Summary
mRNA translation, a highly coordinated affair involving many proteins and RNAs, is generally divided into three steps: initiation, elongation, and termination. Each of these steps serves as a point of regulation to control the amount of protein that is produced. The protein 4E-HP has recently been shown to disrupt recruitment of the translation initiation complex by directly binding the 5’ cap of cellular mRNAs. Recent work has shown elongation rates are likely altered during mitosis and certain types of synaptic transmission. Other work has shown premature termination of mRNAs lacking stop codons appears to repress their translation. Together, these studies highlight the importance of translational control in diverse processes such as development, cancer, and synaptic plasticity.
Introduction
Translation, the decoding of messenger RNA (mRNA) into protein, is a complex process involving mRNA, ribosomes, and a plethora of additional factors that not only promote polypeptide elongation, but regulate this process as well. The overall translation process can be divided into three main steps: initiation, elongation and termination. Each of these steps can be regulated, resulting in the differential synthesis of specific proteins and profound changes in cell physiology. For example, regulated translation controls such diverse phenomena as learning and memory formation, body plan patterning during development, and cancer [1-5]. This review will highlight recent findings of eukaryotic translational control that occur at initiation, elongation and termination. Other more detailed reviews of translation have recently been published [5-7]
A brief overview of translation
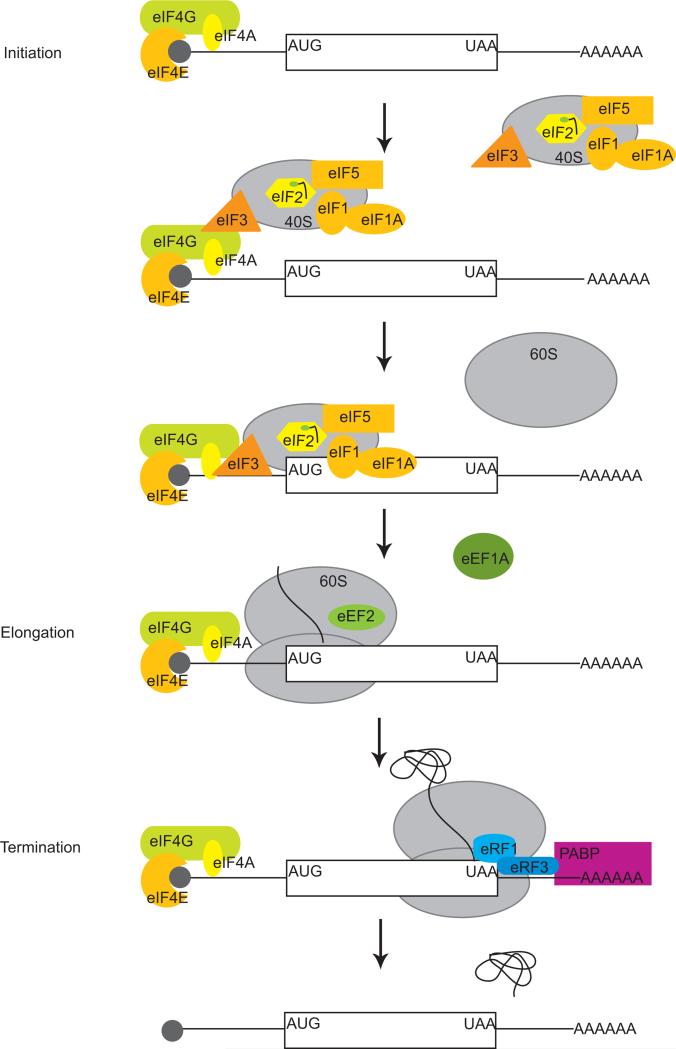
The early events of translation initiation begin with the formation of a ternary complex consisting of GTP, met-tRNA, and the initiation factor eIF2 [8]. Together with additional initiation factors (eIF3, eIF5, eIF1, and eIF1A), the ternary complex associates with the 40S ribosomal subunit to form a 43S preinitiation complex (Figure 1) [9,10]. In mammals, this large agglomeration of factors is recruited to the 5’ end of the mRNA through interactions with the initiation factors eIF4G and eIF3 whereupon it is referred to as the 48S preinitiation complex [11]. The DExH box putative helicase protein DHX29 is also required in this complex for translation of mRNAs with highly structured 5’ UTRs [12]. Proper positioning of the complex on the 5’ end requires the m7G cap-binding complex consisting of eIF4E, eIF4G and eIF4A, which together are called eIF4F [13]. With the aid of the initiation factors eIF1, eIF1A, and DHX29, the 48S complex then scans the mRNA in the 3’ direction until it encounters an AUG initiation codon in the correct context, generally the first AUG, where it is joined by the 60S subunit to begin polypeptide elongation [12,14,15].
Figure 1.
A schematic representation of translation. mRNA translation occurs in 3 steps; initiation, elongation, and termination. During the initiation phase, mRNA is bound by the eIF4F complex, consisting of eIF4G, eIF4E, and eIF4A, through interaction of eIF4E and the 5’ cap of the mRNA. The 43S pre-initiation complex, composed of the 40S ribosome, eIF2-ternary complex, eIF1, eIF1A, eIF5, and eIF3, is recruited to the message through interactions between eIF4G and eIF3, thereby forming the 48S pre-initiation complex. This agglomeration of proteins and the 40S ribosomal subunit scans the mRNA until the initiating AUG codon is reached. The 60S subunit then joins and the initiation factors are released. Translation elongation begins when the peptide chain is elongated. Elongation factor eEF1A brings charged tRNAs to the translating ribosome; eEF2 promotes translocation. Once the termination codon is reached, release factor eRF1 binds in place of tRNA. eRF3 stimulates GTP hydrolysis and the peptide chain is released together with the 40S and 60S subunits. PABP refers to poly(A) binding protein.
In contrast to initiation, elongation is a simpler affair; its requirements are to maintain the reading frame, select and deliver the correct aminoacyl-tRNAs to the 80S ribosome, and form peptide bonds. Only two elongation factors are required for these tasks: eEF1A, which helps bring the charged tRNAs to the ribosome, and eEF2, which promotes translocation of the ribosome along the mRNA [16].
Termination is mediated by the release factor eRF1; it recognizes one of three stop codons and binds to the ribosome in place of a tRNA. This event, along with binding of eRF3, stimulates GTP hydrolysis and release of the peptide chain [17,18].
The complexity of each of these three steps is proportional to the amount of regulation that occurs. That is, most translational control mechanisms described thus far affect initiation, albeit not necessarily at one particular phase of initiation [19]. This makes sense because it is energetically favorable to control the first step in any reaction. Relatively few examples of regulation at elongation are known, although a number are inferred based on the phosphorylation state of eEF2 [20,21]. Finally, with the exception of premature stop codons (which leads to nonsense mediated mRNA decay), there are few clear examples of regulated termination. Here, we discuss recent examples where regulation takes place at each of the three phases of translation, which will serve as useful frameworks as new modes of translational control are uncovered.
Initiation
Translational control can be general, affecting all or most mRNAs, or specific, affecting only a limited number of transcripts. A well-described mechanism that can be both (mostly) general and mRNA-specific affects initiation, particularly at the eIF4E-eIF4G interface (Figure 2A). The now-classical regulator of many mRNAs is the 4E-BP family of proteins, which bind and sequester eIF4E from eIF4G, thereby shutting down most cap-dependent translation (Figure 2B) [5,19,22,23]. In contrast, Maskin, Neuroguidin, Cup, and CYFIP1 are similar to 4E-BP in that they bind eIF4E and prevent its interaction with eIF4G; however, these proteins are also tethered to sequence-specific RNA binding proteins (Figure 2C) [24-27]. They therefore inhibit the translation of only those mRNAs that interact with the RNA binding protein CPEB, in the case of Maskin and neuroguidin, Bruno, in the case of Cup, and the Fragile X mental retardation protein (FMRP) and a small noncoding RNA (BC1) in the case of CYFIP1. Only the binding of Maskin to eIF4E has been shown to be reversible, which is mediated by both Maskin phosphorylation and poly (A) tail lengthening by CPEB and associated factors [28-32]. However, it should be noted that at least partial CYFIP1-mediated translational repression can alleviated by synaptic activity in neurons [27].
Figure 2.
Factors that disrupt eIF4F binding to mRNAs. A. Schematic representation of cellular mRNA and the recruitment of eIF4F (eIF4A, eIF4E, and eIF4G), resulting in translation initiation. PABP refers to poly(A) binding protein. B. Binding of 4E-BP to eIF4E prevents translation in a non-sequence dependent manner. C. CYFIP1, Cup, or Maskin interact with the region of eIF4E that interacts with eIF4G; these proteins also associate with FMRP, Bruno, or CPEB, respectively, which are bound to sequence elements within 3’ UTRs of specific mRNAs leading to translational repression. D. In Drosophila, the Bicoid Binding Regions (BBRs) within the 3’ UTR of caudal mRNA interact with Bicoid protein. Bicoid interacts with 4E-HP, which directly binds the cap, thereby preventing recruitment of the eIF4F complex. E. 4E-HP can also repress hb mRNA translation in Drosophila by binding to a protein complex consisting of Nanos, Brat and Pumillio.
Because the 4E-BPs, Maskin, Neuroguidin, Cup, CYFIP1, and eIF4G all probably bind the same region on eIF4E, it is easy to see how a competition for cap association of translation factors occurs. Most of the eIF4E binding proteins have the motif YXXXXLΦ (where Φ is any hydrophobic amino acid, often a leucine), although Maskin has a T in place of the Y, which is the portion that contacts eIF4E [22]. CYFIP1, however, does not contain this motif, but instead has a short peptide that assumes a “reverse L-shaped” tertiary structure that resembles that of the motif noted above [27, 33]. This finding suggests that there may be additional “noncanonical” eIF4E binding proteins that control translation that have yet to be discovered.
In contrast to the above examples of proteins that disrupt initiation through eIF4E binding, Cho et. al. have characterized a protein that directly binds the 5’ cap of an mRNA, thereby preventing the recruitment of the entire eIF4F complex (Figure 2D) [3]. eIF4E-homologous protein (4E-HP) was identified initially as a homolog of eIF4E; however, because 4E-HP does not interact with eIF4G, its role in translational control was unclear [34]. Cho et al. found that 4E-HP interacted with Bicoid, a Drosophila protein that regulates both transcription and translation [35, 36]. Bicoid was thought to inhibit translation of caudal mRNA, which encodes a protein that regulates body patterning, through binding not only to a specific sequence in the caudal 3’ untranslated region (UTR), but also to eIF4E as well [37]. Thus, Bicoid seemed to have an activity that was a composite of, for example, CPEB and Maskin, Cup and Bruno, and CYFIP1 and FMRP. However, Cho et al. found that Bicoid co-immunoprecipitated with 4EHP, and mutations in Bicoid that abrogated this interaction also resulted in aberrant body patterning [3]. Moreover, mutations in 4E-HP that disrupted its interaction with the cap in vitro also led to Bicoid mutant-like phenotypes in vivo (i.e. inappropriate expression of Caudal in the anterior portion of the Drosophila embryo; Caudal is properly expressed only in the posterior region). Thus, it seems very likely that a Bicoid-4E-HP-cap interaction controls caudal mRNA translation and, as a result, body patterning.
4E-HP also controls the expression of hunchback (hb) mRNA, which like caudal mRNA, is involved in body patterning in Drosophila embryos [4]. Wild type embryos exhibit a gradient of Hb in the embryo; it is high in the anterior and lower in the posterior [38]. Expression of a mutant 4E-HP in embryos elicits a high level of posterior Hb; this molecular phenotype was rescued when wild type 4E-HP was expressed in the mutant 4E-HP-containing embryos, and returned to normal the Hb anterior to posterior gradient. Interestingly, mutations in 4E-HP that abrogated its cap binding activity but that did not affect its interaction with Bicoid also caused increased posterior expression of Hb. This observation suggested that 4E-HP might exert its activity through another protein that bound Hb, but not caudal, mRNA. Indeed, three proteins control Hb expression: Pumilio, Nanos, and Brat [4]. Both Pumilio and Nanos bind RNA while Brat associates with them through direct protein-protein interactions [39, 40]. In vitro experiments with recombinant proteins showed that 4E-HP binds Brat, suggesting that 4E-HP is part of (at least) a quartet of factors that controls Hb mRNA translation (Figure 2E). Thus, through binding alternative 3’ UTR bound protein partners, 4E-HP controls translation of different specific messages by interfering with eIF4F recruitment.
One form of translational control that occurs at initiation but that does not involve the cap or cap-binding factors takes place at the level of 40S-60S joining at the AUG codon. First described for lipoxygenase mRNA that is bound by hnRNP K and hnRNP E1 at its 3’ UTR and subsequently for actin mRNA that is bound by zip code binding protein (ZBP) also at its 3’ UTR, these proteins inhibit translation by preventing the 60S subunit from binding the 40S subunit that is stalled at the AUG codon [41, 42]. While the precise mechanism by which these 3’ UTR binding proteins control 40S-60S joining is unclear, they might resemble the activity of eIF6, an initiation factor purported to control this process [43, 44]. However, this activity of eIF6, which also controls 60S subunit biogenesis, is somewhat controversial [45, 46].
Elongation
There are few clear cases of regulated elongation; it is sometimes inferred from observations that the phosphorylation of elongation factors can affect ribosome transit along an mRNA. It has long been know that protein synthesis decreases as cells enter mitosis, which is at least partly due to eIF4E-4E-BP association [23] However, mitotic cells contain large polysomes that are less translationally active than lighter polysomes. It appears that cells reduce their elongation rates as they prepare to divide, which may allow for rapid protein synthesis once they again enter the G1 phase of the cell cycle [47]. This reduction in elongation rates is likely mediated by phosphorylation of eEF2 by eEF2 kinase, resulting in reduced translation rates [47].
Another instance where changes in elongation rates are inferred occurs in the brain, where repeated stimulation of synapses causes these structures to undergo biochemical and morphological changes [48, 49]. This phenomenon, known as synaptic plasticity probably forms the cellular basis of learning and memory [48]. Some forms of synaptic plasticity require protein synthesis, perhaps at or near synapses (i.e, the synapto-dendritic compartment) [2, 50]. It would therefore seem likely that regulated translation, in response to synaptic stimulation, would underlie learning and memory. While several types of translational control have been reported in neurons, Sutton et al. have found that elongation factor 2 (eEF2) acts as a sensor of synaptic events that results in local repression of translation [1, 51]. Basing their studies on the fact that two different types of synaptic transmission, action potential (AP) and AP-independent miniatures (minis), result in opposite effects on dendritic translation, they set out to identify the cellular translation components that were locally altered under each type of transmission event. By applying a drug inhibitor of AP, they found an increase in eEF2 phosphorylation; blocking of both AP and minis decreased eEF2 phosphorylation [1]. Because phosphorylated eEF2 has a reduced ability to catalyze ribosome translocation, Sutton et al. proposed that changes in elongation were characteristic to each type of stimulation [20, 21]. If eEF2 is a sensor of mini transmissions, then phosphorylation of eEF2 should increase if mini transmission is increased, which indeed was the case [1].
In yeast, the unfolded protein response (UPR) triggered by endoplasmic reticulum (ER) stress results in an increase in the Hac1 transcription factor. Under normal physiological conditions Hac1 mRNA contains stalled ribosomes, which occur when sequences in an unspliced intron basepair with complementary sequences in its 5’ UTR [52]. Alleviation of the base pairing and the stalled ribosomes occurs when the mRNA is fully spliced [52, 53]. Hac1 mRNA, via a specific 3’ UTR element, is recruited to discrete regions of the ER membrane by the ER stress sensor IRE1, a transmembrane kinase/endoribonuclease that splices Hac1 mRNA [54]. When the recruitment element is mutated, Hac1 mRNA is not translated because intron removal does not occur, and ribosomes remained stalled. Efficient splicing of the intron occurs only when the Hac1 mRNA is also translationally repressed. In agreement with these results, heterologous mRNAs containing the Hac1 3’ UTR targeting element are efficiently recruited to IRE1 foci only if they are also translationally repressed [54]. Thus, translational repression both regulates the synthesis of Hac1 protein to induce the UPR and plays a role in the cellular localization, targeting, and processing of Hac1 mRNA.
Termination
Studies from the past 10 years have shown that cells have evolved different pathways to discriminate among mRNAs that have aberrant translation stop signals. One example is mRNAs that lack in-frame stop codons (nonSTOP mRNAs), which can arise from transcriptional pausing, mutations that cause 3’ end formation in coding regions, or from the use of cryptic polyadenylation sites [55-57]. Distinct mechanisms appear to have evolved to deal with nonSTOP mRNAs in different organisms. In eubacteria, translation of nonSTOP mRNAs causes ribosome stalling at the 3’ end of the mRNA. This stalling is relieved by a transfer mRNA (tmRNA), which serves as both a tRNA and mRNA, binding to stalled ribosomes allowing translation to continue using the tmRNA open reading frame [58]. This results in the addition of a short peptide tag that targets the nascent polypeptide for destruction. The stop codon encoded by the tmRNA induces translation termination [59].
Akimitsu et al. have characterized how mammalian cells deal with nonSTOP mRNAs [60]. Using reporter constructs, they found that while protein production from nonSTOP mRNAs was reduced, a corresponding reduction in the level of mRNA or an increased rate of protein destruction was not observed. Because nonSTOP mRNA was associated with polysomes, translation inhibition occurred at a post-initiation step. Experiments measuring 14C-leucine incorporation showed polypeptides of heterogeneous length were produced from nonSTOP mRNA, suggesting premature ribosome termination. In addition, RNase protection assays showed ribosomes stalled at the poly(A) tail at the 3’ end of the mRNA. The authors propose a model whereby stalling of ribosomes at the poly(A) tail leads to increased premature termination of upstream ribosomes, resulting in translational repression of the nonSTOP mRNA.
Generation of 3’UTR isoforms
Although not technically a form of translational control, recent studies have shown that a large diversity in mRNA 3’ UTRs are generated by alternative poly(A) site (i.e., AAUAAA) selection [61-63]. Because many mRNAs are regulated through 3’ UTR elements, it is likely factors such as NOVA, which regulate poly(A) site selection may have a major influence on mRNA translation, stability, and localization [62]. Indeed, consider the studies of Sandberg et al., who showed that in resting or activated T lymphocytes, proliferating cells contained shorter mRNA 3’ UTRs than resting cells [61]. The shorter 3’ UTRs were expressed by preferential usage of upstream polyadenylation sites following T lymphocyte activation. In particular, Sandberg et al. showed that two isoforms of Hip2 mRNA (encoding a ubiquitin conjugating enzyme) arose from alternative polyadenylation; a short form and a longer form that contains miRNA binding sites. While the overall expression of Hip2 RNA was similar in both naïve and activated T lymphocytes, the ratio of the long to short isoform was decreased upon activation, which correlated with an increase in Hip2 protein levels. It appears that the long but not the short form is bound by miRNAs, thereby demonstrating how alternative poly(A) site selection can lean lead to translational control.
Conclusions
A search of PubMed for “translational control” results in over 50,000 hits, ample evidence that this field is large and growing. miRNAs have made surprisingly few contributions to this total, perhaps due to the fact that despite intensive investigation, that field remains in a state of flux. Nonetheless, mechanisms of translational control cannot be divorced from resulting biological consequences. That is, regulated translation gone awry can lead to cancer, infertility, and brain neuropathies. The more we understand the underlying biochemical control of the translation process, the greater will be our repertoire as we seek new therapies to assault human disease.
Acknowledgements
Work in the authors’ laboratory is supported by grants from the National Institutes of Health. RG is supported by an Institutional postdoctoral training grant (5T32HD007312-25).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sutton MA, Taylor AM, Ito HT, Pham A, Schuman EM. Postsynaptic decoding of neural activity: eEF2 as a biochemical sensor coupling miniature synaptic transmission to local protein synthesis. Neuron. 2007;55:648–661. doi: 10.1016/j.neuron.2007.07.030. [This paper shows that the phosphorylation state of eEF2 changes depending on the type of synaptic transmission (that is action potential-dependent or independent (miniature synaptic transmissions). Moreover, there was a tight correlation between the amount of eEF2 phosphorylation and translation of a reporter mRNA.] [DOI] [PubMed] [Google Scholar]
- 2.Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- 3.Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–423. doi: 10.1016/j.cell.2005.02.024. [Together with Ref [4], this paper illustrates translation inhibition of a specific mRNA in Drosophila by 4E-HP simultaneously binding both the cap of the mRNA and protein bound to sequences in the mRNA 3' UTR.] [DOI] [PubMed] [Google Scholar]
- 4.Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [See annotation to Ref. [3].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 6.Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 7.Mathews MB, Sonenberg N, Hershey JWB. Origins and Principles of Translational Control. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 1–40. [Google Scholar]
- 8.Pestova TV, Lorsch JR, Hellen CU. The Mechanism of Translation Initiation in Eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 9.Majumdar R, Bandyopadhyay A, Maitra U. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40 S preinitiation complex. J Biol Chem. 2003;278:6580–6587. doi: 10.1074/jbc.M210357200. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhuri J, Chowdhury D, Maitra U. Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40 S ribosomal preinitiation complex. J Biol Chem. 1999;274:17975–17980. doi: 10.1074/jbc.274.25.17975. [DOI] [PubMed] [Google Scholar]
- 11.Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 12.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation Initiation on Mammalian mRNAs with Structured 5′UTRs Requires DExHBox Protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [This paper shows the protein DHX29, a putative DExH helicase, is part of the preinitiation complex, which is necessary for 48S complex scanning of mRNAs with highly structured 5′ UTRs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 14.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 15.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor DJ, Frank J, Kinzy TG. Structure and Function of the Eukaryotic Ribosome and Elongation Factors. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 59–86. [Google Scholar]
- 17.Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, Thompson SR, Pestova TV, Bedwell DM. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrenberg M, Hauryliuk V, Crist CG, Nakamura Y. Translation Termination, the Prion [PSI+], and Ribosomal Recycling. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 173–196. [Google Scholar]
- 19.Raught B, Gingras AC. Signaling to Translation Initiation. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 369–400. [Google Scholar]
- 20.Redpath NT, Price NT, Severinov KV, Proud CG. Regulation of elongation factor-2 by multisite phosphorylation. Eur J Biochem. 1993;213:689–699. doi: 10.1111/j.1432-1033.1993.tb17809.x. [DOI] [PubMed] [Google Scholar]
- 21.Ryazanov AG, Shestakova EA, Natapov PG. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature. 1988;334:170–173. doi: 10.1038/334170a0. [DOI] [PubMed] [Google Scholar]
- 22.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 23.Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- 25.Jung MY, Lorenz L, Richter JD. Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol Cell Biol. 2006;26:4277–4287. doi: 10.1128/MCB.02470-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 27.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [This paper describes a new 4E-BP, CYFIP1, which interacts with both eIF4E and 3′ UTR bound FMRP to repress translation of FMRP bound mRNAs. It also suggests how CYFIP1 can bind eIF4E even though it lacks the canonical YXXXXLΦ motif.] [DOI] [PubMed] [Google Scholar]
- 28.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Cao Q, Kim JH, Richter JD. CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat Struct Mol Biol. 2006;13:1128–1134. doi: 10.1038/nsmb1169. [DOI] [PubMed] [Google Scholar]
- 30.Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 32.Barnard DC, Cao Q, Richter JD. Differential phosphorylation controls Maskin association with eukaryotic translation initiation factor 4E and localization on the mitotic apparatus. Mol Cell Biol. 2005;25:7605–7615. doi: 10.1128/MCB.25.17.7605-7615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 34.Rom E, Kim HC, Gingras AC, Marcotrigiano J, Favre D, Olsen H, Burley SK, Sonenberg N. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J Biol Chem. 1998;273:13104–13109. doi: 10.1074/jbc.273.21.13104. [DOI] [PubMed] [Google Scholar]
- 35.Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 36.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring WJ, Jackle H. RNA binding and translational suppression by bicoid. Nature. 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 37.Niessing D, Blanke S, Jackle H. Bicoid associates with the 5′-cap-bound complex of caudal mRNA and represses translation. Genes Dev. 2002;16:2576–2582. doi: 10.1101/gad.240002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- 39.Edwards TA, Wilkinson BD, Wharton RP, Aggarwal AK. Model of the brain tumor-Pumilio translation repressor complex. Genes Dev. 2003;17:2508–2513. doi: 10.1101/gad.1119403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [This paper was the first to describe the inhibition of 60S subunit joining to the 40S by proteins bound to the 3′ UTR of an mRNA.] [DOI] [PubMed] [Google Scholar]
- 42.Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 43.Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, Biffo S: Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 44.Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature. 2008;455:684–688. doi: 10.1038/nature07267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basu U, Si K, Warner JR, Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol. 2001;21:1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 47.Sivan G, Kedersha N, Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol. 2007;27:6639–6646. doi: 10.1128/MCB.00798-07. [This paper reports that during mitosis, translation elongation rates are reduced, likely through phosphorylation of eEF2.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collingridge GL, Isaac JT, Wang YT. Receptor trafficking and synaptic plasticity. Nat Rev Neurosci. 2004;5:952–962. doi: 10.1038/nrn1556. [DOI] [PubMed] [Google Scholar]
- 49.Malenka RC, Nicoll RA. Long-term potentiation--a decade of progress? Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- 50.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 51.Richter JD, Klann E. Making Synaptic Plasticity and Memory Last: Mechanisms of Translational Regulation. Genes and Development. 2008 doi: 10.1101/gad.1735809. In press. [DOI] [PubMed] [Google Scholar]
- 52.Ruegsegger U, Leber JH, Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- 53.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 54.Aragon T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2008 doi: 10.1038/nature07641. [This paper shows that translational repression and a 3′ UTR targeting element within Hac1 mRNA, a key mediator of the unfolded protein response, is critical for correct splicing and expression.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui Y, Denis CL. In vivo evidence that defects in the transcriptional elongation factors RPB2, TFIIS, and SPT5 enhance upstream poly(A) site utilization. Mol Cell Biol. 2003;23:7887–7901. doi: 10.1128/MCB.23.21.7887-7901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sparks KA, Dieckmann CL. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 1998;26:4676–4687. doi: 10.1093/nar/26.20.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Temperley RJ, Seneca SH, Tonska K, Bartnik E, Bindoff LA, Lightowlers RN, Chrzanowska-Lightowlers ZM. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Hum Mol Genet. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- 58.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 59.Haebel PW, Gutmann S, Ban N. Dial tm for rescue: tmRNA engages ribosomes stalled on defective mRNAs. Curr Opin Struct Biol. 2004;14:58–65. doi: 10.1016/j.sbi.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 60.Akimitsu N, Tanaka J, Pelletier J. Translation of nonSTOP mRNA is repressed post-initiation in mammalian cells. Embo J. 2007;26:2327–2338. doi: 10.1038/sj.emboj.7601679. [This paper investigates the mammalian response to aberrant nonSTOP mRNAs and suggests premature ribosome fall off represses translation of these mRNAs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [This paper shows that proliferating and resting cells express short and long forms, respectively, of many mRNAs depending on poly(A) site selection.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [This paper uses a technique to map protein-RNA binding sites in vivo on a genome-wide scale to show that Nova regulates alternative splicing and alternative polyadenylation in the brain.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [This paper uses deep sequencing and bioinformatic analysis to show that mRNA isoforms are more diverse than previously thought, even between cell lines and related tissues.] [DOI] [PMC free article] [PubMed] [Google Scholar]