Abstract
The strictly anaerobic Gram-negative bacteria Butyricimonas species have recently been described in human faeces and have to our knowledge not been isolated in infectious clinical materials. We report the first case of Butyricimonas virosa bacteraemia in a 72-year-old man with colon adenocarcinoma, who underwent aortic aneurysm replacement surgery.
Keywords: Anaerobes, bacteraemia, Butyricimonas, gut microbiota, Porphyromonadaceae
Case report
Anaerobic bacteraemia, accounting for 1–17% of all bacteraemia, mostly originates from endogenous microbiota, especially in patients with malignancies (particularly colon cancer), haematological disorders, immunodeficiency, recent gastrointestinal or gynaecological surgery and advanced age. The mortality rate of those infections remains high (15–35%), so early diagnosis and appropriate treatment of these infections are critical [1,2].
We described a case of Butyricimonas virosa bacteraemia in a 72-year-old man. The patient with a previously diagnosed colonic adenocarcinoma underwent aortic aneurysm surgery and suffered fever (37.8 °C) 24 days after surgery. Blood and urine were collected for culture, then, meropenem plus colistin was administered empirically. The patient was transferred to the intensive care unit because of septic shock on the following day.
Gram-negative rods isolated from anaerobic blood culture bottles after a 72-h incubation period were obligate anaerobes, catalase-producing, inhibited on Bacteroides bile aesculin agar, and resistant to vancomycin (5 μg), kanamycin (1 mg) and colistin sulphate (10 μg), suggesting that the isolate was a Prevotella species. On Rapid ID 32A (bioMérieux, Marcy l’Etoile, France), the isolate was positive for β-galactosidase, N-acetyl-β-glucosaminidase, glutamic acid decarboxylase, indole, alkaline phosphatase, leucyl glycine, alanine, glutamyl glutamic acid, arylamidases and pyroglutamic acid arylamidase. However; both Rapid ID 32A and matrix-assisted laser desorption/ionization time-of-flight (VITEK MS; bioMérieux) were insufficient for identification. The 16S rRNA gene sequence of the strain showed 99% nucleotide identity to that of the strain Butyricimonas virosa isolated from rat faeces (GenBank accession no. NR_041691.1). Investigation of the next most closely related species showed that our strain exhibited 97%, 96% and 94% sequence similarity with Butyricimonas paravirosa, Butyricimonas faecihominis and Butyricimonas synergistica, respectively [3,4], (Fig. 1).
Fig. 1.
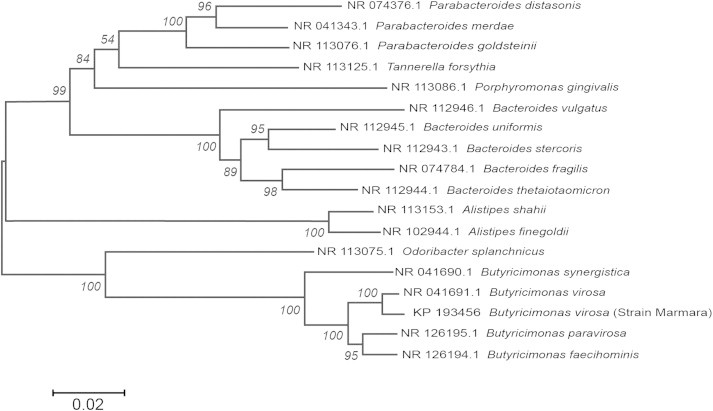
Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences, showing the relationships between STRAIN MARMARA and some related taxa. GenBank accession numbers are shown in parentheses. Numbers at nodes indicate bootstrap percentages (based on 1000 replicates). Bar represents 0.02 substitutions per nucleotide position.
The bacterium did not produce β-lactamase when tested with a nitrocefin disc (bioMérieux). Antibiotic susceptibilities were determined by E-test (bioMérieux) on Brucella agar (Oxoid, Basingstoke, UK) supplemented with 5% defibrinated sheep blood. The results showed sensitivity to ampicillin, sulbactam-ampicillin, amoxicillin-clavulanic acid, piperacillin-tazobactam, imipenem, meropenem, clindamycin and metronidazole.
Sakamoto et al. [3] first characterized the species B. virosa, a butyric acid-producing bacterium in the family Porphyromonadaceae isolated from rat faeces. Having also 99% 16S rRNA gene sequence similarity to uncultured bacterial clones obtained from human faeces suggested that the isolate was part of the patient's intestinal microbiota and his underlying bowel malignancy had created a predisposition for bacteraemia [5,6].
This is the first case of human infection due to B. virosa (GenBank accession number: BankIt1777481 Seq KP193456) in the world literature. However, it may not reflect the actual result because phenotypic characteristics resembling Prevotella sp. may lead to misidentification and identification systems have an insufficient database for this microorganism.
The patient's fever disappeared 3 days after administration of antibiotic therapy. His clinical condition improved relatively and he was followed up in the intensive care unit; however, he died 4 weeks later as the result of Acinetobacter septicaemia and a potentially fatal condition resulting from multi-organ failure.
In conclusion, to discover the actual prevalence of B. virosa infection, routine laboratories should re-evaluate the isolates that are identified as Prevotella species.
Conflict of interest
None declared.
References
- 1.Brook I. The role of anaerobic bacteria in bacteremia. Anaerobe. 2010;16:183–189. doi: 10.1016/j.anaerobe.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Ani C., Farshidpanah S., Bellinghausen Stewart A., Nguyen H.B. Variations in organism-specific severe sepsis mortality in the United States: 1999–2008. Crit Care Med. 2014:16. doi: 10.1097/CCM.0000000000000555. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Sakamoto M., Takagaki A., Matsumoto K., Kato Y., Goto K., Benno Y. Butyricimonas synergistica gen. nov., sp. nov. and Butyricimonas virosa sp. nov., butyric acid-producing bacteria in the family 'Porphyromonadaceae' isolated from rat faeces. Int J Syst Evol Microbiol. 2009;59:1748–1753. doi: 10.1099/ijs.0.007674-0. [DOI] [PubMed] [Google Scholar]
- 4.Sakamoto M., Tanaka Y., Benno Y., Ohkuma M. Butyricimonas faecihominis sp. nov. and Butyricimonas paravirosa sp. nov., isolated from human faeces, and emended description of the genus Butyricimonas. Int J Syst Evol Microbiol. 2014;64:2992–2997. doi: 10.1099/ijs.0.065318-0. [DOI] [PubMed] [Google Scholar]
- 5.Li M., Wang B., Zhang M., Rantalainen M., Wang S., Zhou H. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker A.W., Sanderson J.D., Churcher C., Parkes G.C., Hudspith B.N., Rayment N. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]


