Abstract
Background
The renin-angiotensin-aldosterone-system (RAAS) is critical for regulation of blood pressure and fluid balance and influences cardiovascular remodeling. Dysregulation of the RAAS contributes to cardiovascular and renal morbidity. The genetic architecture of circulating RAAS components is incompletely understood.
Methods and Results
We meta-analyzed genome-wide association data for plasma renin activity (n=5,275), plasma renin concentrations (n=8,014) and circulating aldosterone (n=13,289) from up to four population-based cohorts of European and European-American ancestry, and assessed replication of the top results in an independent sample (n=6,487).
Single nucleotide polymorphisms (SNPs) in two independent loci displayed associations with plasma renin activity atgenome-wide significance (p<5×10-8). A third locus was close to this threshold (rs4253311 in kallikrein B [KLKB1], p=5.5×10-8). Two of these loci replicated in an independent sample for both plasma renin and aldosterone concentrations (SNP rs5030062 in kininogen 1 [KNG1]: p=0.001 for plasma renin, p=0.024 for plasma aldosterone concentration; rs4253311 with p<0.001 for both plasma renin and aldosterone concentration). SNPs in the NEBL gene reached genome-wide significance for plasma renin concentration in the discovery sample (top SNP rs3915911, p= 8.81×10-9), but did not replicate (p=0.81). No locus reached genome-wide significance for aldosterone. SNPs rs5030062 and rs4253311 were not related to blood pressure or renal traits; in a companion study, variants in the kallikrein B locus were associated with B-type natriuretic peptide concentrations in African-Americans.
Conclusions
We identified two genetic loci (kininogen 1 and kallikrein B) influencing key components of the RAAS, consistent with the close interrelation between the kallikrein-kinin system and the RAAS.
Keywords: renin angiotensin system, aldosterone, Genome Wide Association Study
Introduction
The renin-angiotensin-aldosterone system (RAAS) is a central pathway in cardiovascular and renal physiology. Through a series of enzymatic reactions, the liver-derived protein angiotensinogen is transformed into angiotensin I (this conversion is catalyzed by renin) and subsequently into angiotensin II, which is the key effector of the RAAS, mediating multiple biological effects in the kidneys, the heart and the vasculature through its local and systemic effects.1-4 Angiotensin II is a potent vasoconstrictor and adversely impacts cardiac and vascular remodeling. 1-3 On a parallel note, angiotensin II stimulates the production of aldosterone in the adrenal glands, which enhances sodium and water reabsorption in the kidneys.1-2 Thus, the RAAS is a major determinant of fluid and electrolyte hemostasis, blood pressure (BP) regulation and cardiovascular remodeling.1, 3 Consequently, RAAS activity influences the development and progression of cardiovascular disease,1-2 and pharmacological inhibition of the RAAS has been shown to improve patient outcomes in a variety of clinical settings.2 Moreover, recent studies suggested a cross talk between adipose tissue and the adrenal gland, and high aldosterone concentrations were reported to be associated with the metabolic syndrome and type 2 diabetes.5
Despite its clinical significance, the genetic architecture of the RAAS, as evident in circulating levels of its components, is incompletely understood. Prior studies have established different circulating components of the RAAS as heritable traits.6-7 To improve our understanding of the genetic determinants of the RAAS, we conducted a genome-wide association analysis of plasma renin (investigated by either its activity or concentration) and circulating aldosterone concentrations in up to four population-based cohorts with replication in a fifth independent cohort. Given the reported heritability of RAAS components,6-7 we hypothesized that these circulating RAAS biomarkers will be associated with common genetic variation in the general population.
Methods
Please find more detailed information regarding sample description and biomarker measurements in the supplementary material.
Study Samples
Framingham Heart Study (FHS) samples
RAAS biomarkers were measured at examination cycle 6 (1995–1998) of the Framingham Offspring cohort (Generation 2, Gen 2)8 and at the first examination cycle (2002-2005) of the Third Generation cohort (Generation 3, Gen 3).9 At each Heart Study visit, participants were comprehensively characterized with respect to cardiovascular risk factors and subclinical disease measures. All participants provided written informed consent, and the study protocol was approved by the institutional review board at the Boston University Medical Center.
Cooperative Health Research in the Region of Augsburg (KORA) sample
KORA comprises several population-based cohort studies in the region of Augsburg, Southern Germany.10 The present analysis includes data from the follow-up examination KORA F4 (2006-2008) of the KORA S4 survey (1999/2000).10 Participants with missing genotype or phenotype data were excluded from the present analyses, as were participants reporting intake of diuretics or participants with a renin or aldosterone concentration of more than 1000 ng/L. The final study sample comprised 1,786 participants. The studies were approved by the ethics committee of the Bavarian Medical Association (Bayerische Landesärztekammer) and all study participants gave written informed consent.
Study of Health in Pomerania (SHIP) sample
SHIP is a population-based cohort study in West Pomerania, the north-east area of Germany.11
The first follow-up examination (SHIP-1) was conducted from March 2003 to July 2006.11 Participants were comprehensively phenotyped with respect to cardiometabolic traits and subclinical disease burden, as detailed elsewhere.11 All participants gave written informed consent. The study protocol was approved by the Ethics Committee of the University of Greifswald.
Supplémentation en Vitamines et Minéraux Antioxydants (SUVIMAX) study sample
Participants of the SUVIMAX study were healthy volunteers free of hypertension, cardiovascular disease or cancer at baseline.12 The recruitment of the SUVIMAX cohort was performed in metropolitan France and all individuals were of European descent. RAAS biomarkers and genetic information were available in a subsample of 1,518 participants.
Prevention of REnal and Vascular ENd-stage Disease (PREVEND) sample
PREVEND is a general population sample from the city of Groningen (the Netherlands). This longitudinal study was primarily designed to assess the association between urinary albumin excretion and cardiovascular and renal disease.13 The study design has been described in detail elsewhere.14 Basic vascular risk factors were determined on two occasions in the examination center.14 The sample for the current analysis comprised 6,487 participants.
RAAS measurements
Measurements of renin and aldosterone
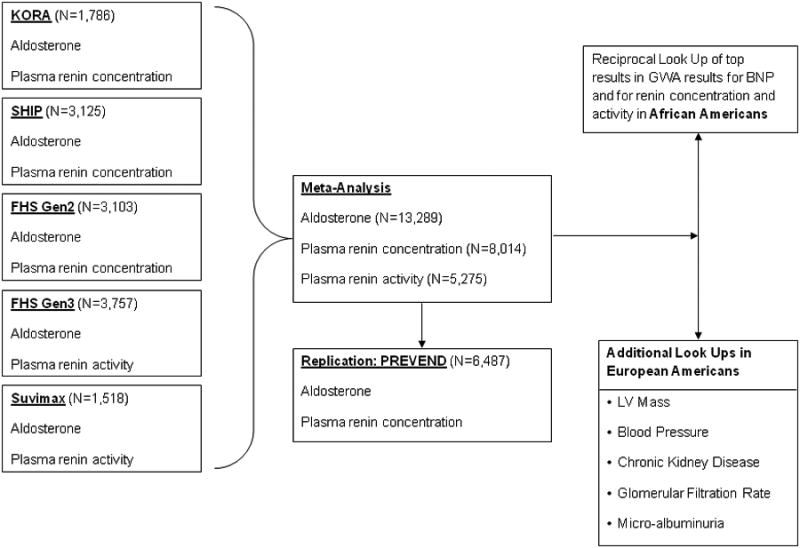
In FHS, KORA and SUVIMAX, blood samples were taken in the early morning from seated participants after an overnight fast. In PREVEND, fasting blood samples were taken between 8 AM and 4 PM. In SHIP, blood was drawn throughout the day (between 8 AM and 8 PM) in non-fasting participants while they were taking their regular medication. Aldosterone measurements were available in all cohorts (FHS Gen 2 and Gen 3, KORA, SHIP, SUVIMAX, PREVEND; Figure 1); plasma renin concentrations were measured in the FHS Gen 2, KORA, SHIP and PREVEND samples; plasma renin activity was determined in the FHS Gen 3 and SUVIMAX samples (Figure 1). Details on the methods to determine RAAS biomarkers in the different samples are provided in the supplementary material.
Figure 1. Basic study design.
Genotyping
All cohorts were genotyped using genome-wide arrays from Affymetrix or Illumina. Details on the specific arrays used and the quality control measures applied to genetic data are detailed in the supplementary material.
Statistical analysis
Meta-analysis of genome-wide association data and replication
The basic study design is displayed in Figure 1. The biomarkers were natural logarithmically transformed prior to analyses. Within each cohort, the association between approximately 2.5 million SNPs (exposure) and biomarkers of the RAAS were assessed using linear regression models adjusted for age and sex, assuming an additive genetic model. Aldosterone levels, plasma renin concentration, and plasma renin activity served as dependent variables (each biomarker considered separately). In the FHS sample, a linear mixed effects model was used to adjust for familial correlation.15 For aldosterone levels, genome-wide association results from Gen2 and Gen3 cohorts were combined by using Obrien's method,16 because aldosterone measurements were correlated between Gen2 and Gen3. The GWAS data from up to four cohorts (FHS, KORA, SHIP, SUVIMAX) were meta-analyzed with a sample size-weighted approach (combining sample size weighted Z statistics from each cohort) using the METAL software.17
Corrections for genomic controls were performed within each cohort, and after the meta-analyses. To graphically display the association results from the meta-analyses for each biomarker, we plotted the p-value of each SNP vs. its chromosomal position (Manhattan plot). Furthermore, we also plotted the observed vs. the expected p-value distribution under the null hypothesis of no association between biomarker and SNPs (quantile-quantile plots) for each biomarker. Genetic variants with a minor allele frequency >0.05, imputation quality ratio >0.75, and a p-value ≤5×10-8 for any of the three biomarkers were assessed for replication in the PREVEND sample. In PREVEND, only plasma renin concentration and plasma aldosterone levels were available. Thus, the top loci associated with each RAAS biomarker in the meta-analyses of FHS, KORA, SHIP, and SUVIMAX were tested for association with plasma renin concentrations and aldosterone levels in PREVEND using a linear regression model adjusting for age and sex (Figure 1). Furthermore, since plasma renin concentration and aldosterone levels were also available in the discovery samples (Figure 1), we performed a ‘look-up’ of the top renin activity SNPs with respect to renin and aldosterone concentrations in the individual discovery cohorts.
Finally, we performed fine mapping of the top regions associated with plasma renin activity (since these regions reached or were close to genome-wide significance) and of the candidate genes REN and CYP11B2 (+/- 500 kb around the gene) based on imputations to the 1000 genome dataset. These 1000 genome imputations were available in FHS, SHIP and KORA.
Look-up of top SNPs in African Americans
We performed a “look-up” of our top loci, associated with plasma renin activity or plasma renin concentrations in Europeans, to assess their associations with plasma renin activity, renin concentration and B-type natriuretic peptide (BNP) levels in African Americans in the Jackson Heart Study.18
Association of top SNPs with BP traits, renal traits and echocardiographic left ventricular mass in Europeans
In secondary analyses, we tested the successfully replicated SNPs from our GWAS (rs5030062 and rs4253311) for associations with traits known to be influenced by the RAAS, i. e., BP, left ventricular mass and renal traits. For BP, we related our top SNPs to systolic and diastolic BP in the International Consortium for Blood Pressure Genome-Wide Association Studies.19 We also assessed the association of these two SNPs with left ventricular mass in the EchoGen Consortium,20 and with renal traits in the CKDGen Consortium21-22 (Figure 1). In the latter consortium, we tested specifically the association of these SNPs with the estimated glomerular filtration rate (eGFR), the urinary albumin-to-creatinine ratio (UACR), and with chronic kidney disease (CKD, as binary trait).21-22
Pathway analyses
Pathway analyses provide a potential route to investigate the collective effects of multiple genetic variants on biological systems. For each genetic variant, we assigned an overall SNP score to indicate its association with RAAS-related traits. This SNP score was equivalent to the most significant p-value among the three RAAS traits examined (plasma renin concentration, plasma renin activity, circulating aldosterone levels). The genetic variants with the score assigned were then mapped back to the human reference genome (NCBI Build 36, 2006) and we examined their locations relative to RefSeq genes (Mar 17, 2013), and gene scores were obtained. The gene score was defined as the most significant variant that was located within 110kb upstream and 40kb downstream of the gene's most extreme transcript boundaries. We selected these boundaries because they are expected to encompass the majority of cis-eQTLs based on expression data.23 Similar boundaries have been used in earlier studies.24-25 Of the 23,696 genes evaluated, 548 reached a score less than 1.0 × 10-4. These genes were then imported into Ingenuity for pathway analyses (IPA; Ingenuity Systems, Redwood, CA). Fisher's exact test was used to justify the enrichment of each of the canonical pathways.
Results
The discovery sample size comprised 13,289 participants for circulating aldosterone concentrations, 8,014 participants for plasma renin concentrations and 5,275 participants for plasma renin activity. Baseline clinical and biochemical characteristics of the study sample are shown in Table 1.
Table 1. Baseline characteristics of the contributing cohorts.
| Characteristics | FHS: Gen 2 | FHS: Gen 3 | KORA | SHIP | SUVIMAX | PREVEND |
|---|---|---|---|---|---|---|
|
| ||||||
| (N=3,103) | (N=3,757) | (N=1,786) | (N=3,125) | (N=1,518) | (N=6,487) | |
| Age, years | 59 (10) | 40 (9) | 61 (9) | 54 (15) | 51 (6) | 50 (13) |
| Female (%) | 53.2% | 53.3% | 51.3% | 51.2% | 60.4% | 50.5% |
| SBP, mmHg | 128 (19) | 117 (14) | 125 (19) | 133 (20) | 121 (12) | 130 (21) |
| DBP, mmHg | 75 (9) | 75 (10) | 76 (10) | 82 (11) | 78 (8) | 74 (10) |
| Hypertension (%) | 30.4% | 16.5% | 45.8% | 50.7% | 18.5% | 34.7% |
| Diabetes (%) | 10.9% | 3.0% | 10.4% | 10.9% | 0.3% | 3.7% |
| Prevalent CVD (%) | 10.8% | 0.9% | 3.6% | 7.9% | 0.0% | 4.0% |
| eGFR, mL/min/1.73m2 | 91 (78) | 99 (18) | 82 (18) | 85 (21) | NA | 80 (14) |
| eGFR <60 mL/min/1.73m2 (%) | 8.4% | 0.5% | 7.4% | 9.8% | NA | 6.3% |
| Aldosterone (ng/L) | 115.9 (73.9) | 128.2 (73.3) | 45.7 (31.8) | 51.5 (38.2) | 132 (89) | 132.6 (67.7) |
| PRC, ng/L | 28.5 (113.6) | NA | 23.0 (50.3) | 16.5 (35.0) | NA | 24.8 (31.8) |
| PRA, ng (angiotensin)/mL/hr | NA | 2.38 (3.56) | NA | NA | 1.97 (1.30) | NA |
FHS, Framingham Heart Study; Gen 2, Offspring cohort; Gen 3, Third Generation cohort; KORA, Cooperative Health Research in the Region of Augsburg; SHIP, Study of Health in Pomerania; SUVIMAX, Supplémentation en Vitamines et Minéraux Antioxydants; PREVEND, Prevention of REnal and Vascular ENd-stage Disease; SBP, systolic blood pressure; DBP, diastolic blood pressure; CVD, cardiovascular disease; eGFR, estimated glomerular filtration rate; PRA, plasma renin activity; PRC, plasma renin concentration. Data are mean (SD) or percent. NA, not available
GWAS meta-analysis and replication for plasma renin activity, plasma renin concentrations and circulating aldosterone levels
We identified two independent loci that were associated with plasma renin activity at a genome-wide significant level (p<5 × 10-8). A third locus was very close to this threshold (tagged by rs4253311; p=5.5 × 10-8). The respective Manhattan plot and the quantile-quantile plot for plasma renin activity are shown in Figures 2 A and B. The quantile-quantile plot for plasma renin activity in the FHS Generation 3 sample only is provided in supplementary Figure 1. The top SNPs were rs12374220, an intronic variant in the TENM3 gene, rs5030062 in intron 6 of the kininogen 1 [KNG1] gene and rs4253311 in intron 11 of the kallikrein B[KLKB1] gene (Table 2). The two latter SNPs could successfully be replicated in an independent sample (PREVEND), i. e. they showed nominal statistically significant associations with plasma renin concentrations and circulating aldosterone levels in PREVEND (Table 2; plasma renin activity was not available in PREVEND). However, except for an association of rs5030062 with aldosterone levels in KORA, SNPs rs12374220, rs5030062 and rs4253311 provided no evidence for association with renin concentrations (supplementary Table 1) and aldosterone levels (supplementary Table 2) in the individual discovery cohorts.
Figure 2. Manhattan plot (Panel A) and quantile-quantile plot (Panel B) for the genome-wide analysis for plasma renin activity.
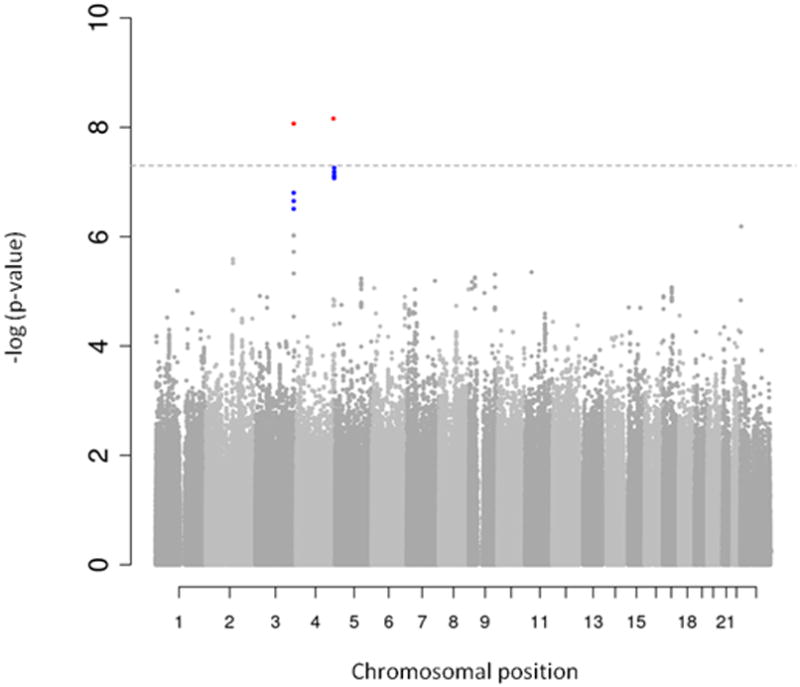
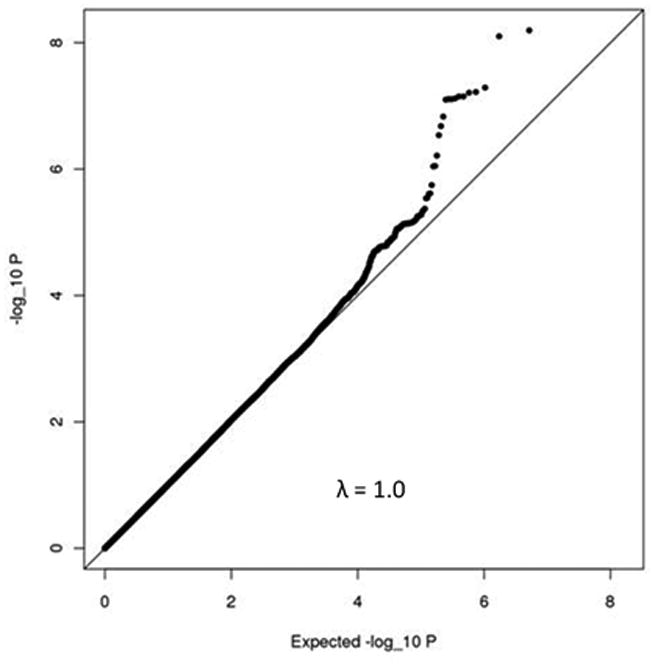
Table 2.
Lead SNPs of the top loci associated with plasma renin activity (PRA) in the discovery cohorts. Replication with respect to circulating aldosterone levels and plasma renin concentration (PRC) in the PREVEND sample.
| Meta-analysis of the discovery samples (FHS Gen 3+SUVIMAX) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| SNP | Position | Trait | Chr | MAF | Coded allele | Direction | P-value | Gene | ||
| rs12374220 | 183677641 | PRA | 4 | 0.0651 | T | -- | 6.92×10-9 | TENM3 | ||
| rs5030062 | 187936874 | PRA | 3 | 0.3889 | A | -- | 8.57×10-9 | KNG1 | ||
| rs4253311 | 187411677 | PRA | 4 | 0.4828 | A | -- | 5.51×10-8 | KLKB1 | ||
|
| ||||||||||
| Replication sample (PREVEND) | ||||||||||
|
| ||||||||||
| SNP | Trait | N | Coded allele | Beta | P | Trait | N | Coded allele | Beta | P |
|
| ||||||||||
| rs12374220 | Aldosterone | 5,894 | T | -0.026 | 0.126 | PRC | 6325 | T | 0.0046 | 0.886 |
| rs5030062 | Aldosterone | 5,865 | A | 0.017 | 0.024 | PRC | 6296 | A | -0.046 | 0.001 |
| rs4253311 | Aldosterone | 5,875 | A | 0.028 | <0.001 | PRC | 6308 | A | -0.065 | <0.001 |
SNP, single nucleotide polymorphism; FHS, Framingham Heart Study; Gen 3, Third Generation cohort; SUVIMAX, Supplémentation en Vitamines et Minéraux Antioxydants; PREVEND, Prevention of REnal and Vascular ENd-stage Disease; KNG1, kininogen 1; KLKB1, kallikrein B; Chr, chromosome; MAF, minor allele frequency; TENM3, teneurin transmembrane protein 3
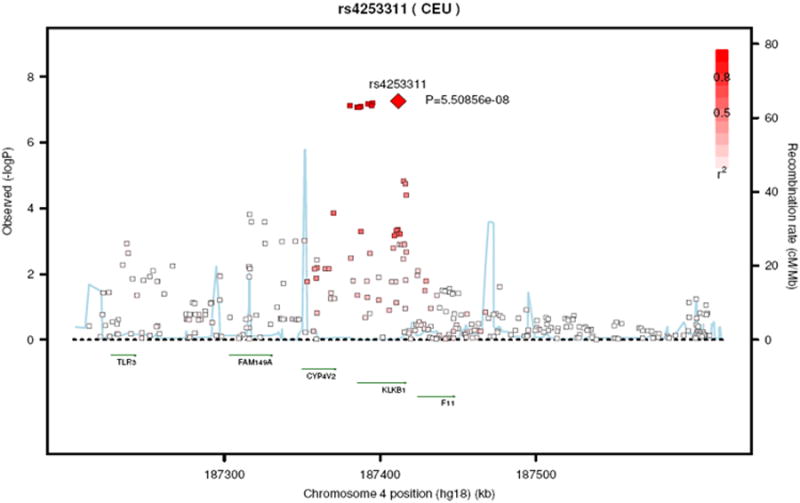
Regional plots of the kallikrein B locus and the kininogen 1 locus are shown in Figure 3A and Figure 3B, respectively. Analyzing these two regions using a dataset imputed to the 1000 genome data26 revealed essentially similar results. Regional plots based on the 1000 genome imputations are shown in supplementary Figure 2A and 2B.
Figure 3. Regional plot of rs4253311 in exon 11 of the kallikrein B gene (Panel A) and of rs5030062 in intron 6 of the kininogen 1 gene (Panel B).
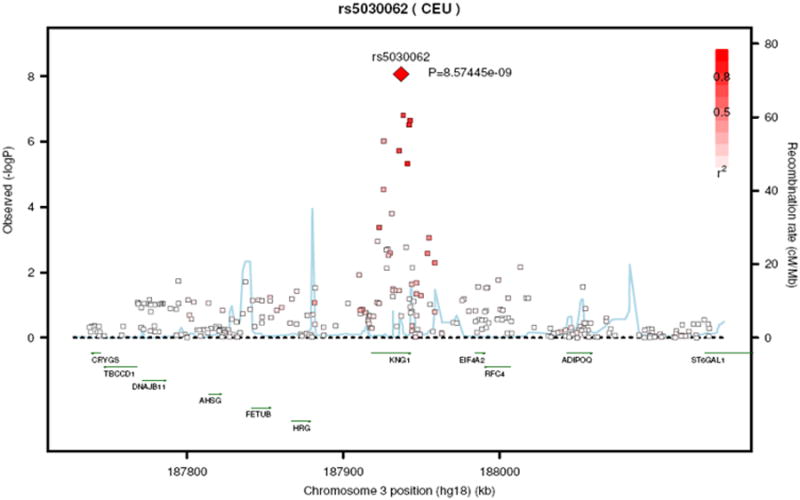
SNP rs5030062 and rs4253311 explained 0.85% and 0.87% of plasma renin activity variance, respectively. Plasma renin activity, stratified by rs5030062 genotype (Panel A) and stratified by rs4253311 genotype (Panel B), are displayed in supplementary Figure 3. Top loci associated with plasma renin activity stratified by sample, e. g., in the FHS Generation 3 sample and in the SUVIMAX sample, are provided in supplementary Table 3. The association results for all three SNPs were consistent in directionality in FHS and SUVIMAX but reached a higher level of statistical significance in FHS than in SUVIMAX, possibly due to the much larger sample size in FHS.
The Manhattan plot (Panel A) and the quantile-quantile plot (Panel B) for the genome-wide analysis for plasma renin concentration are shown in supplementary Figure 4. SNPs in the NEBL gene reached genome-wide significance for plasma renin concentration in the discovery sample (top SNP rs3915911, meta-analytical p= 8.81 × 10-9), but this observationwas not replicated in the PREVEND cohort (p=0.81; supplementary Table 4).
No SNP reached genome-wide significance (p≤5 × 10-8) for circulating aldosterone concentrations (supplementary Figure 5, Panel A and B). The top SNP associated with circulating aldosterone levels was SNP rs6986428 on chromosome 8 with a meta-analytical p=4.01 × 10-6. The top four loci associated with circulating aldosterone levels and the replication results are displayed in supplementary Table 5.
Look-up of top SNPs in African Americans
In the Jackson Heart Study, rs5030062 (associated with plasma renin activity in our sampleand in the replication sample PREVEND) was associated with plasma renin concentration (beta=0.100; p=0.017) and SNP rs3915911 (associated with plasma renin concentration in our discovery sample, but not in the replication cohort; supplementary Table 4) displayed evidence for association with plasma renin activity in African Americans (beta=0.14; p=0.003). Furthermore, rs3733402, highly correlated with rs4253311 (r2=0.91), and also located in the KLKB1 gene, was associated with circulating BNP levels in African Americans (companion manuscript by Dr. Musani et al.).
Association of top loci with systolic and diastolic BP, renal traits and left ventricular mass
In secondary analyses, rs5030062 and rs4253311 (the two SNPs that reached or were very close to genome-wide significance for plasma renin activity in the discovery sample and replicated in PREVEND) were assessed for their association with systolic and diastolic BP. In the International Consortium for Blood Pressure Genome-Wide Association Studies19 neither SNP was associated with systolic or diastolic BP (Table 3). Furthermore, no statistically significant associations could be observed for these two SNPs with three renal traits (i.e., estimated glomerular filtration rate, urinary albumin-to-creatinine ratio and chronic kidney disease; Table 3). SNP rs4253311 displayed some evidence for association with left ventricular mass (p=0.03; Table 3).
Table 3. Association of rs5030062 and rs4253311 with BP in the International Consortium for Blood Pressure Genome-Wide Association Studies;19 with renal traits in the CKDGen consortium;21 and with echocardiographically determined left ventricular mass in the EchoGen Consortium20.
| rs5030062_A | rs4253311_A | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Study | Ethnicity | Trait | beta (SE) | p | beta (SE) | P |
| ICBP19 | European descent | SBP | -0.165 (0.101) | 0.10 | 0.059 (0.097) | 0.54 |
| ICBP19 | European descent | DBP | -0.077 (0.064) | 0.23 | 0.082 (0.061) | 0.18 |
| EchoGen20 | European descent | LV mass | -0.662 (0.444) | 0.14 | -0.966 (0.440) | 0.03 |
| CKDGen21 | European descent | eGFR | 0.0001 (0.001) | 0.96 | -0.002 (0.001) | 0.19 |
| CKDGen21 | European descent | UACR | 0.013 (0.009) | 0.16 | -0.002 (0.009) | 0.80 |
| CKDGen21 | European descent | CKD | -0.014 (0.022) | 0.53 | 0.023 (0.021) | 0.29 |
LV mass, left ventricular mass, echocardiographically determined; eGFR, estimated glomerular filtration rate, based on serum creatinine measurements; UACR, urinary albumin-to-creatinine ratio; CKD, chronic kidney disease; ICBP, International Consortium for Blood Pressure Genome-Wide Association Studies; SBP, systolic blood pressure; DBP, diastolic blood pressure
Association of the genetic variation at the renin (REN) locus and at the CYP11B2 locus with RAAS biomarkers
Our genome-wide data set included 13 genetic variants in or close to the REN gene (encoding renin) and seven SNPs at the CYP11B2 locus (encoding the aldosterone synthase). Genetic variants in the REN gene were not associated with plasma renin activity or concentrations, nor were SNPs at the CYP11B2 locus associated with circulating aldosterone concentrations (supplementary Table 6a-6c). Additional analyses based on the 1000 genome imputation (restricted to variants with a MAF ≥1%) revealed rs72745753 (p= 0.0022; MAF, 1%), rs189709785 (p=0.001687; MAF, 4%) and rs34617726 (p= 0.00128; MAF, 14%) as the SNPs in the candidate regions most significantly associated PRC, PRA, and aldosterone, respectively. However, given the number of SNPs tested in the CYP11B2 (n=4275) and the REN (n=3167 [PRC-related analyses] and n=2979 [PCA-related analyses]) regions, these associations were not considered to be statistically significant.
Pathway analyses
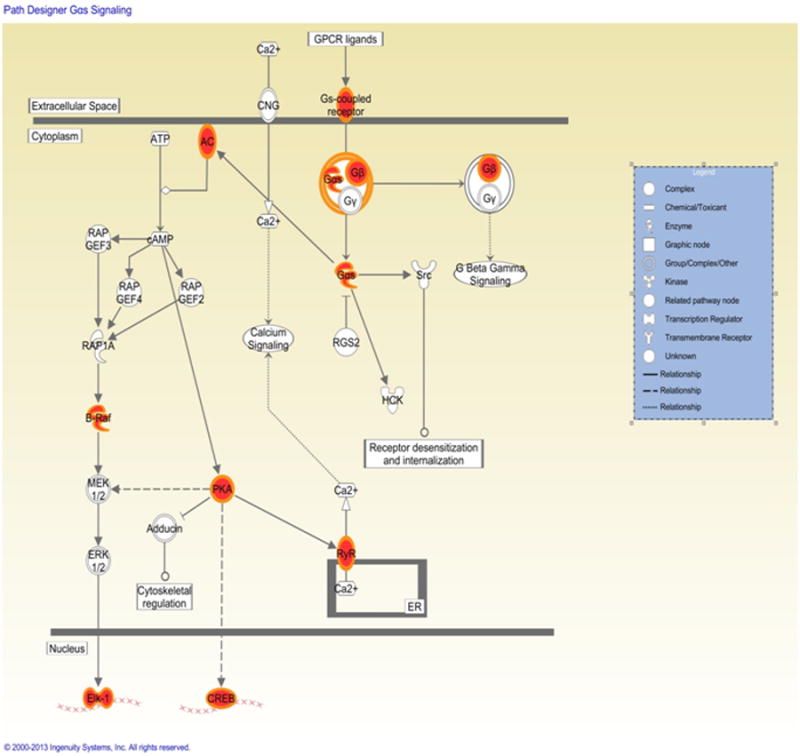
Our pathway analyses revealed that two canonical pathways were significantly enriched (P<1×10-3) with RAAS-related genes, including the G-protein αs-subunits (Gαs) signaling pathway (p=1.2×10-4), and the protein kinase A (PKA) signaling pathway (p=5.5×10-4) (Table 4; Figure 4).
Table 4. Top results from the pathway analysis. Top five enriched canonical pathways.
| Name | p-value | Ratio |
|---|---|---|
| Gαs Signaling | 1.2×10-4 | 10/116 (0.086) |
| Protein Kinase A Signaling | 5.5×10-4 | 19/383 (0.05) |
| GNRH Signaling | 1.8×10-3 | 9/140 (0.064) |
| Corticotropin Releasing Hormone Signaling | 2.8×10-3 | 8/122 (0.066) |
| CCR5 Signaling in Macrophages | 3.7×10-3 | 6/87 (0.069) |
Gαs, G-protein αs-subunits; GNRH, gonadotropin-releasing hormone; CCR5, C-C chemokine receptor type 5
Figure 4. G-protein αs-subunits (Gαs) signaling pathway. Red nodes are genes related to RAAS traits.
Discussion
Using genome-wide association data from up to four population-based cohorts with replication in an independent large fifth cohort, we identified two genetic loci that displayed statistically significant associations with clinically relevant hormones of the RAAS. The main findings of our analyses are summarized below. First, genetic variations in the kininogen 1 and in the kallikrein B genes were associated with plasma renin activity in the discovery sample and with plasma renin concentration and circulating aldosterone concentrations in the replication sample (where plasma renin activity was not available). These variants, however, were not associated with plasma renin concentration or aldosterone levels in the individual discovery cohorts. SNP rs5030062 (in the kininogen 1 gene) was also associated with plasma renin concentration in African Americans. Second, the top SNPs in these genes were not related to BP or renal traits. Third, pathway analyses identified two canonical pathways that were significantly enriched for RAAS-related genes: the Gαs signaling pathway and the PKA signaling pathway. Fourth, no genetic variant was associated with circulating aldosterone levels in a genome-wide significant fashion. Fifth, genetic variation in genes encoding renin and the aldosterone synthase, respectively, was not related to plasma renin concentration or activity or to aldosterone levels.
In the context of the published literature
Possible mechanism for the observed association between SNPs in the kallikrein-kinin system and RAAS biomarkers
As a key finding, we observed that genetic variation within the kallikrein-kinin system (in the kininogen 1 gene and in the kallikrein B gene) was associated with key biomarkers of the RAAS. The kininogen 1 gene encodes both high and low molecular weight kininogen, the precursors of bradykinin and kallidin (Lys-Bradykinin), respectively.27 The kallikrein B gene encodes plasma prekallikrein, a serine protease that, upon transformation to kallikrein, catalyzes the conversion of high molecular weight kininogen to bradykinin (Figure 5)27 and possibly other factors such as adrenomedullin and endothelin-1.28
Figure 5. Graphical display of some key elements of the kallikrein-kinin system and the RAAS system4, 27.
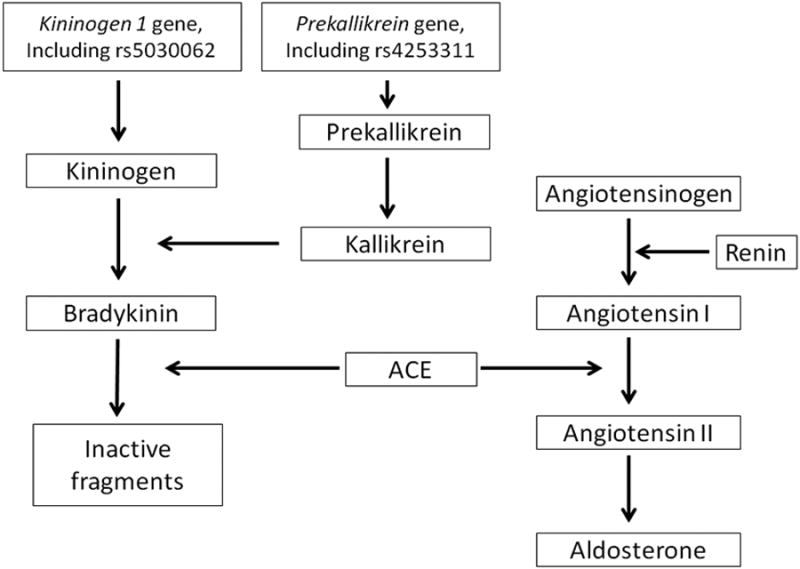
Thus, genetic variation in the precursor substance of bradykinin and in the precursor substance of the enzyme (kallikrein) that catalyzes the conversion of high molecular weight kininogen to bradykinin was related to biomarkers of the RAAS in our genome-wide analysis.
These observations are consistent with the concept that the kallikrein-kinin system and the RAAS are tightly interrelated.29 A prime example for this interaction is the angiotensin-converting enzyme (ACE), which catalyzes on one hand the conversion of angiotensin I to angiotensin II, and on the other hand, degrades bradykinin.29-30 In other words, the ACE affects the concentrations of key effectors of the RAAS (i.e. angiotensin II) and the kallikrein-kinin system, (i.e. bradykinin) in opposite directions. Alsoboth the systems interact at the receptor level. The AT1 receptor and the bradykinin 2 receptor have been shown to physically interact by forming heterodimers, and this heterodimerization affects downstream signaling.29 Rodents lacking the bradykinin 2 receptor gene display reduced renin mRNA expression as compared to wild-type animals, underscoring that kinins influence renin synthesis through activation of the bradykinin 2 receptor.29, 31 Our genetic-epidemiological data add support to this concept by indicating that genetic variation in the kallikrein-kinin system influences inter-individual variation of plasma renin activity.
Our analytical sample was restricted to participants of European descent. However, our top SNPs associated with plasma renin activity on a genome-wide scale also displayed some evidence for association with related traits in participants of African American ancestry: SNP rs5030062 (in the kininogen 1 gene) displayed evidence for association with renin concentration in participants of the Jackson Heart Study; and in an companion paper, Dr. Musani and colleagues report that a close proxy of SNP rs4253311 (rs3733402; r2=0.91) in the kallikrein B gene was associated with circulating BNP levels in African Americans in a genome-wide significant fashion. Overall, these data lend support to the concept that the kallikrein-kinin system is involved in regulating pathways with high relevance for the cardiovascular system, including the RAAS and the BNP pathway. Furthermore, these interrelations seem to be relevant in both individuals of European and of African descent, even though the association findings are not identical in these two ethnicities (but display important overlap). Experimental data further support the significance of the two successfully replicated SNPs for the cardiovascular system. SNP rs5030062 (in the kininogen 1 gene) was predicted to disrupt the binding site of a forkhead box transcription factor,32 which has been shown to be relevant for several cardiovascular traits, including the development of atherosclerotic plaque lesions.33 SNP rs4253311 is in high linkage disequilibrium (r2=0.91) with a missense mutation, rs3733402, which is associated with plasma prekallikrein deficiency.34
Association of top SNPs with BP, renal traits and left ventricular mass
Since the RAAS is an important modulator of acute and chronic changes in BP and a key determinant of renal function, we assessed the association of the top (genome-wide significant) SNPs in the kininogen 1 gene and in the kallikrein B gene (rs5030062 and rs4253311) with BP and renal traits. Both SNPs were neither related to systolic or diastolic BP nor to renal traits, including urinary albumin-to-creatinine ratio, estimated glomerular filtration rate or chronic kidney disease (as a binary trait) in large international consortia.19, 21-22 SNP rs4253311 provided evidence for association with left ventricular mass (p=0.03), but this association would not remain statistically significant upon correction for the multiple “look-ups” of a number of intermediate cardiovascular traits. Thus, in our sample, genetic variation associated with RAAS biomarkers did not translate into observable associations with BP or renal traits. We submit, though, that BP and renal function are complex traits that are regulated by multiple genetic, environmental and lifestyle factors. Thus, the impact of single genetic variants, primarily identified through modest associations with the RAAS pathway, on intermediate cardiovascular disease traits (such as BP or renal function), would not be expected to be prominent.
Association of genetic variation in REN and CYP11B2 with circulating renin and aldosterone levels
In prior studies, genetic variation in the renin (REN) gene itself has been associated with plasma renin activity35-36 and the risk of incident hypertension.35-36 On a parallel note, genetic variation in the CYP11B2 gene, which encodes the aldosterone synthase (MIM 124080), has been related to the ratio of plasma aldosterone concentration/plasma renin activity.37 In our analyses, however, a limited number of SNPs in these two genes were not associated with plasma renin concentration, plasma renin activity or circulating aldosterone levels, respectively. This is consistent with results from a prior GWAS in Japanese individuals (n=936).38
Pathway analyses
Pathway analyses identified the Gαs signaling pathway and the PKA signaling pathway as being overrepresented with RAAS-related genes. G-proteins play a central role in signal transduction. Gαs is a stimulatory G-protein subunit activating the adenylate cyclase, which – upon such activation – increases intracellular cAMP levels.39-41 cAMP is an important second messenger binding e.g. to PKA, which in turn influences the transcription of multiple genes.40-41 As reviewed in detail by Kim et al., the Gαs/cAMP/PKA pathway plays a central role in the regulation of renin secretion, one of the key biomarkers of the RAAS.42 Mice with targeted deletion of Gαs in juxtaglomerular cells display lower basal renin secretion and expression42 and a blunted response to chronic ACE inhibition or AT1 receptor blockade (impaired “feedback loop”),43 indicating that Gαs signaling is highly relevant to renin secretion and expression under steady-state and dynamic conditions.42
Co-administration of ACE inhibitors or AT1 antagonists with inhibitors of the adenylate cyclase likewise substantially reduces renin expression as compared to ACE-inhibitors or AT1 antagonists alone.42-43 Thus, cell-specific disruption of the Gαs gene in juxtaglomerular cells and inhibition of the adenylate cyclase leads to impaired renin secretion in response to chronic blockade of the RAAS.42-43 In addition, it is well established that the sympathetic nervous system is an important determinant of renin secretion and these effects are likewise mediated via the cAMP/PKA pathway.42 Thus, multiple lines of evidence link the RAAS to the Gαs/cAMP/PKA axis and our pathway analyses are in agreement with these experimental data, supporting the significance of Gαs/PKA signaling for the regulation of the RAAS.
Strengths and limitations
To our knowledge, the present analysis is the first comprehensive genome-wide association study analyzing the main circulating biomarkers of the RAAS conjointly. Additional strengths include the considerable samples size, the careful phenotyping and genotyping in the contributing cohorts as well as their population-based character. As an apparent limitation, plasma renin activity was measured in only two cohorts (total n=5,275) of the discovery sample, and not in the replication sample. Therefore, the genome-wide significant hits for plasma renin activity from the discovery sample had to be tested for association with plasma renin concentrations and circulating aldosterone levels in the replication sample. However, in the absence of large changes in plasma angiotensinogen, measurements of plasma renin activity and concentration investigate the same biomarker, circulating “active” renin, almost exclusively of renal origin, and both biomarkers (renin concentration and renin activity) are strongly correlated.44 In population studies, the control of the physiological factors which influence plasma renin, mainly posture and sodium intake, are less well controlled than in metabolic studies, and this may lead to an underestimation of the genetic association. However, we do not expect major changes in angiotensinogen in an epidemiological setting. Furthermore, since these three biomarkers are tightly inter related and part of the same pathway, the key finding of our investigation, that genetic variation in the kallikrein-kinin systems is associated with RAAS biomarker levels, is supported by the replication analyses. As an additional limitation, the top SNPs associated with plasma renin activity in the discovery cohorts were associated with circulating renin concentrations and aldosterone levels in an independent sample (PREVEND) but not in the individual discovery cohorts themselves. Potential explanations for this discrepancy include the smaller sample sizes and the larger coefficients of variation for renin and aldosterone measurements in some individual discovery cohorts as compared to the PREVEND sample, resulting in larger measurement errors and reduced statistical power to detect modest associations in the discovery as compared to the replication cohort.
In conclusion, we present data from a meta-analysis of large genome-wide association studies for three biomarkers of the RAAS in up to 13,289 Europeans and European Americans. We observed that genetic variation in the kallikrein-kinin system was associated with plasma renin activity at a genome-wide significant level. This is consistent with experimental data linking the RAAS to the kallikrein-kinin system. Furthermore, pathway analyses identified two canonical pathways that were significantly enriched for RAAS-related genes: the Gαs signaling pathway and the PKA signaling pathway which influence renal renin release. Our observations require replication in other independent cohorts, including samples from other ethnicities and age groups, to address the generalizability of our findings. Furthermore, the significant SNPs warrant further experimental exploration in order to elucidate in detail the molecular mechanisms underlying the observed associations.
Supplementary Material
Acknowledgments
Funding Sources: A detailed description of funding sources is provided in the supplementary material.
Conflict of Interest Disclosures: Dr. Wang, Dr. Vasan and Dr. Wilson received NIH funding related to this manuscript. Details about the funding are provided in the supplementary material. Dr. de Boer received lecture fees from Novartis and Medcon.
References
- 1.Weir MR, Dzau VJ. The renin-angiotensin-aldosterone system: A specific target for hypertension management. Am J Hypertens. 1999;12:205S–213S. doi: 10.1016/s0895-7061(99)00103-x. [DOI] [PubMed] [Google Scholar]
- 2.Givertz MM. Manipulation of the renin-angiotensin system. Circulation. 2001;104:E14–18. doi: 10.1161/hc3001.094733. [DOI] [PubMed] [Google Scholar]
- 3.Heeneman S, Sluimer JC, Daemen MJ. Angiotensin-converting enzyme and vascular remodeling. Circ Res. 2007;101:441–454. doi: 10.1161/CIRCRESAHA.107.148338. [DOI] [PubMed] [Google Scholar]
- 4.Eriksson U, Danilczyk U, Penninger JM. Just the beginning: Novel functions for angiotensin-converting enzymes. Curr Biol. 2002;12:R745–752. doi: 10.1016/s0960-9822(02)01255-1. [DOI] [PubMed] [Google Scholar]
- 5.Sowers JR, Whaley-Connell A, Epstein M. Narrative review: The emerging clinical implications of the role of aldosterone in the metabolic syndrome and resistant hypertension. Ann Intern Med. 2009;150:776–783. doi: 10.7326/0003-4819-150-11-200906020-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinck WJ, Fagard RH, Vlietinck R, Lijnen P. Heritability of plasma renin activity and plasma concentration of angiotensinogen and angiotensin-converting enzyme. J Hum Hypertens. 2002;16:417–422. doi: 10.1038/sj.jhh.1001410. [DOI] [PubMed] [Google Scholar]
- 7.Newton-Cheh C, Guo CY, Gona P, Larson MG, Benjamin EJ, Wang TJ, et al. Clinical and genetic correlates of aldosterone-to-renin ratio and relations to blood pressure in a community sample. Hypertension. 2007;49:846–856. doi: 10.1161/01.HYP.0000258554.87444.91. [DOI] [PubMed] [Google Scholar]
- 8.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The framingham offspring study. Am J Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 9.Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, et al. The third generation cohort of the national heart, lung, and blood institute's framingham heart study: Design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 10.Wichmann HE, Gieger C, Illig T. Kora-gen--resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl 1):S26–30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 11.Volzke H, Alte D, Schmidt CO, Radke D, Lorbeer R, Friedrich N, et al. Cohort profile: The study of health in pomerania. Int J Epidemiol. 2011;40:294–307. doi: 10.1093/ije/dyp394. [DOI] [PubMed] [Google Scholar]
- 12.Hercberg S, Galan P, Preziosi P, Bertrais S, Mennen L, Malvy D, et al. The su.Vi.Max study: A randomized, placebo-controlled trial of the health effects of antioxidant vitamins and minerals. Arch Intern Med. 2004;164:2335–2342. doi: 10.1001/archinte.164.21.2335. [DOI] [PubMed] [Google Scholar]
- 13.de Boer RA, Schroten NF, Bakker SJ, Mahmud H, Szymanski MK, van der Harst P, et al. Plasma renin and outcome in the community: Data from prevend. Eur Heart J. 2012;33:2351–2359. doi: 10.1093/eurheartj/ehs198. [DOI] [PubMed] [Google Scholar]
- 14.Hillege HL, Janssen WM, Bak AA, Diercks GF, Grobbee DE, Crijns HJ, et al. Microalbuminuria is common, also in a nondiabetic, nonhypertensive population, and an independent indicator of cardiovascular risk factors and cardiovascular morbidity. J Intern Med. 2001;249:519–526. doi: 10.1046/j.1365-2796.2001.00833.x. [DOI] [PubMed] [Google Scholar]
- 15.Chen MH, Yang Q. Gwaf: An r package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Wu H, Guo CY, Fox CS. Analyze multivariate phenotypes in genetic association studies by combining univariate association tests. Genet Epidemiol. 2010;34:444–454. doi: 10.1002/gepi.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuqua SR, Wyatt SB, Andrew ME, Sarpong DF, Henderson FR, Cunningham MF, et al. Recruiting african-american research participation in the jackson heart study: Methods, response rates, and sample description. Ethn Dis. 2005;15:S6–18. 29. [PubMed] [Google Scholar]
- 19.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasan RS, Glazer NL, Felix JF, Lieb W, Wild PS, Felix SB, et al. Genetic variants associated with cardiac structure and function: A meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–178. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boger CA, Chen MH, Tin A, Olden M, Kottgen A, de Boer IH, et al. Cubn is a gene locus for albuminuria. J Am Soc Nephrol. 2011;22:555–570. doi: 10.1681/ASN.2010060598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, et al. High-resolution mapping of expression-qtls yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fox ER, Musani SK, Barbalic M, Lin H, Yu B, Ogunyankin KO, et al. Genome-wide association study of cardiac structure and systolic function in african americans: The candidate gene association resource (care) study. Circ Cardiovasc Genet. 2013;6:37–46. doi: 10.1161/CIRCGENETICS.111.962365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreau ME, Garbacki N, Molinaro G, Brown NJ, Marceau F, Adam A. The kallikrein-kinin system: Current and future pharmacological targets. J Pharmacol Sci. 2005;99:6–38. doi: 10.1254/jphs.srj05001x. [DOI] [PubMed] [Google Scholar]
- 28.Verweij N, Mahmud H, Mateo Leach I, de Boer RA, Brouwers FP, Yu H, et al. Genome-wide association study on plasma levels of midregional-proadrenomedullin and c-terminal-pro-endothelin-1. Hypertension. 2013;61:602–608. doi: 10.1161/HYPERTENSIONAHA.111.203117. [DOI] [PubMed] [Google Scholar]
- 29.Shen B, El-Dahr SS. Cross-talk of the renin-angiotensin and kallikrein-kinin systems. Biol Chem. 2006;387:145–150. doi: 10.1515/BC.2006.019. [DOI] [PubMed] [Google Scholar]
- 30.Tschope C, Schultheiss HP, Walther T. Multiple interactions between the renin-angiotensin and the kallikrein-kinin systems: Role of ace inhibition and at1 receptor blockade. J Cardiovasc Pharmacol. 2002;39:478–487. doi: 10.1097/00005344-200204000-00003. [DOI] [PubMed] [Google Scholar]
- 31.Yosipiv IV, Dipp S, El-Dahr SS. Targeted disruption of the bradykinin b(2) receptor gene in mice alters the ontogeny of the renin-angiotensin system. Am J Physiol Renal Physiol. 2001;281:F795–801. doi: 10.1152/ajprenal.2001.281.5.F795. [DOI] [PubMed] [Google Scholar]
- 32.Ward LD, Kellis M. Haploreg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bot PT, Grundmann S, Goumans MJ, de Kleijn D, Moll F, de Boer O, et al. Forkhead box protein p1 as a downstream target of transforming growth factor-beta induces collagen synthesis and correlates with a more stable plaque phenotype. Atherosclerosis. 2011;218:33–43. doi: 10.1016/j.atherosclerosis.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 34.Katsuda I, Maruyama F, Ezaki K, Sawamura T, Ichihara Y. A new type of plasma prekallikrein deficiency associated with homozygosity for gly104arg and asn124ser in apple domain 2 of the heavy-chain region. Eur J Haematol. 2007;79:59–68. doi: 10.1111/j.1600-0609.2007.00871.x. [DOI] [PubMed] [Google Scholar]
- 35.Sun B, Williams JS, Pojoga L, Chamarthi B, Lasky-Su J, Raby BA, et al. Renin gene polymorphism: Its relationship to hypertension, renin levels and vascular responses. J Renin Angiotensin Aldosterone Syst. 2011;12:564–571. doi: 10.1177/1470320311405873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasimu B, Nakayama T, Mizutani Y, Izumi Y, Asai S, Soma M, et al. Haplotype analysis of the human renin gene and essential hypertension. Hypertension. 2003;41:308–312. doi: 10.1161/01.hyp.0000049762.77830.89. [DOI] [PubMed] [Google Scholar]
- 37.Tamaki S, Iwai N, Tsujita Y, Kinoshita M. Genetic polymorphism of cyp11b2 gene and hypertension in japanese. Hypertension. 1999;33:266–270. doi: 10.1161/01.hyp.33.1.266. [DOI] [PubMed] [Google Scholar]
- 38.Hiura Y, Tabara Y, Kokubo Y, Okamura T, Miki T, Tomoike H, et al. A genome-wide association study of hypertension-related phenotypes in a japanese population. Circ J. 2010;74:2353–2359. doi: 10.1253/circj.cj-10-0353. [DOI] [PubMed] [Google Scholar]
- 39.Zurita AR, Birnbaumer L. The same mutation in gsalpha and transducin alpha reveals behavioral differences between these highly homologous g protein alpha-subunits. Proc Natl Acad Sci U S A. 2008;105:2363–2368. doi: 10.1073/pnas.0712261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lalli E, Sassone-Corsi P. Signal transduction and gene regulation: The nuclear response to camp. J Biol Chem. 1994;269:17359–17362. [PubMed] [Google Scholar]
- 41.Castrop H, Hocherl K, Kurtz A, Schweda F, Todorov V, Wagner C. Physiology of kidney renin. Physiol Rev. 2010;90:607–673. doi: 10.1152/physrev.00011.2009. [DOI] [PubMed] [Google Scholar]
- 42.Kim SM, Briggs JP, Schnermann J. Convergence of major physiological stimuli for renin release on the gs-alpha/cyclic adenosine monophosphate signaling pathway. Clin Exp Nephrol. 2012;16:17–24. doi: 10.1007/s10157-011-0494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Kim SM, Eisner C, Oppermann M, Huang Y, Mizel D, et al. Stimulation of renin secretion by angiotensin ii blockade is gsalpha-dependent. J Am Soc Nephrol. 2010;21:986–992. doi: 10.1681/ASN.2009030307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toffelmire EB, Slater K, Corvol P, Menard J, Schambelan M. Response of plasma prorenin and active renin to chronic and acute alterations of renin secretion in normal humans. Studies using a direct immunoradiometric assay. J Clin Invest. 1989;83:679–687. doi: 10.1172/JCI113932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


