversion of HMW kininogen to bradykinin and thereby treats the disease during acute episodic attacks of HAE. Icatibant is a competitive antagonist selective for the bradykinin B2 receptor, with an affinity similar to bradykinin. Icatibant inhibits bradykinin from binding the B2 receptor and thereby treats the clinical symptoms of an acute, episodic attack of HAE [4].
CAPS refer to rare genetic syndromes generally caused by mutations in the NLRP-3 gene. Cryopyrin regulates the protease caspase-1 and controls the activation of IL-1β. Mutations in NLRP-3 result in an overactive inflammasome resulting in excessive release of activated IL-1β that drives inflammation. Canakinumab is a human monoclonal anti-human IL-1β antibody of the IgG1/κ isotype. Canakinumab binds to human IL1β and neutralizes its activity by blocking its interaction with IL-1 receptors. Rilonacept blocks IL-1β signaling by acting as a soluble decoy receptor that binds IL-1β and prevents its interaction with cell surface receptors [4].
A genetic mutation in patients with paroxysmal nocturnal hemoglobinuria (PNH) leads to the generation of populations of abnormal red blood cells (RBCs), known as PNH cells, that are deficient in terminal complement inhibitors, rendering PNH RBCs sensitive to persistent terminal complement-mediated destruction. The destruction and loss of these PNH cells (intravascular hemolysis) results in low RBC counts (anemia), and also fatigue, difficulty in functioning, pain, dark urine, shortness of breath and blood clots. Eculizumab is a monoclonal antibody that specifically binds to the complement protein C5 with high affinity, thereby inhibiting its cleavage to C5a and C5b and preventing the generation of the terminal complement complex C5b-9 [4].
Milgustat for type 1 Gaucher's disease is an inhibitor of glucosylceramide synthase that was originally developed as an antiviral agent because most enveloped viruses use the same pathway for glycoprotein synthesis in infected cells. The goal of treatment with miglustat is to reduce the rate of glycosphingolipid biosynthesis so that the amount of glycosphingolipid substrate is reduced to a level which allows the residual activity of the deficient glucocerebrosidase enzyme to be more effective (substrate reduction therapy) [4,30].
N-acetylglutamate synthase (NAGS) deficiency is a rare inborn error of metabolism affecting ammonia detoxification in the urea cycle. The product of NAGS is N-acetylglutamate, which is the absolutely required allosteric activator of the first urea cycle enzyme carbamoylphosphate synthetase 1. In defects of NAGS, the urea cycle function can be severely affected resulting in fatal hyperammonemia in neonatal patients or at any later stage in life. NAGS deficiency can be treated with a structural analog of N-acetylglutamate, N-carbamyl-l-glutamate, carglumic acid [32]. Carglumic acid is approved for the treatment of acute hyperammonemia associated with NAGs deficiency.
Ivacaftor is a potentiator of the CFTR protein and approved for cystic fibrosis. The CFTR protein is a chloride channel present at the surface of epithelial cells in multiple organs. Ivacaftor facilitates increased chloride transport by potentiating the channel-open probability (or gating) of the G551D-CFTR protein. In vitro, ivacaftor increased CFTR-mediated transepithelial current (IT) in rodent cells expressing G551D-CFTR protein following addition of a cAMP agonist [4].
Nitisinone is approved for hereditary tyrosinemia type 1. It is a competitive inhibitor of 4-hydroxyphenyl-pyruvate dioxygenase, an enzyme upstream of fumarylacetoacetate hydrolase in the tyrosine catabolic pathway. By inhibiting the normal catabolism of tyrosine in patients with hereditary tyrosinemia type 1, nitisinone prevents the accumulation of the catabolic intermediates maleylacetoacetate and fumarylacetoacetate. In patients with hereditary tyrosinemia type 1, these catabolic intermediates are converted to the toxic metabolites succinylacetone and succinylacetoacetate, which are responsible for the observed liver and kidney toxicity. Nitisinone was originally discovered as an herbicide, but early mechanism-of-action studies in rats revealed that the compound caused a marked increase in plasma tyrosine concentrations [22].
In common with all these case studies is that the mutation caused a gain or loss of function that was associated with the disease phenotype in the context of an understanding of the physiological regulation. This knowledge led to drug discovery strategies that enabled the identification of drug candidates. Gain-of-function mutations leading to the symptoms for HAE, CAPS and PNH were blocked via targeting enzymes in the pathways with either small molecules or biologics. The pathopysiology for loss-of-function mutations were reduced by the discovery of activators/potentiators that either worked directly on the protein as is the case of ivacaftor for CFTR or as a replacement in the case of carglumic acid.
Physiologic
The approaches that used nongenetic mechanistic understanding were categorized into phenotypic and target for small molecules and biologics for the recombinant enzymes and monoclonal antibodies. PDD lead to 11 first-in-class NMEs and compared with seven for target-based. There were 11 first-in-class biologics approved during this period (Table 2 & Figure 2).
Table 2.
First-in-class new molecular entities with orphan diseases designation approved by the US FDA between 1999 and 2012 that were informed by physiology.
| NME | Year | Discovery strategy | Therapeutic area | Indication | MMOA | Ref. |
|---|---|---|---|---|---|---|
| Omacetaxine | 2012 | Phenotypic | Cancer | CML | Inhibition of translation of short-lived proteins | [33] |
| Bedaquiline | 2012 | Phenotypic | Anti-infective | Tuberculosis | Inhibition of ATP synthase | [34] |
| Vorinostat | 2006 | Phenotypic | Cancer | Cutaneous T-cell lymphoma | HDAC inhibitor | [35,36] |
| Nelarabine | 2005 | phenotypic | Cancer | T-cell acute lymphoblastic leukemia | Nucleoside prodrug incorporated into DNA leading to inhibition of DNA synthesis | [37–41] |
| Plerixafor | 2008 | Phenotypic | Cancer | Hematopoietic stem cell transplantation | CXCR4 inhibitor | [42] |
| Rufinamide | 2008 | Phenotypic | CNS | Seizures associated with Lennox–Gastaut syndrome | Prolongation of the inactive state of the sodium channel | [43,44] |
| Artemether/lumefantrine | 2009 | Phenotypic | Anti-infective | Malaria | Activation of reactive intermediates | [45,46] |
| Dalfampridine | 2010 | Phenotypic | CNS | Multiple sclerosis | Kv channel-blocker greatest affinity for slowly inactivating channels. | [47] |
| Cinacalcet | 2004 | Phenotype | Metabolic | Secondary hyperparathyroidism | Allosteric activator | [48] |
| Azacitidine | 2004 | Phenotype | Cancer | Myelodysplastic syndromes | Nucleoside | [49,50] |
| Nitazoxanide | 2002 | Phenotypic | Anti-infective | Antiprotazoan | Interference with pyruvate:ferredoxin oxidoreductase | [51,52] |
| Lomitapide | 2012 | Target | Metabolic | Homozygous familial hypercholesterolemia | Binds to microsomal triglyceride transfer protein | [53,54] |
| Vigabatrin | 2009 | Target | CNS | Seizures | Irreversble GABA inhibitor | [4] |
| Eltrombopag | 2008 | Target | Vascular | ITP | Thrombopoietin receptor agonist | [55] |
| Sorafenib | 2005 | Target | Cancer | Renal cell carcinoma | Multikinase inhibitor VEGFR/PDGFR/KIT | [56] |
| Bortezomib | 2003 | Target | Cancer | Multiple myelomas | Proteasome | [57] [58] |
| Bosentan | 2001 | Target | Cardiovascular | Pulmonary arterial hypertension | Endothenlin-1 receptor antagonists | [59] |
| Pasireotide | 2012 | Target | Metabolic | Cushing's disease | Somatostatin receptor agonists | [60] |
| Raxibacumab | 2012 | Biologic | Anti-infective | Anthrax | Human mAb against Bacillus anthracis toxin P | [61] |
| Teduglutide | 2012 | Biologic | GI | Short bowel syndrome orphan diseases | GLP-2 agonist resistant to dipeptidyl peptidase IV | [62] |
| Glucarpidase | 2012 | Biologic | Cancer | Toxic plasma methotrexate concentrations | Enzyme metabolizes methotrexate | [63] |
| Brentuximab vedotin | 2011 | Biologic | Cancer | Hodgkin's lymphoma and ALCL | Toxic warhead conjugated to ADC for selectivity | [4] |
| Yervoy | 2011 | Biologic | Cancer | Melanoma | mAb | [4] |
| Ofatumumab | 2009 | Biologic | Cancer | CLL | mAb | [4] |
| Romiplostim | 2008 | Biologic | Vascular | Idiopathic immune thrombocytopenic purpura | Fc-peptide fusion protein (peptibody) agonist at thrombopoietin receptor | [64] |
| Pegvisomant | 2003 | Biologic | Metabolic | Acromegaly | Growth hormone receptor | [65–67] |
| Alemtuzumab | 2001 | Biologic | Cancer | B-cell chronic lymphocytic leukemia | CD52 cytolytic antibody B-cells | [68] |
| Gemtuzumab | 2000 | Biologic | Cancer | Cancer | CD33 conjugate | [69] |
| Denileukin diftitox | 1999 | Biologic | Cancer | Cutaneous T-cell lymphoma | CD25-directed cytotoxin | [70] |
ADC: Antibody drug conjugate; ALCL: Anaplastic large cell lymphoma; CLL: Chronic lymphocytic leukemia; CML: Chronic myelogenous leukemia; GI: Gastrointestinal; HDAC: Histone deacetylases; ITP: Chronic immune thrombocytopenia; mAb: Monoclonal antibody; NME: New molecular entity.
Phenotypic
The molecules that were discovered with phenotypic assays in the majority of cases employed assays that demonstrated a phenotype related to the disease, including cell death for cancer and infectious disease (omacetaxine, bedaquiline, artemisinin, nelarabine, azacitidine and nitazoxamide), calcium regulation (cinacalcet) and epilepsy (rufinamide). Two NMEs were identified serendipitously plerixafor and vorinostat. Some representative examples are listed below.
Omacetaxine was originally identified as a novel plant alkaloid with antitumor properties. Its mechanism of action was found to be inhibition of protein translation. Its clinical development was halted with the introduction of imatinib and related tyrosine kinase inhibitors but restarted to address resistance to tyrosine kinase inhibitors in patients with chronic myeloid leukemia [33].
Bedaquiline is the first molecule approved for tuberculosis (TB) in decades. Drug hunters at Johnson & Johnson sought new anti-TB compounds by selecting prototypes of different chemical series and testing them for inhibition of multiple-cycle growth of Mycobacterium smegmatis. A whole-cell assay was preferred because of its ability to concurrently assess multiple targets. They subsequently used resistance mutations to identify ATP synthase as the site of action [34].
Artemisinin is the most effective current therapy for the treatment of malaria. The discovery of artemisinin is attributed to You-You Tu, at the Institute of Chinese Materia Medica, China Academy of Traditional Chinese Medicine in the early 1970s. An Artemisia extract showed a promising degree of inhibition against parasite growth, consistent with activity which had been reported for this species in “A Handbook of Prescriptions for Emergencies” by Ge Hong (Jin Dynasty, 284–346 AD). It is reported that the extracts were evaluated in mouse malaria P. berghei [45].
Artemisinin is selectively activated to a reactive species with selective cytotoxicity. Recently, Mercer and coworkers demonstrated that artemisinin endoperoxide bioactivation to carbon-centered radicals only results in cytotoxicity when it is mediated by heme or a heme-containing protein [46]. They hypothesized that artemisinin radicals act locally within the mitochondria to modify heme or heme containing proteins, which results in the generation of reactive oxygen species (ROS) via electron transport chain dysfunction and the induction of cell death via apoptosis [46].
The pharmacological activity of bicyclam molecules was first identified in the search for new agents to treat HIV. Plerixafor evolved through an intensive medicinal chemistry effort from these early alkyl-linked bicyclams. Mechanistic studies suggested that the bicyclams acted at the early stages of the HIV infection process, but it was not until the discovery that HIV required a chemokine coreceptor, either CCR5 or CXCR4, along with CD4, for host cell entry, that the molecular target for plerixafor was discovered. Plerixafor was shown to Specifically inhibit infection of CXCR4-using (X4) virus, thus pointing to antagonism of the CXCR4 chemokine receptor as the target. Plerixafor was subsequently investigated in clinical trials. In a Phase I study, a single subcutaneous injection of plerixafor resulted in a marked and rapid increase in circulating white blood cells (WBCs). Initially this was attributed to demargination but subsequently, as described below, this was found to be due to cell mobilization. Although plerixafor was shown to be able to reduce X4-viral load in HIV patients, it was the pharmacological action as a mobilizer of hematopoietic stem cells (HSCs) that was the subsequent focus for clinical development. HSCs are the stem cells from which all blood cells are derived, a process known as hematopoiesis. HSC have the ability to self-renew and are capable of generating every cell lineage of the hematopoietic system including erythrocytes, platelets, lymphoid and myeloid cells. An early step in hematopoiesis is the differentiation of a HSC to a hematopoietic progenitor cell (HPC) that is further committed to differentiation down a particular hematopoietic lineage pathway. As HPCs can be easily quantified by colony-forming assays, they are frequently used as a measure of HSC [42].
Target
The medicines identified through TDD used standard target validation approaches followed by optimization.
A potentially effective therapy for homozygous familial hypercholesterolemia would be to reduce low-density lipoprotein (LDL) production. The MTP is responsible for transferring triglycerides onto ApoB within the liver in the assembly of very-low-density lipoprotein (VLDL), the precursor to LDL. In the absence of functional MTP, as in the rare recessive genetic disorder abetalipoproteinemia, the liver cannot secrete VLDL, leading to the absence of all lipoproteins containing ApoB in the plasma. Thus, the pharmacologic inhibition of MTP might be a strategy for reducing LDL production and plasma LDL cholesterol levels. Preclinical studies in animal models lacking LDL receptors had shown that the inhibition of MTP significantly reduces serum cholesterol levels [53]. Lomitapide whose precursors were discovered in a high through put screen [54], directly binds and inhibits MTP, which resides in the lumen of the endoplasmic reticulum, thereby preventing the assembly of apoB-containing lipoproteins in enterocytes and hepatocytes. This inhibits the synthesis of chylomicrons and VLDL. The inhibition of the synthesis of VLDL leads to reduced levels of plasma LDL-C [4].
Pasireotide is an injectable cyclohexapeptide somatostatin analogue approved for the treatment of Cushing's disease were surgery is not an option. Pasireotide exerts its pharmacological activity via binding to somatostatin receptors (ssts). Five human sst subtypes are known: hsst1, 2, 3, 4 and 5. These receptor subtypes are expressed in different tissues under normal physiological conditions. Corticotroph tumor cells from Cushing's disease patients frequently overexpress hsst5 whereas the other receptor subtypes are often not expressed or are expressed at lower levels. Natural somatostatins bind with high affinity to all five human ssts receptor subtypes, hsst1–5. However, the therapeutic use of peptides is limited by the rapid proteolytic degradation in plasma. Pasireotide was discovered in a search for short, metabolically stable peptidomimetics with improved properties. It binds and activates the hsst receptors resulting in inhibition of ACTH secretion, which leads to decreased cortisol secretion [4,60].
Biologics
Biologics use a number of innovative mechanisms to address unmet medical need. This included a monoclonal antibody to anthrax (raxibacumab) and a recombinant enzyme to degrade toxic levels of the anticancer drug methotrexate (glucarpidase) and a number of fusion proteins that conjugate toxic molecules to a protein to direct them to a Specific location (brentuximab vedotin, gentuzumab and dinileukin difutox). Pegvisomant is an analog of growth hormone.
Raxibacumab is a monoclonal antibody that binds free protective antigen of anthrax. The anthrax toxin is a tripartite toxin that comprises lethal factor and edema factor as the enzymatic moieties of the toxin and protective antigen as the binding moiety. Blocking the binding of protective antigen to its host receptors can counter the deleterious effects of the anthrax toxin and provides the basis for vaccine and passive immunization with antiprotective antigen immunoglobulin. Raxibacumab inhibits the binding of protective antigen to its cellular receptors, preventing the intracellular entry of the anthrax lethal factor and edema factor, the enzymatic toxin components responsible for the pathogenic effects of anthrax toxin [4,61].
Glucarpidase is a recombinant bacterial enzyme that hydrolyzes the carboxylterminal glutamate residue from folic acid and classical antifolates such as methotrexate. Methotrexate, an antifolate agent, has been used for over 60 years in the treatment of various cancers. High-dose methotrexate has been particularly useful in the treatment of leukemias and lymphomas. However, even with aggressive hydration and urine alkalinization, such regimens can lead to acute renal dysfunction, as indicated by decreases in urine production and concomitant increases in blood urea nitrogen and serum creatinine levels. Because methotrexate is largely excreted by the kidneys, this can greatly potentiate tissue damage. Toxic levels of blood methotrexate can be rapidly and effectively decreased by intravenous administration of glucarpidase. Glucarpidase is a recombinant form of carboxypeptidase G2, a bacterial enzyme that rapidly cleaves methotrexate to form the amino acid glutamate and 2,4-diamino-N(10)-methylpteroic acid. Catabolites of methotrexate are much less toxic than the parent compound, and are primarily excreted by hepatic mechanisms [4,63].
Brentuximab vedotin is an antibody drug conjugate (ADC). The antibody is a chimeric IgG1 directed against CD30. The small molecule, MMAE, is a microtubule disrupting agent. MMAE is covalently attached to the antibody via a linker. Nonclinical data suggest that the anticancer activity of brentuximab vedotin is due to the binding of the ADC to CD30-expressing cells, followed by internalization of the ADC–CD30 complex, and the release of MMAE via proteolytic cleavage. Binding of MMAE to tubulin disrupts the microtubule network within the cell, subsequently inducing cell cycle arrest and apoptotic death of the cells [4].
The cumulative distribution of NMEs by genetics versus physiology is shown in Figure 3B. This includes both first in class and followers. The overall rate of approvals by physiology informed discovery is consistently greater than that for genetics informed discovery.
MMOA
Further in-depth analysis of the MMOAs demonstrated the diversity through which the NMEs interacted with physiology to provide therapeutically useful pharmacological responses. Enzyme replacement therapy replaced physiological molecules and accordingly, retained the physiological MMOAs (Table 1). This is obviously a preferred approach, but has its challenges including delivery to the pathological location. The kinases for cancer had gain-of-function mutations that were addressed with inhibitors. However, it is not that simple since inhibitors to ATP binding site must compete with high endogenous concentrations of ATP (∼1 mM). Discovery of the first kinase inhibitors (including imatinib) was assisted by phenotypic assays to that identified early candidate molecules that blocked kinase activation. Target informed phenotypic assays were required to identify this therapeutically useful MMOA. Other MMOAs for genetic informed discoveries also used a target informed phenotypic assay including the potentiator for the CTFR mutant protein, ivacaftor. As noted above carglumic acid is a structural analog of an essential allosteric activator and product of the deficient enzyme.
Consistent with our previous analysis there was a diversity of MMOAs for the medicines discovered were discovery was primarily based on physiological knowledge (Table 2). MMOAs included agonists such a pasireotide, a somatastatin analog, allosteric activators such as cinacalcet (calcium receptor), irreversible inhibitors such as vigabatrin (GABA), reversible inhibitors such as vorinostat (HDAC), channel modulators such as rufinamide, nucleoside analogs (azacitidine and nelarabine) that work via incorporation into DNA leading to inhibition of DNA synthesis or irreversibly inactivation DNA methyltransferases, respectively. Some interesting biological MMOAs included ADCs that conjugate war heads to proteins/antibodies that target Specific cell types. These include denileukin diftitox, brentuximab vendotin and alemtuzumab. Glucarpidase is an enzyme that degrades methotrexate to non-toxic metabolites and raxibacumab is a monoclonal antibody to anthrax. The exact MMOA of artemisinin is only now being understood. One obvious conclusion from these examples is that “one size does not fit all” in terms of MMOAs that are therapeutically useful.
Discussion
This work was initiated to provide a knowledge basis for addressing how to increase success in drug discovery for rare diseases. It is clear that drug discovery will use available knowledge to inform strategy.
First-in-class medicines accounted for 45 of the 102 medicines. Perhaps the most striking finding is that this addresses only a small fraction of the approximately 6800 rare diseases. The relatively high numbers of followers (51) was also surprising due to the small patient populations and large unmet medical need across all disease area. However, a number of the followers were addressing unmet medical need caused by resistance to previously approved drugs (e.g., ponatinib for abl kinase).
The drug discovery strategies for first in class distributed between phenotypic (33%), target-based (27%) and biologics (40%) (Figure 2A) is similar compared with what was observed for first-in-class medicines approved for all disease between 1999 and 2008 [2]. In that analysis the distribution, not including modified natural substances, was similar (target-based 24%, phenotypic 40% and biologics 36%) except that biologics had a higher ratio than phenotype. This is presumably due to the number of biologic medicines approved as enzyme replacement therapy. It is somewhat surprising that TDD is not more successful for rare diseases due to the potential greater understanding of the genes that cause the diseases.
In our previous analysis across all diseases we had suggested that the success of phenotypic screening was in part due to the ability to identify an optimal MMOA. Another way to look at this is that the knowledge of the pathophysiology is incomplete, therefore discovery strategies based on phenotypic biomarkers had a greater chance of success. The results from this analysis are consistent with this conclusion. In the previous analysis we observed that for followers, the molecular, target-based approach was more successful than phenotypic, by a ratio of 83 NMEs to 30 NMEs, which was concluded to support the idea that more complete knowledge of the MMOA led to greater success; there is more knowledge to guide follower drug discovery. With the orphan drugs the trend was similar but much less dramatic (Figure 2B). The ratio of target-based to phenotypic was only 18 to 13. If followers are considered as best in class, it is clear that in with large patient populations there is a need to evolve medicines to best in class and to have followers that can address the heterogeneity of the patient population. In contrast with rare diseases, the small patient populations do not provide incentive for followers (best in class) and in many cases in which there is a very small patient population most patients are evaluated with the original medicine (truly personalized medicine) so there is limited need for a follower.
In this work we analyzed the impact of genetic knowledge on successful drug discovery since over 80% of rare diseases are genetic [1]. A consideration for the inclusion of the genetic analysis was to evaluate how this knowledge would impact the drug discovery strategy. As discussed above, it was proposed that the lack of success of target-based drug discovery was, in part, due to the inability to a priori predict the MMOA that would yield a useful therapeutic index. Selecting a target from genomic data was not Sufficient to ensure success, translational assays were required. With genetic diseases the cause of the disease is clearly known, however that does not necessarily provide the knowledge required for a treatment, unless gene therapy will one day become successful.
We observed that the contribution of genetic association to the successful discovery strategies was under-represented (25%) with respect to the number of genetic diseases (>80%) (Figure 2). Knowledge of associated genotypes provided the greatest contribution to the discovery of new medicine for cancers associated with Specific mutations to protein kinases and new medicines to treat in born errors in metabolism (Figure 4). A detailed evaluation of the drug discovery strategy for each of the genetically informed medicines showed that there was practical knowledge that contributed to the discovery regardless of the disease strategy (phenotypic, target or biologic). For example, biochemical solutions were readily apparent. It is clear that inhibition of a kinase activity will block the gain of function. Enzyme replacement therapy is proven approach for in-born errors in metabolism.
Not only was there information to facilitate the discovery strategy in these areas, patients are easier to identify and diagnose, and regulatory precedent for the disease exist, all of which makes designing and conducting drug development programs more feasible. More basic and translational research has been carried out in these therapeutic areas than for other genetic disorders.
The observation that genotypic knowledge was not Sufficient to ensure drug discovery success may be the most compelling finding from this analysis. New medicines were discovered when the genotypic knowledge was combined with other physiologic knowledge and technical feasibility. Drug discovery is an iterative knowledge-gaining endeavor in which the knowledge will always be incomplete (Figure 1). Integrating knowledge and approaches from different sources, basic research involving genetics, biology, medicine and physiology, is required to understand translational biomarkers, clinical pharmacology to design clinical trials and chemistry, biology, toxicology, pharmacology and biopharmaceutics for discovery.
But this also highlights the huge challenge for rare disease drug discovery. Realistically there is not and never will be Sufficient funding and time to acquire all the knowledge for all the diseases. How do you proceed effectively with incomplete knowledge?
The success of phenotypic screening provides some clues. With phenotypic screening Sufficient knowledge must be acquired to determine a translational biomarker and phenotype related to the disease. With that knowledge a program can be executed empirically. One way to think about how to proceed in a knowledge-insufficient area is to use available knowledge to explore solutions empirically.
Future perspective
How can the unmet medical need for treatments for rare diseases be addressed without an exponential increase in funding?
Because of the need and the unavailability of complete knowledge rare disease drug discovery will require new strategies that will be more efficient and effective. Those solutions may come from increased funding, increased drug discovery efficiency or new approaches.
Increased funding
It is important to increase funding for rare diseases. It is also important to fund aspects that will lead to greater success. The greatest importance should be given to discovery and validation of translational biomarkers used for clinical testing and guiding drug discovery. All drug discovery strategies require a translational biomarker.
Increased drug discovery efficiency
Drug discovery strategies must be employed that provide a greater chance for clinical success. Regardless if the chosen strategies involve small molecules or biologics they need to use available knowledge and require a translational biomarker. Empirical strategies will help to create success when the knowledge is incomplete. The starting point for empirical strategies can be with phenotypic assays as well as targets and biologics.
New approaches
There are always new technologies being developed that have promise and are over-hyped. At first glance rare diseases seem to be a great place to test all the new ideas. But can they afford the failure rates? For the genetic diseases, gene therapy would be the most effective since it is replacing the gene that is mutated.
The many breakthroughs in new genetic technologies such as next-generation sequencing help to identify the genes that contribute to the disease. The challenge is to relate those genes or causes to a cure. It is analogous to knowing the parts to an engine or computer but it is difficult to fix unless you replace the part exactly. There are a number of new techniques that are being developed including RNAi and stem cell replacement that have promise. Perhaps in the future these will provide opportunities for rare diseases.
To conclude, unfortunately, our knowledge of most diseases and the underlying molecular causes are incomplete. An additional challenge is that knowledge of the cause, for instance a genetic defect or multiple genetic defects, rarely provides a Specific molecular solution, despite our hopes. Plenge, Scholnick and Altshuler recently noted most preclinical discovery programs have incomplete supporting material to inform the drug discovery strategy [71]. Empirical solutions that can be effective with the limited knowledge available are needed. This creates a huge challenge and opportunity in the current knowledge driven approach to medical research.
Supplementary Material
Key Terms.
Biologics
New medicines that are proteins and generally prepared using recombinant technologies. The most common therapeutic biologics are enzymes and monoclonal antibodies.
First in class
When possible we use the classification reported by the US FDA. A drug new medicine was considered first in class if no other approved medicines worked the same way; when the target is known the first modulator will be first-in-class; medicines that provide therapeutic differentiation via mechanistic differentiation, such as the partial agonist aripiprazole; and when the mechanism or target is unknown they are first in class unless there exists a chemical analog of a previously approved medicine for the same indication.
Follower
Not a first-in-class agent.
Target-based drug discovery
Molecular, reductionist strategy associated with an isolated gene product known as a target.
Phenotypic drug discovery
Empirical strategy associated with a phenotype that translated to the desired clinical response.
Molecular mechanism of action
Connects specific molecular interactions between the therapeutic treatment and the biological target that yields the pharmacological response.
Genetic contributions
Most are caused by defects in a single gene (e.g., α1-antitrypsin deficiency), whereas others are due to mutations in several genes (e.g., oncology indications).
Executive summary.
Purpose of analysis
Provide new knowledge to inform drug discovery in rare diseases by addressing the question, “What are the drug discovery strategies that produce new medicines (NMEs)?”
Findings
There were 102 orphan medicines approved between 1999 and 2012. Cancer indications comprised 44% (43 of 97) of the approved NMEs, followed by metabolic diseases 23% (22 of 97%) There were nine NMEs each for inflammation/immunomodulation, neurologic and vascular/cardiovascular. Forty six were first-in-class NMEs and 51 were followers; five others were imaging agents or chelators.
Overall, 30 NMEs were discovered using target-based approaches, 27 NMEs were discovered using phenotypic-based approaches, eight NMEs were based on modifications of natural substances and 30 of the agents were biologics.
First-in-class medicines were discovered with phenotypic assays (15), target-based approaches (12) and biologic strategies (18).
Since >80% of rare diseases are of genetic origin we also analyzed the contribution of known genetic mutations to success. The discovery strategies for 26 NMEs were directly linked to the underlying genetic mutations, eight were for kinases (five for BCR-ABL) and nine NMEs were enzyme replacement therapy to treat genetic in-born errors in metabolism. The remaining nine NMEs associated with specific mutations benefited from biological systems understandings to identify the drug discovery strategies.
Identification of genetic causes in areas with more basic and translational research, for example, cancer and in-born errors in metabolism, contributed to success regardless of discovery strategy.
Conclusion
Greater knowledge increases the chance of success and empirical solutions can be effective when knowledge is incomplete. This creates a huge challenge and opportunity in the current knowledge-driven approach to medical research.
Acknowledgments
During the review of this manuscript, Dr Shuangluo Xia passed away. Dr Xia was an invaluable colleague and friend to many. He made many significant scientific contributions during his highly productive career and will be deeply missed.
Footnotes
Supplementary data: To view the supplementary data that accompany this paper please visit the journal website at: www.future-science com/10.4155/FMC.14.65
Financial & competing interests disclosure: The authors were employed by the nonprofit Institute for Rare and Neglected Diseases Drug Discovery and partially supported by NIH grant RO1-AI103476. DC Swinney has also consulted or received honoraria for lectures from GSK and Vertex. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Melnikova I. Rare diseases and orphan drugs. Nat Rev Drug Discov. 2012;11(4):267–268. doi: 10.1038/nrd3654. [DOI] [PubMed] [Google Scholar]
- 2.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10(7):507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 3.Bashaw ED, Huang SM, Cote TR, et al. Clinical pharmacology as a cornerstone of orphan drug development. Nat Rev Drug Discov. 2011;10(11):795–796. doi: 10.1038/nrd3595. [DOI] [PubMed] [Google Scholar]
- 4.FDA drug approvals. www.accessdata.fda.gov/scripts/cder/drugsatfda.
- 5.Mesa RA, Yasothan U, Kirkpatrick P. Ruxolitinib. Nat Rev Drug Discov. 2012;11(2):103–104. doi: 10.1038/nrd3652. [DOI] [PubMed] [Google Scholar]
- 6.Shaw AT, Yasothan U, Kirkpatrick P. Crizotinib. Nat Rev Drug Discov. 2011;10(12):897–898. doi: 10.1038/nrd3600. [DOI] [PubMed] [Google Scholar]
- 7.Bollag G, Tsai J, Zhang J, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11(11):873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 8.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov. 2002;1(7):493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 9.Keller G, Schafhausen P, Brummendorf TH. Bosutinib. Recent Results Cancer Res. 2010;184:119–127. doi: 10.1007/978-3-642-01222-8_9. [DOI] [PubMed] [Google Scholar]
- 10.Zhou T, Commodore L, Huang WS, et al. Structural mechanism of the pan-BCR-ABL inhibitor ponatinib (AP24534): lessons for overcoming kinase inhibitor resistance. Chem Biol Drug Des. 2011;77(1):1–11. doi: 10.1111/j.1747-0285.2010.01054.x. [DOI] [PubMed] [Google Scholar]
- 11.Cortes JE, Kantarjian H, Shah NP, et al. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med. 2012;367(22):2075–2088. doi: 10.1056/NEJMoa1205127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weisberg E, Manley P, Mestan J, Cowan-Jacob S, Ray A, Griffin JD. AMN107 (nilotinib): a novel and selective inhibitor of BCR-ABL. Br J Cancer. 2006;94(12):1765–1769. doi: 10.1038/sj.bjc.6603170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jabbour E, Cortes J, Kantarjian H. Dasatinib for the treatment of Philadelphia chromosome-positive leukaemias. Expert Opin Invest Drugs. 2007;16(5):679–687. doi: 10.1517/13543784.16.5.679. [DOI] [PubMed] [Google Scholar]
- 14.Nilederiwieser A, Curtius HC. Inherited Dieseases of Amino Acid Metabolism. Georg Thieme; Stuttgart, Germany: 1985. pp. 104–121. [Google Scholar]
- 15.Kure S, Hou DC, Ohura T, et al. Tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency. J Pediatr. 1999;135(3):375–378. doi: 10.1016/s0022-3476(99)70138-1. [DOI] [PubMed] [Google Scholar]
- 16.Muntau AC, Roschinger W, Habich M, et al. Tetrahydrobiopterin as an alternative treatment for mild phenylketonuria. N Engl J Med. 2002;347(26):2122–2132. doi: 10.1056/NEJMoa021654. [DOI] [PubMed] [Google Scholar]
- 17.Blau N, Erlandsen H. The metabolic and molecular bases of tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency. Mol Genet Metab Rep. 2004;82(2):101–111. doi: 10.1016/j.ymgme.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Muenzer J, Wraith JE, Beck M, et al. A Phase II/III clinical study of enzyme replacement therapy with idursulfase in mucopolysaccharidosis II (Hunter syndrome) Genet Med. 2006;8(8):465–473. doi: 10.1097/01.gim.0000232477.37660.fb. [DOI] [PubMed] [Google Scholar]
- 19.Hopwood JJ, Bate G, Kirkpatrick P. Galsulfase. Nat Rev Drug Discov. 2006;5(2):101–102. doi: 10.1038/nrd1962. [DOI] [PubMed] [Google Scholar]
- 20.Kakkis ED, Muenzer J, Tiller GE, et al. Enzyme-replacement therapy in mucopolysaccharidosis I. N Engl J Med. 2001;344(3):182–188. doi: 10.1056/NEJM200101183440304. [DOI] [PubMed] [Google Scholar]
- 21.Eng CM, Guffon N, Wilcox WR, et al. Safety and efficacy of recombinant human alpha-galactosidase A – replacement therapy in Fabry's disease. N Engl J Med. 2001;345(1):9–16. doi: 10.1056/NEJM200107053450102. [DOI] [PubMed] [Google Scholar]
- 22.Lock EA, Ellis MK, Gaskin P, et al. From toxicological problem to therapeutic use: the discovery of the mode of action of 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC), its toxicology and development as a drug. J Inherit Metab Dis. 1998;21(5):498–506. doi: 10.1023/a:1005458703363. [DOI] [PubMed] [Google Scholar]
- 23.Kavana M, Moran GR. Interaction of (4-hydroxyphenyl) pyruvate dioxygenase with the specific inhibitor 2-[2-nitro-4-(trifluoromethyl)benzoyl]-1,3-cyclohexanedione. Biochemistry. 2003;42(34):10238–10245. doi: 10.1021/bi034658b. [DOI] [PubMed] [Google Scholar]
- 24.Brownlee JM, Johnson-Winters K, Harrison DH, Moran GR. Structure of the ferrous form of (4-hydroxyphenyl) pyruvate dioxygenase from Streptomyces avermitilis in complex with the therapeutic herbicide, NTBC. Biochemistry. 2004;43(21):6370–6377. doi: 10.1021/bi049317s. [DOI] [PubMed] [Google Scholar]
- 25.Davis PB, Yasothan U, Kirkpatrick P. Ivacaftor. Nat Rev Drug Discov. 2012;11(5):349–350. doi: 10.1038/nrd3723. [DOI] [PubMed] [Google Scholar]
- 26.Jih KY, Hwang TC. Vx-770 potentiates CFTR function by promoting decoupling between the gating cycle and ATP hydrolysis cycle. Proc Natl Acad Sci USA. 2013;110(11):4404–4409. doi: 10.1073/pnas.1215982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matis LA, Rollins SA. Complement-specific antibodies: designing novel anti-inflammatories. Nat Med. 1995;1(8):839–842. doi: 10.1038/nm0895-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas TC, Rollins SA, Rother RP, et al. Inhibition of complement activity by humanized anti-C5 antibody and single-chain Fv. Mol Immunol. 1996;33(17–18):1389–1401. doi: 10.1016/s0161-5890(96)00078-8. [DOI] [PubMed] [Google Scholar]
- 29.Parker CJ, Kar S, Kirkpatrick P. Eculizumab. Nat Rev Drug Discov. 2007;6(7):515–516. doi: 10.1038/nrd2369. [DOI] [PubMed] [Google Scholar]
- 30.Platt FM, Neises GR, Dwek RA, Butters TD. N-butyldeoxynojirimycin is a novel inhibitor of glycolipid biosynthesis. J Biol Chem. 1994;269(11):8362–8365. [PubMed] [Google Scholar]
- 31.Pastores GM, Barnett NL. Substrate reduction therapy: miglustat as a remedy for symptomatic patients with Gaucher disease type 1. Expert Opin Invest Drugs. 2003;12(2):273–281. doi: 10.1517/13543784.12.2.273. [DOI] [PubMed] [Google Scholar]
- 32.Haberle J. Role of carglumic acid in the treatment of acute hyperammonemia due to N-acetylglutamate synthase deficiency. Ther Clin Risk Manag. 2011;7:327–332. doi: 10.2147/TCRM.S12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wetzler M, Segal D. Omacetaxine as an anticancer therapeutic: what is old is new again. Curr Pharm Design. 2011;17(1):59–64. doi: 10.2174/138161211795049778. [DOI] [PubMed] [Google Scholar]
- 34.Andries K, Verhasselt P, Guillemont J, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307(5707):223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 35.Richon VM, Webb Y, Merger R, et al. Second generation hybrid polar compounds are potent inducers of transformed cell differentiation. Proc Natl Acad Sci USA. 1996;93(12):5705–5708. doi: 10.1073/pnas.93.12.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25(1):84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 37.Krenitsky TA, Koszalka GW, Tuttle JV, Rideout JL, Elion GB. An enzymatic synthesis of purine D-arabinonucleotides. Carbohydr Res. 1981;97(1):139–146. [Google Scholar]
- 38.Lambe CU, Averett DR, Paff MT, Reardon JE, Wilson JG, Krenitsky TA. 2-amino-6-methoxypurine arabinoside: an agent for T-cell malignancies. Cancer Res. 1995;55(15):3352–3356. [PubMed] [Google Scholar]
- 39.Rodriguez CO, Jr, Stellrecht CM, Gandhi V. Mechanisms for T-cell selective cytotoxicity of arabinosylguanine. Blood. 2003;102(5):1842–1848. doi: 10.1182/blood-2003-01-0317. [DOI] [PubMed] [Google Scholar]
- 40.Parker WB, Secrist JA, 3rd, Waud WR. Purine nucleoside antimetabolites in development for the treatment of cancer. Curr Opin Invest Drugs. 2004;5(6):592–596. [PubMed] [Google Scholar]
- 41.Gandhi V, Keating MJ, Bate G, Kirkpatrick P. Nelarabine. Nat Rev Drug Discov. 2006;5(1):17–18. doi: 10.1038/nrd1933. [DOI] [PubMed] [Google Scholar]
- 42.Fricker SP. Physiology and pharmacology of plerixafor. Transfus Med Hemother. 2013;40(4):237–245. doi: 10.1159/000354132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jain KK. An assessment of rufinamide as an anti-epileptic in comparison with other drugs in clinical development. Expert Opin Invest Drugs. 2000;9(4):829–840. doi: 10.1517/13543784.9.4.829. [DOI] [PubMed] [Google Scholar]
- 44.Rogawski MA. Diverse mechanisms of antiepileptic drugs in the development pipeline. Epilepsy Res. 2006;69(3):273–294. doi: 10.1016/j.eplepsyres.2006.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao F. Discovery of artemisinin (qinghaosu) Molecules. 2009;14(12):5362–5366. [Google Scholar]
- 46.Mercer AE, Copple IM, Maggs JL, O'neill PM, Park BK. The role of heme and the mitochondrion in the chemical and molecular mechanisms of mammalian cell death induced by the artemisinin antimalarials. J Biol Chem. 2011;286(2):987–996. doi: 10.1074/jbc.M110.144188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodman AD, Stone RT. Enhancing neural transmission in multiple sclerosis (4-aminopyridine therapy) Neurotherapeutics. 2013;10(1):106–110. doi: 10.1007/s13311-012-0156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nemeth EF. Misconceptions about calcimimetics. Ann NY Acad Sci. 2006;1068:471–476. doi: 10.1196/annals.1346.044. [DOI] [PubMed] [Google Scholar]
- 49.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 50.Issa JP, Kantarjian HM, Kirkpatrick P. Azacitidine. Nat Rev Drug Discov. 2005;4(4):275–276. doi: 10.1038/nrd1698. [DOI] [PubMed] [Google Scholar]
- 51.Rossignol JF, Maisonneuve H. Nitazoxanide in the treatment of Taenia saginata and Hymenolepis nana infections. Am J Trop Med Hyg. 1984;33(3):511–512. doi: 10.4269/ajtmh.1984.33.511. [DOI] [PubMed] [Google Scholar]
- 52.Hemphill A, Mueller J, Esposito M. Nitazoxanide, a broad-spectrum thiazolide anti-infective agent for the treatment of gastrointestinal infections. Expert Opin Pharmacother. 2006;7(7):953–964. doi: 10.1517/14656566.7.7.953. [DOI] [PubMed] [Google Scholar]
- 53.Cuchel M, Bloedon LT, Szapary PO, et al. Inhibition of microsomal triglyceride transfer protein in familial hypercholesterolemia. N Engl J Med. 2007;356(2):148–156. doi: 10.1056/NEJMoa061189. [DOI] [PubMed] [Google Scholar]
- 54.Jamil H, Gordon DA, Eustice DC, et al. An inhibitor of the microsomal triglyceride transfer protein inhibits apoB secretion from HepG2 cells. Proc Natl Acad Sci USA. 1996;93(21):11991–11995. doi: 10.1073/pnas.93.21.11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuter DJ. New thrombopoietic growth factors. Blood. 2007;109(11):4607–4616. doi: 10.1182/blood-2006-10-019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilhelm S, Carter C, Lynch M, et al. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5(10):835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 57.Goldberg A. Cancer Drug Discovery and Development: Proteasome Inhibitors in Cancer Therapy. Humana; NJ, USA: 2004. [Google Scholar]
- 58.Stein RL, Ma YT, Brand S. US5693617. 1995
- 59.Remuzzi G, Perico N, Benigni A. New therapeutics that antagonize endothelin: promises and frustrations. Nat Rev Drug Discov. 2002;1(12):986–1001. doi: 10.1038/nrd962. [DOI] [PubMed] [Google Scholar]
- 60.Bruns C, Lewis I, Briner U, Meno-Tetang G, Weckbecker G. SOM230: a novel somatostatin peptidomimetic with broad somatotropin release inhibiting factor (SRIF) receptor binding and a unique antisecretory profile. Eur J Endocrinol. 2002;146(5):707–716. doi: 10.1530/eje.0.1460707. [DOI] [PubMed] [Google Scholar]
- 61.Migone TS, Subramanian GM, Zhong J, et al. Raxibacumab for the treatment of inhalational anthrax. N Engl J Med. 2009;361(2):135–144. doi: 10.1056/NEJMoa0810603. [DOI] [PubMed] [Google Scholar]
- 62.Jeppesen PB, Sanguinetti EL, Buchman A, et al. Teduglutide (ALX-0600), a dipeptidyl peptidase IV resistant glucagon-like peptide 2 analogue, improves intestinal function in short bowel syndrome patients. Gut. 2005;54(9):1224–1231. doi: 10.1136/gut.2004.061440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Green JM. Glucarpidase to combat toxic levels of methotrexate in patients. Ther Clin Risk Manag. 2012;8:403–413. doi: 10.2147/TCRM.S30135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Broudy VC, Lin NL. AMG531 stimulates megakaryopoiesis in vitro by binding to Mpl. Cytokine. 2004;25(2):52–60. doi: 10.1016/j.cyto.2003.05.001. [DOI] [PubMed] [Google Scholar]
- 65.Fuh G, Cunningham BC, Fukunaga R, Nagata S, Goeddel DV, Wells JA. Rational design of potent antagonists to the human growth hormone receptor. Science. 1992;256(5064):1677–1680. doi: 10.1126/science.256.5064.1677. [DOI] [PubMed] [Google Scholar]
- 66.Ross RJ, Leung KC, Maamra M, et al. Binding and functional studies with the growth hormone receptor antagonist, B2036-PEG (pegvisomant), reveal effects of pegylation and evidence that it binds to a receptor dimer. J Clin Endocrinol Metabol. 2001;86(4):1716–1723. doi: 10.1210/jcem.86.4.7403. [DOI] [PubMed] [Google Scholar]
- 67.Kopchick JJ, Parkinson C, Stevens EC, Trainer PJ. Growth hormone receptor antagonists: discovery, development, and use in patients with acromegaly. Endocrine Rev. 2002;23(5):623–646. doi: 10.1210/er.2001-0022. [DOI] [PubMed] [Google Scholar]
- 68.Keating MJ, Flinn I, Jain V, et al. Therapeutic role of alemtuzumab (Campath-1H) in patients who have failed fludarabine: results of a large international study. Blood. 2002;99(10):3554–3561. doi: 10.1182/blood.v99.10.3554. [DOI] [PubMed] [Google Scholar]
- 69.Bernstein ID. Monoclonal antibodies to the myeloid stem cells: therapeutic implications of CMA-676, a humanized anti-CD33 antibody calicheamicin conjugate. Leukemia. 2000;14(3):474–475. doi: 10.1038/sj.leu.2401663. [DOI] [PubMed] [Google Scholar]
- 70.Olsen E, Duvic M, Frankel A, et al. Pivotal Phase III trial of two dose levels of denileukin diftitox for the treatment of cutaneous T-cell lymphoma. J Clin Oncol. 2001;19(2):376–388. doi: 10.1200/JCO.2001.19.2.376. [DOI] [PubMed] [Google Scholar]
- 71.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12(8):581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.