Abstract
Cell migration is a fundamental process underlying diverse (patho)physiological phenomena. The classical understanding of the molecular mechanisms of cell migration has been based on in vitro studies on two-dimensional substrates. More recently, mounting evidence from intravital studies has shown that during metastasis, tumor cells must navigate complex microenvironments in vivo, including narrow, pre-existing microtracks created by anatomical structures. It is becoming apparent that unraveling the mechanisms of confined cell migration in this context requires a multi-disciplinary approach through integration of in vivo and in vitro studies, along with sophisticated bioengineering techniques and mathematical modeling. Here, we highlight such an approach that has led to discovery of a new model for cell migration in confined microenvironments (i.e., the Osmotic Engine Model).
Introduction
Cell migration plays a key role in both cell physiology, including embryonic development, wound healing, and the immune response, and in development of pathological conditions. For example, in cancer metastasis, cells migrate away from the primary tumor, through the surrounding microenvironment, and to the microvessels, where they can invade into the blood and/or lymphatic circulation for metastasis to distal sites [1–3]. After traveling in the circulation, the tumor cells extravasate from a blood vessel and migrate to the site where a secondary tumor will form. Recent in vivo intravital microscopy studies suggest that the metastatic cascade involves migration of tumor cells through extremely complex microenvironments [4–8], and it is becoming increasingly evident that physical forces are at play during multiple steps of metastasis [3,9]. To achieve migration through such microenvironments, cells are required to either degrade matrix to create their own migration tracks [10] or find preexisting tracks [11,12] through which to migrate. Interestingly, recent intravital microscopy studies reveal that cells preferentially migrate along very narrow pre-existing tracks in vivo [4,8]. These tracks vary from <3 µm to ~30 µm in width and are 100–600 µm in length [13]. The microtrack width modestly increases during perimuscular invasion [7], which may be attributed to limited matrix metalloproteinase (MMP)-dependent proteolysis or outward pushing exerted by invading cells. It is noteworthy that no significant changes in track width are detected during migration through collagen networks, fat tissue, or perineural space [7]. Hence, invading tumor cells not only preferentially follow pre-existing tissue tracks, but also adapt their shape to the space available without significant tissue remodeling or degradation. This may partly explain why MMP inhibitors have largely failed clinically in cancer patients [14].
Cell migration through confined spaces plays important roles in both physiological and pathological cell migration events [8,15–17]. During the past decades, in vitro cell migration studies have been mainly performed on unconfined two-dimensional (2D) surfaces such as glass or plastic; while we have learned an extensive amount about how cells migrate from these 2D assays [18–21], they fail to recapitulate the in vivo microenvironment. A number of assays have been developed to provide additional information, such as how cells respond to biochemical [22,23], adhesive [24], topographical [25], mechanical [26–32], and dimensional [33–36] cues; however, each of these assays faces its own limitations (Fig. 1). Only relatively recently have microfabrication techniques been used to simulate microtracks in vitro. The fundamental question now is whether cells utilize the same machinery and mechanisms for confined versus unconfined migration, and how the biochemical and mechanical properties of the microenvironment affect these mechanisms. Answering this question will most likely require a multi-disciplinary approach through integration of in vivo and in vitro studies, along with mathematical modeling.
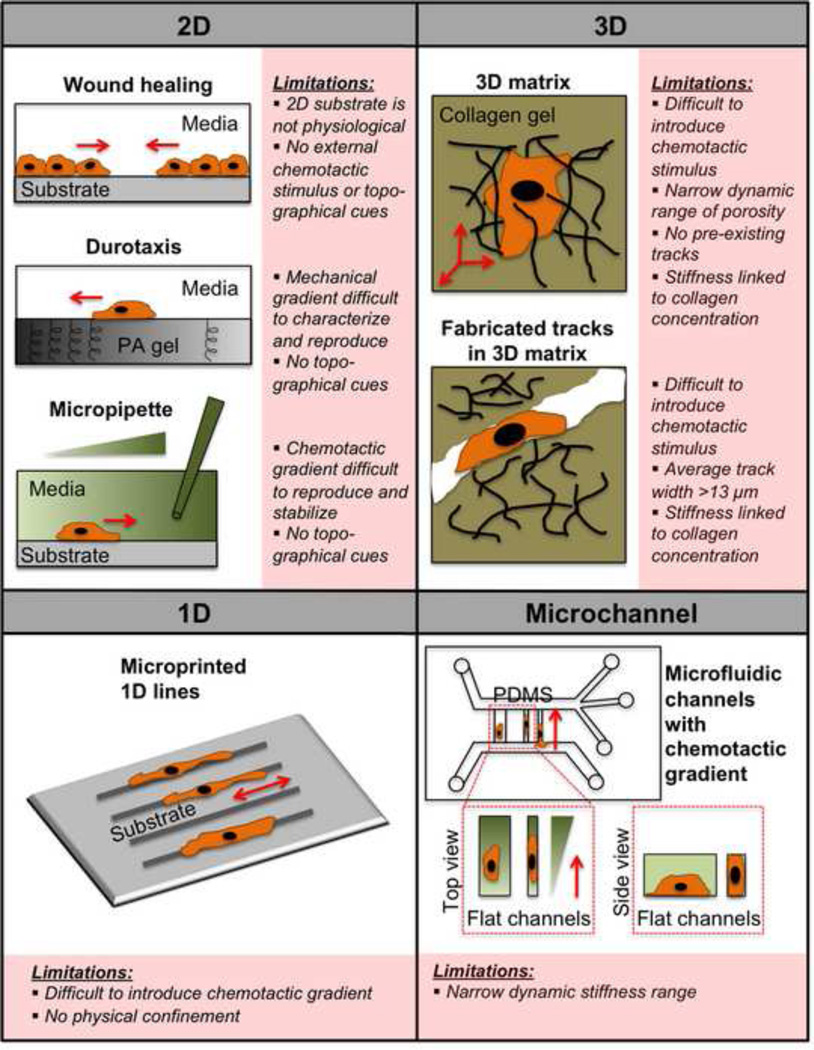
Figure 1.
Overview of 2D, 3D, 1D, and microchannel cell migration assays and their limitations. In the wound healing assay, a monolayer of cells is scratched, or a physical barrier is removed, and the cells subsequently migrate towards each other to close the wound. In the durotaxis assay, a gradient of substrate stiffness is created by placing two drops of polymerizing polyacrylamide (PA) of different stiffnesses next to each other, and covering the solutions with a glass coverslip. Cells are then induced to migrate in response to the mechanical gradient of stiffness. In the micropipette assay, cells respond to a chemotactic gradient created by a chemoattractant-filled micropipette. In a 3D matrix, cells must enzymatically degrade the surrounding matrix in order to move, while in fabricated tracks within a 3D matrix, preexisting tracks are created in a collagen gel via microfabrication techniques. In the assay with microprinted 1D lines, cells adhere selectively to 1D protein lines of specific width. In the microchannel assay, cells are induced to migrate into confined or unconfined microchannels in response to a chemoattractant gradient. Some parts of the figure are adapted with permission from [9].
Engineering the cellular microenvironment
Given the physiological relevance of cell migration through confined spaces in vivo [4,7], it is necessary to create appropriate in vitro systems that enable understanding of cell migration in this context. Reconstituted three-dimensional (3D) collagen gels have been extensively used to study the mechanisms of random 3D migration in vitro [13,37–39]. However, these 3D assays fail to recapitulate the longitudinal tracks and the dynamic range of collagen-free pore sizes encountered by cells in vivo [7,13]. To circumvent the limitations presented by traditional 2D and reconstituted gel migration assays, engineering techniques such as microfabrication have recently allowed researchers to evaluate the effects of physical confinement on cell migration [40–50] (Fig. 1). For instance, the microfabrication technology has been applied to create in vitro models of cellular intravasation [41], which represents a form of migration in a confined space, as cells must squeeze between endothelial cell-cell junctions in order to enter a blood vessel. Microfabrication techniques have also been employed to generate surfaces, wells, or molds with adhesive areas of varying size and shape in order to evaluate the effects of spatial confinement on cellular differentiation [51], proliferation [52], angiogenesis [53], and protein expression [52]. Recently, perfusable engineered vascular channels have been developed [54] by 3D printing of rigid filament networks of carbohydrate glass, which served as a template for the casting of either a synthetic or natural extracellular matrix containing cells around the lattice. Upon dissolving the carbohydrate glass away, endothelial cells are introduced into the vascular architecture and perfused with media to simulate blood flow and the physiological endothelial cell function. This microfabrication approach could also be used to create microtracks to investigate cell migration in confined microchannels.
We and others have developed PDMS-based microfluidic devices where physical cues (e.g., microchannel cross-sectional area and topography) and biochemical cues (e.g., chemoattractant gradient and surface protein presentation) can be simultaneously varied within the same device [45–47,55–57]. Furthermore, we have used this device to investigate the molecular mechanisms and signaling pathways involved in cell migration in unconfined versus confined spaces [47,55]. While this PDMS-based device is likely relevant in the context of stiffer in vivo microtracks such as those which might be found along muscle and nerve fibers [7], a limitation is its narrow range of tunable stiffness (Fig. 1). To address this limitation, a polyacrylamide gel-based device has been fabricated consisting of 3-wall microchannels of varying width (10–40 µm) and stiffness (0.4 kPa to 120 kPa) [42,43]. However, this device cannot replicate a truly confined microenvironment, as it is comprised of 3- rather than 4- wall microchannels of 10 µm or larger in width, nor does it incorporate a chemotactic gradient. To circumvent these limitations, we are currently developing a new model of our chemotaxis-based device in which the stiffness of the narrow (3 µm wide) microchannel walls can also be manipulated systematically; this will allow for modeling of softer microtracks, such as those that would occur between bundles of collagen fibers within the ECM [4].
Using the microchannel device, two classes of cell responses have been discovered – (1) those that utilize a Rho/Rac crosstalk mechanism (e.g., in normal fibroblasts, fibroblast-like cells, primary murine T-cells, and α4-expressing A375 melanoma cells), where Rho-mediated cell contractility is necessary for migration in confined spaces [55], and (2) those that do not require actomyosin during migration in confined spaces (e.g, metastatic cells such as murine S180 sarcoma cells, human MDA-MB-231 breast tumor and CH2879 chondrosarcoma cells) [45,47] (Fig. 2).
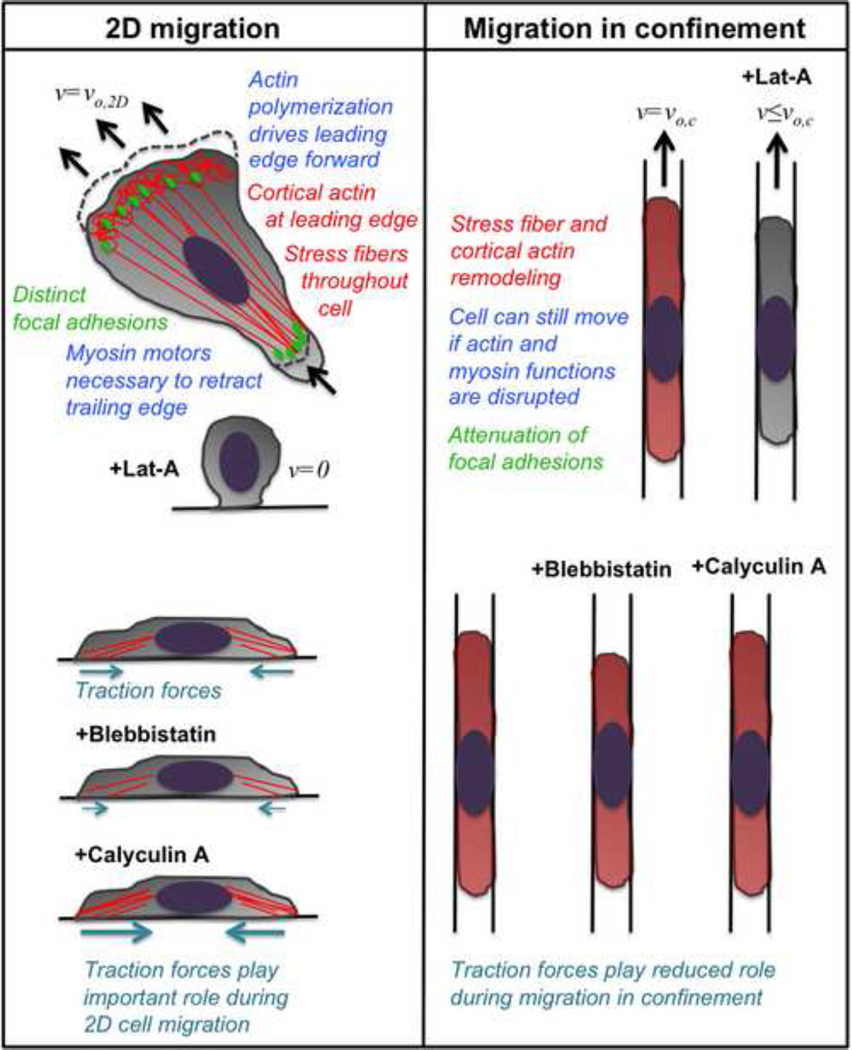
Figure 2.
Comparison of major differences between 2D migration and migration through confined spaces (i.e., microchannels). In 2D migration, actin polymerization drives the leading edge forward, and both cortical actin and stress fibers are evident within the cell. Myosin motors are necessary to retract the cell’s trailing edge. Distinct focal adhesions help anchor the cell and traction forces are generated through these focal adhesions. When the actin-disrupting drug latrunculin-A is added to cells in 2D, they lose attachment to the substrate, round up, and cell velocity goes to zero. Blebbistatin, which inhibits myosin II function and decreases cell contractility, decreases cell traction forces in 2D; meanwhile, calyculin A, which inhibits protein phosphatases and increases cell contractility, increases cell traction forces in 2D. During migration in confined microchannels, the cell undergoes dramatic stress fiber and cortical actin remodeling, with both becoming more diffuse throughout the cell. Attenuation of focal adhesion size is also observed in microchannels. In contrast to 2D, the cell can still move in confined microchannels if actin and myosin functions are disrupted. Furthermore, neither blebbistatin nor calyculin A has any effect on the magnitude of cell traction forces in confinement, indicating that cell traction forces play a reduced role during migration through confined spaces. In confined spaces, the net direction of forces is towards the chemoattractant, though appreciable forces are also directed towards the side walls of the microchannels.
By incorporating a bed of micropillars onto the bottom wall of the microchannels within the microfluidic device, cellular traction forces have been measured during migration in confined and unconfined spaces [46]. This assay has revealed that traction forces exerted by cells in confined microchannels are lower than those in unconfined (2D) channels. These observations are in line with studies demonstrating that cells exert lower traction forces on 1D micropatterned lines in comparison with 2D substrates [36]. As expected, treatment of human osteosarcoma (HOS) cells by blebbistatin, which suppresses myosin II-mediated contractility, or calyculin A, which increases cell contractility, decreases or increases cell traction forces, respectively, in wide channels (i.e., unconfined spaces) [46]. Remarkably, neither blebbistatin nor calyculin A has any effect on cell traction forces in narrow channels (i.e., confined spaces). Thus, myosin-mediated cell contractility appears to play reduced role in HOS cell confined migration, as in other metastatic cell lines [45,47]. In agreement with these observations, tumor cells have recently been shown to exert less frictional forces along channel walls in comparison with normal cells [58].
Tumor cells also display an altered actin cytoskeleton, with fewer stress fibers [59,60], and increased deformability [58,61,62]. Similar to observations in 3D collagen gels [38], focal adhesions are also suppressed in tumor cells within narrow microchannels [45]. Furthermore, physical confinement induces F-actin remodeling, such that stress fibers are drastically diminished in physically confined spaces [45]. Moreover, actin appears to be concentrated on the leading and trailing edges of cells migrating in narrow channels [45]. In line with our observations, HL60 neutrophil-like cells chemotactically migrating in confining microchannels (5×5 µm2) form a “slab” of actin that fills the entire cross-section of the channel at the cell’s leading edge, rather than assembling thin ~200 nm-thick actin-rich lamellipodia at the leading edge, as occurs on 2D surfaces [56]. Our findings, along with experimental observations [45] and a mathematical model [63] showing that tumor cells are able to migrate even in the absence of integrin-mediated adhesion, may help explain the reduced magnitude of traction forces measured in confined relative to unconfined migration [46]. The marked decrease in the formation of stress fibers and focal adhesions may also explain why inhibition of cell contractility via blebbistatin has no effect on tumor cell migration in confined spaces [45–47]. In contrast, normal fibroblast-like cells intrinsically displaying a higher level of stress fibers and focal adhesions demonstrate decreased confined migration upon blebbistatin treatment [55]. Intriguingly, tumor cells are still able to undergo confined migration in the presence of latrunculin-A, which disrupts actin polymerization, even though the same treatment completely abrogates migration in wide channels, as it does on a 2D surface. It is thus becoming increasingly apparent that the cellular mechanisms utilized during tumor cell migration in confined spaces can be fundamentally different from migration in unconfined spaces (i.e. 2D planar surfaces).
New Model for Cell Migration
Recently, we discovered a new model for confined cell migration that is driven by water permeation through the cell membrane [47]. This mechanism, termed the “Osmotic Engine Model” of cell migration, requires the coordinated activity of ion channels and aquaporins and is based on water flux into the cell at the leading edge and water flux out of the cell at the trailing edge (Fig. 3A). Ion pumps and aquaporins have previously been implicated in 2D cell migration [64–66], however their function has been both underappreciated and not well understood. Their role as cellular migration machinery has mostly been associated with the cytoskeleton. For example, the Sodium Hydrogen Exchanger-1 (NHE-1) physically interacts with the actin cytoskeleton, and in turn, the actin cytoskeleton regulates the activity of ion channels [67–69]. Indeed, inhibition of NHE-1 decreases 2D migration speed in several cell types [47,70,71]. Furthermore, Aquaporin 5 (AQP5) is overexpressed in lung and breast tumor cells [72,73] and acts to facilitate actin polymerization [64] while stabilizing microtubules [74], thus supporting 2D cell migration. In addition, aquaporins crosstalk to cell-matrix adhesion molecules (i.e., integrins) during cell migration, especially in renal cells and cancer cells that abundantly express aquaporins [75–77].
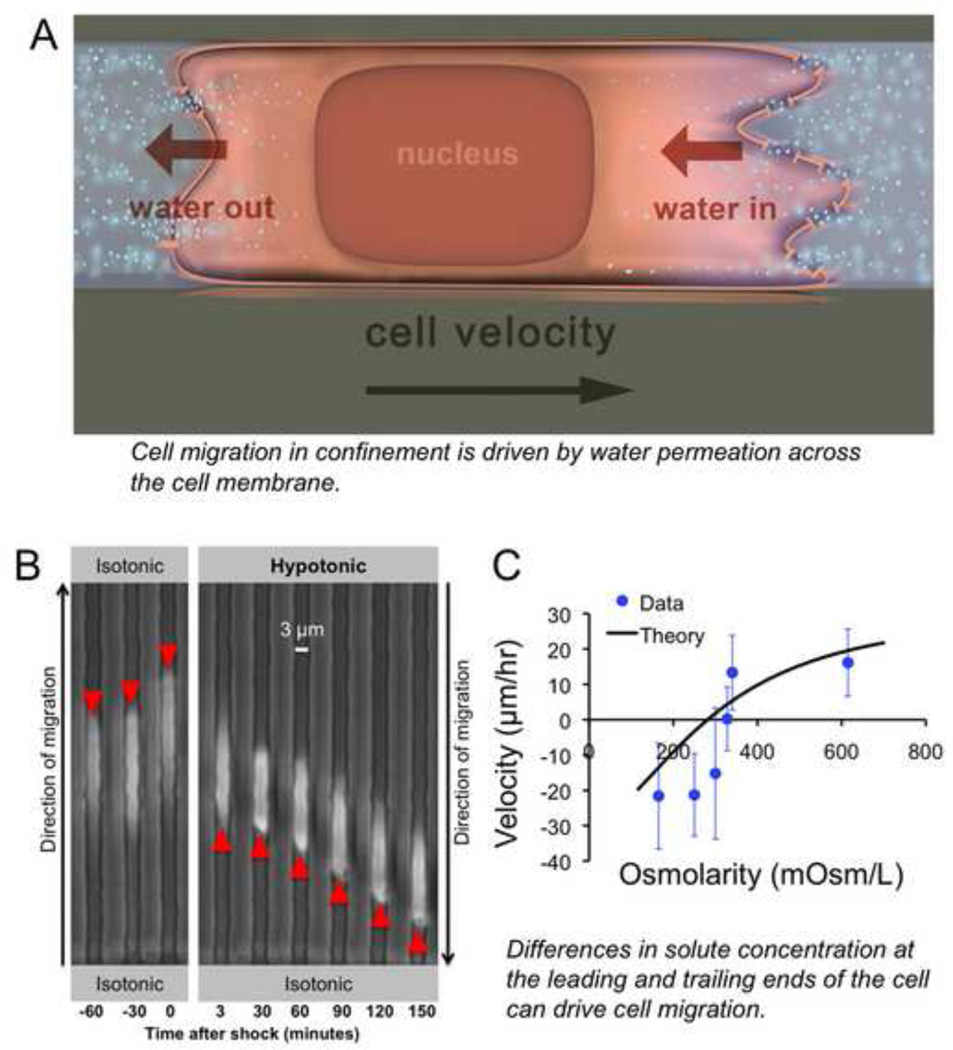
Figure 3.
Overview of the Osmotic Engine Model of cell migration. (A) Cell migration in confined spaces is driven by water permeation across the cell membrane. Water flows in at the cell’s leading edge, which allows the front of the cell to extend forward, and water flows out at the cell’s trailing edge, which allows the back of the cell to retract. This results in translocation of the cell body forward with little change in cell length (or volume). (B) The Osmotic Engine Model can be tested by applying osmotic shocks to the cell’s leading (or trailing) edge. Here, a hypotonic shock is introduced within the microfluidic device at the cell’s leading edge, causing the cell to reverse direction and migrate away from the chemoattractant gradient. (C) The velocity of cell migration depends on the magnitude of osmolarity of the extracellular medium at the cell’s leading edge. The reversal of cell migration direction in response to a hypotonic shock at the cell’s leading edge is predicted by the theoretical framework of the Osmotic Engine Model. Thus, differences in solute concentration at the leading and trailing ends of the cell can drive cell migration. In the absence of osmotic shocks, the cell’s ion channels and aquaporins must be polarized in order to sustain migration. Figures reproduced with permission from [79].
Our work provides a new context in which ion channels and aquaporins (e.g., NHE-1 and AQP5) are not only required but can also drive migration through confined spaces when the function of actin polymerization is disrupted [47]. By integrating theory and experiments, we derived an analytical expression for cell migration velocity in confined spaces. The mathematical model takes into account the kinetics of water, kinetics and diffusion of ions, flow of the cell cytoplasm, and mechanics of the cell cortex (i.e., the friction between the cell and channel wall and between the cell cortex and cytoplasm) [78,79]. Importantly, the model predicts that a nonzero cell velocity can be achieved even without actin polymerization and myosin II activity in confined spaces, which aligns with experimental observations, and distinguishes our model from previous mathematical frameworks [18,63,80–88]. For instance, although prior work has modeled the cell as a soft, fluid-infiltrated sponge surrounded by a water permeable barrier capable of taking in water across the cell membrane, it couples hydraulics and cytoskeleton-dependent cellular mechanics [89]. Both theoretical and experimental data from our Osmotic Engine Model reveal that confined cell migration depends on osmotic and hydrostatic pressure differences across the cell membrane at both the cell and trailing edges. For instance, application of a hypotonic shock at the cell leading edge or of a hypertonic shock at the trailing edge causes a rapid reversal in the direction of cell migration (Fig. 3B–C). In both cases, cells repolarize to migrate towards the higher osmolarity regions.
During cell entry into narrow microchannels, a highly polarized distribution of ion pumps (i.e., NHE-1) and aquaporins (i.e., AQP5) is detected along the longitudinal surface of the cell with an intense signal at the cell leading edge [47]. This polarized, spatial distribution of ion pumps and aquaporins is required for sustained migration through confined spaces when actin polymerization is disrupted, as also suggested by the theoretical framework. In contrast, the ion pumps and aquaporins are more randomly distributed on the surface of migrating cells on 2D substrates and act in coordination with the actin cytoskeleton to help drive cell protrusion at the leading edge. As such, interfering with actin polymerization is sufficient to abrogate 2D, but not confined, migration. Of note, knockdown or inhibition of ion pumps and aquaporins markedly suppresses both unconfined and confined migration [47,70,71]. It is also noteworthy that actin polymerization appears to be necessary to set up the polarization of ion pumps and aquaporins during cell entry into confined channels; however, once this polarization is established, actin polymerization is dispensable for confined cell migration [79]. Interestingly, actin is required for the re-polarization of aquaporins and ion channels in cells migrating inside narrow channels following the application of an osmotic shock. If actin polymerization is disrupted, the cells fail to repolarize aquaporins and ion channels to the post-shock leading edge, and are unable to sustain migration [79]. Collectively, the Osmotic Engine Model can predict cellular movement in confined spaces even in the case where actin polymerization is not the driving force, and it thus offers a new perspective into how we view cell migration.
In the classical model of cell migration, cell protrusion at the leading edge is driven by actin polymerization and is stabilized by integrin-dependent adhesion to the substrate, while de-adhesion at the rear is facilitated by cellular contractile forces. Cells likely use this mechanism when migrating in vivo in situations where they are not laterally confined. However, cells may possess multiple power sources for motility, and it appears that tumor cells have evolved to be capable of using different mechanisms (i.e. actomyosin-based and water permeation-based) depending on the specific properties of the microenvironment. Of course, we cannot eliminate the possibility that normal cells also use the Osmotic Engine Model of migration, but the overexpression of aquaporins and ion pumps in numerous metastatic tumor cells [72,73,90] may cause the Osmotic Engine Model to be more evident in these tumor cells.
In confined spaces, water flux through the cell membrane is directed along a single axis longitudinally through the confined cell, allowing water permeation to be a major mechanism driving cell migration within the microchannel [79]. This mode of migration cannot be detected on 2D surfaces due to the lack of biological (e.g., ion channels and aquaporins) and geometrical (e.g., pill-shaped) polarization. Thus, in the 2D setting, actin polymerization is indispensable to guide the protrusions and drive migration. We speculate that Rho-associated kinase 1 (ROCK1), which phosphorylates myosin light chains to induce actomyosin contractility and is an upstream activator of NHE-1 [91], may serve as a linker between the actin-driven and water permeation-based mechanisms. NHE-1 is also involved in regulation of intracellular pH due to its role in exchanging Na+ and H+ ions; NHE-1 recruitment to cellular invadopodia is promoted by cortactin phosphorylation, thereby regulating both cellular pH and invasive capability [92]. Cellular pH could thus be incorporated along with osmolarity in future refinements of the Osmotic Engine Model. Prior modeling work suggests that the relative significance of hydraulics and cytoskeletal dynamics (i.e., actin-, microtubule-, or intermediate filament-based) on cellular morphology depends on the time-, length-, and force-scales involved [89]. In light of this model, we postulate that physical confinement alters cellular parameters such as the cytoskeletal mesh structure, membrane permeability, local contractility, and adhesion, all of which could heterogeneously alter cellular hydraulics and thus cellular migration in confined spaces.
A recent microfluidic study has also suggested that cells push water in confined spaces, where “barotaxis” can override chemotaxis in asymmetric hydraulic microenvironments [57]. Specifically, cells reaching an intersection “decide” to follow the path of least hydraulic resistance, which was manipulated experimentally by adjusting the length or width of the downstream channel far from a bifurcation (i.e., increasing channel length or reducing channel width increased hydraulic resistance). Interestingly, cell velocity is identical to the flow velocity of suspended 500-nm fluorescent polystyrene beads (and therefore also the bulk velocity), suggesting that these differentiated HL60 (dHL60) cells “push” fluid as they migrate forward. This mechanism may depend primarily on actin, given that actin-based motility is faster than the Osmotic Engine Model [47] and that dHL60 cells are fast-moving in comparison with tumor cells; however, the molecular constituents involved in this process have yet to be delineated. This phenomenon may occur in tandem with the Osmotic Engine Model, especially in slower-moving tumor cells, since cell water uptake may be limited by the number of aquaporins and ion channels on the cell surface. The question that remains to be unraveled is whether other conditions besides physical confinement encourage cells to favor the Osmotic Engine Model of migration.
Outlook
Cutting edge bioengineering techniques, such as microfabrication, in vivo and in vitro imaging, in combination with molecular biology, have led to new insights to the mechanisms by which normal and pathological cells migrate in heterogeneous microenvironments. These tools have allowed researchers to unravel the distinct effects of physical and biochemical cues, including confinement and dimensionality, matrix stiffness, topography, chemoattractant gradients, soluble factors, and matrix-bound adhesion proteins, on cell migration. Furthermore, we wish to emphasize the importance of mathematical modeling in combination with both in vitro and in vivo experimental analyses in order to help explain non-intuitive cellular behaviors. An integrated theoretical and multifaceted experimental approach can lead to discovery of new mechanisms for cell migration, just as the Osmotic Engine Model revealed novel roles for cellular machinery including ion pumps and aquaporins.
Open questions include how forces induced by the cellular microenvironment direct the distinct machinery cells use to move. For example, how does the force of physical confinement lead tumor cells to utilize an “Osmotic Engine” mode in addition to actin polymerization-based migration? We hypothesize that physical confinement induces biochemical signaling pathways within cells, similar to how cells transduce signals from biomechanical stimuli (e.g., matrix stiffness, fluid shear stress) [93–96]. Furthermore, the precise mechanisms by which physical confinement leads to polarization of ion pumps and aquaporins remain to be defined. Moving forward, it will also be critical to unravel the interplay between various mechanisms of migration (e.g., actomyosin-based and water permeation-based) and identify the specific microenvironments that promote one mechanism to become more dominant over the other; we predict it may depend on the type of cell (i.e., normal versus tumor), cell mechanics (e.g., traction forces, actin organization), and/or protein expression (e.g., Rho/Rac crosstalk, aquaporins, ion pumps), as well as the physical and biochemical properties of the microenvironment. An effective strategy may be to perform such analyses on the highly migratory population of tumor cells selectively isolated from a primary tumor that bear a gene signature predictive of cancer metastasis [97]. Engineering technology, combined with mathematical modeling, cutting-edge imaging and biological approaches, and in vivo studies will likely need to be integrated holistically in order to attack these questions.
Acknowledgments
This work was supported by awards from the National Science Foundation (NSF-1159823 to KK), National Cancer Institute (R01-CA186286 to KK, U54-CA143868 to KK, SXS and F32-CA177756 to KMS), and the Kleberg Foundation (to KK, SXS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
*of interest
**of special interest
- 1.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nature Reviews Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reymond N, d'Agua BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nature Reviews Cancer. 2013;13:858–870. doi: 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- 3.Wirtz D, Konstantopoulos K, Searson PC. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nature Reviews Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedl P, Alexander S. Cancer Invasion the Microenvironment: Plasticity and Reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P. Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochemistry and cell biology. 2008;130:1147–1154. doi: 10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- 6.Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nature Reviews Cancer. 2003;3:921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- 7. Weigelin B, Bakker G-J, Friedl P. Intravital third harmonic generation microscopy of collective melanoma cell invasion. IntraVital. 2012;1:32–43. doi: 10.4161/intv.21223. Presents concrete intravital imaging evidence that collective melanoma cell invasion in vivo follows pre-existing tissue tracks without immediate tissue remodeling or destruction
- 8. Alexander S, Weigelin B, Winkler F, Friedl P. Preclinical intravital microscopy of the tumour-stroma interface: invasion, metastasis, and therapy response. Curr Opin Cell Biol. 2013;25:659–671. doi: 10.1016/j.ceb.2013.07.001. A comprehensive review with new in vivo evidence for cell migration at the tumor-stroma interface
- 9.Stroka KM, Konstantopoulos K. Physical Biology in Cancer. 4. Physical cues guide tumor cell adhesion and migration. Am J Physiol Cell Physiol. 2014;306:C98–C109. doi: 10.1152/ajpcell.00289.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nature Cell Biology. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 11.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nature Cell Biology. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 12.Lammermann T, Sixt M. Mechanical modes of 'amoeboid' cell migration. Current opinion in cell biology. 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Seminars in cell & developmental biology. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 15.Ritsma L, Steller EJ, Beerling E, Loomans CJ, Zomer A, Gerlach C, Vrisekoop N, Seinstra D, van Gurp L, Schafer R, et al. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Science translational medicine. 2012;4 doi: 10.1126/scitranslmed.3004394. 158ra145. [DOI] [PubMed] [Google Scholar]
- 16.Stolp B, Imle A, Coelho FM, Hons M, Gorina R, Lyck R, Stein JV, Fackler OT. HIV-1 Nef interferes with T-lymphocyte circulation through confined environments in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18541–18546. doi: 10.1073/pnas.1204322109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tozluoglu M, Tournier AL, Jenkins RP, Hooper S, Bates PA, Sahai E. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nature Cell Biology. 2013;15:751–762. doi: 10.1038/ncb2775. Presents a cell migration model that explores the theoretical requirements (e.g., cell-to-matrix adhesion and cellular contractility) for cell migration in different matrix geometries, including confined and unconfined spaces
- 18.Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 20.Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 21.Palecek SP, Huttenlocher A, Horwitz AF, Lauffenburger DA. Physical and biochemical regulation of integrin release during rear detachment of migrating cells. J Cell Sci. 1998;111(Pt 7):929–940. doi: 10.1242/jcs.111.7.929. [DOI] [PubMed] [Google Scholar]
- 22.Parent CA. Making all the right moves: chemotaxis in neutrophils and Dictyostelium. Curr Opin Cell Biol. 2004;16:4–13. doi: 10.1016/j.ceb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 24.Lin L, Chu YS, Thiery JP, Lim CT, Rodriguez I. Microfluidic cell trap array for controlled positioning of single cells on adhesive micropatterns. Lab Chip. 2013;13:714–721. doi: 10.1039/c2lc41070b. [DOI] [PubMed] [Google Scholar]
- 25. Le Berre M, Liu YJ, Hu J, Maiuri P, Benichou O, Voituriez R, Chen Y, Piel M. Geometric friction directs cell migration. Phys Rev Lett. 2013;111:198101. doi: 10.1103/PhysRevLett.111.198101. Demonstrates that topographical cues (i.e., tilted micropillars) can direct cell migration independently of adhesion
- 26.Lo C-M, Wang H-B, Dembo M, Wang Y-L. Cell movement is guided by the rigidity of the substrate. Biophysical Journal. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 28.Stroka KM, Aranda-Espinoza H. Neutrophils display biphasic relationship between migration and substrate stiffness. Cell Motil Cytoskeleton. 2009;66:328–341. doi: 10.1002/cm.20363. [DOI] [PubMed] [Google Scholar]
- 29.Stroka KM, Aranda-Espinoza H. Endothelial cell substrate stiffness influences neutrophil transmigration via myosin light chain kinase-dependent cell contraction. Blood. 2011;118:1632–1640. doi: 10.1182/blood-2010-11-321125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Guo WH, Rape A, Wang YL. Micropatterning cell adhesion on polyacrylamide hydrogels. Methods Mol Biol. 2013;1066:147–156. doi: 10.1007/978-1-62703-604-7_13. [DOI] [PubMed] [Google Scholar]
- 31.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bangasser BL, Odde DJ. Master equation-based analysis of a motor-clutch model for cell traction force. Cell Mol Bioeng. 2013;6:449–459. doi: 10.1007/s12195-013-0296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 34.Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14:633–639. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 35.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. Journal of Cell Biology. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang SS, Guo WH, Kim Y, Wang YL. Guidance of Cell Migration by Substrate Dimension. Biophysical Journal. 2013;104:313–321. doi: 10.1016/j.bpj.2012.12.001. Investigates how cells are able to sense dimension of the microenvironment as a guidance cue and reveals that traction stresses are significantly lower along 1D micropatterned lines in comparison with 2D substrates
- 37.Fraley SI, Feng YF, Giri A, Longmore GD, Wirtz D. Dimensional and temporal controls of three-dimensional cell migration by zyxin and binding partners. Nature Communications. 2012;3 doi: 10.1038/ncomms1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraley SI, Feng YF, Krishnamurthy R, Kim DH, Celedon A, Longmore GD, Wirtz D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nature Cell Biology. 2010;12:U598–U169. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu PH, Giri A, Sun SX, Wirtz D. Three-dimensional cell migration does not follow a random walk. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1318967111. The first demonstration that 3D cell migration does not follow a random walk
- 40.Polacheck WJ, German AE, Mammoto A, Ingber DE, Kamm RD. Mechanotransduction of fluid stresses governs 3D cell migration. Proc Natl Acad Sci U S A. 2014;111:2447–2452. doi: 10.1073/pnas.1316848111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13515–13520. doi: 10.1073/pnas.1210182109. Powerful three-dimensional microfluidic cell migration assay mimicking the in vivo tumor-vascular interface reveals different tumor microenvironments can significantly alter the tumor-endothelial interactions.
- 42.Pathak A, Kumar S. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10334–10339. doi: 10.1073/pnas.1118073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pathak A, Kumar S. Transforming potential and matrix stiffness co-regulate confinement sensitivity of tumor cell migration. Integrative Biology. 2013;5:1067–1075. doi: 10.1039/c3ib40017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mak M, Reinhart-King CA, Erickson D. Microfabricated physical spatial gradients for investigating cell migration and invasion dynamics. PLoS One. 2011;6:e20825. doi: 10.1371/journal.pone.0020825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, Martin SS, Konstantopoulos K. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. Demonstrates that metastatic breast tumor cells are able to migrate in confined spaces even in the absence of actin polymerization, myosin II-mediated contractility, or integrin-mediated adhesion
- 46.Raman PS, Paul CD, Stroka KM, Konstantopoulos K. Probing cell traction forces in confined microenvironments. Lab Chip. 2013;13:4599–4607. doi: 10.1039/c3lc50802a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stroka KM, Jiang H, Chen S-H, Tong Z, Wirtz D, Sun SX, Konstantopoulos K. Water permeation drives tumor cell migration in confined microenvironments. Cell. 2014;157:611–623. doi: 10.1016/j.cell.2014.02.052. Introduces the Osmotic Engine Model, where water permeation through the cell membrane drives cell migration in confined microenvironments; this model requires not only the function of but also the polarization of ion channels and aquaporins
- 48.Tong ZQ, Balzer EM, Dallas MR, Hung WC, Stebe KJ, Konstantopoulos K. Chemotaxis of Cell Populations through Confined Spaces at Single-Cell Resolution. Plos One. 2012;7 doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kraning-Rush CM, Carey SP, Lampi MC, Reinhart-King CA. Microfabricated collagen tracks facilitate single cell metastatic invasion in 3D. Integr Biol. 2013;5:606–616. doi: 10.1039/c3ib20196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PCDP, Ivanovska IL, Discher DE. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. Journal of Cell Biology. 2014;204:669–682. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wozniak MA, Cheng CQ, Shen CJ, Gao L, Olarerin-George AO, Won KJ, Hogenesch JB, Chen CS. Adhesion regulates MAP kinase/ternary complex factor exchange to control a proliferative transcriptional switch. Curr Biol. 2012;22:2017–2026. doi: 10.1016/j.cub.2012.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baranski JD, Chaturvedi RR, Stevens KR, Eyckmans J, Carvalho B, Solorzano RD, Yang MT, Miller JS, Bhatia SN, Chen CS. Geometric control of vascular networks to enhance engineered tissue integration and function. Proc Natl Acad Sci U S A. 2013;110:7586–7591. doi: 10.1073/pnas.1217796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DH, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hung WC, Chen SH, Paul CD, Stroka KM, Lo YC, Yang JT, Konstantopoulos K. Distinct signaling mechanisms regulate migration in unconfined versus confined spaces. J Cell Biol. 2013;202:807–824. doi: 10.1083/jcb.201302132. Elucidates the relative contributions of the negative cross-talk mechanism between Rho and Rac in unconfined versus confined migration
- 56.Wilson K, Lewalle A, Fritzsche M, Thorogate R, Duke T, Charras G. Mechanisms of leading edge protrusion in interstitial migration. Nat Commun. 2013;4:2896. doi: 10.1038/ncomms3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Prentice-Mott HV, Chang CH, Mahadevan L, Mitchison TJ, Irimia D, Shah JV. Biased migration of confined neutrophil-like cells in asymmetric hydraulic environments. Proc Natl Acad Sci U S A. 2013;110:21006–21011. doi: 10.1073/pnas.1317441110. Demonstrates asymmetric hydraulic pressure can alter the cellular decision for migration direction, suggesting that cells can push water during migration in confined spaces
- 58. Byun S, Son S, Amodei D, Cermak N, Shaw J, Kang JH, Hecht VC, Winslow MM, Jacks T, Mallick P, et al. Characterizing deformability and surface friction of cancer cells. Proc Natl Acad Sci U S A. 2013;110:7580–7585. doi: 10.1073/pnas.1218806110. Introduces a device that enables the precise measurement of a cell’s size, given its buoyant mass, the entry velocity into a constricting microchannel, and the transit velocity as it passes through the constriction
- 59.Rao JY, Hemstreet GP, 3rd, Hurst RE, Bonner RB, Min KW, Jones PL. Cellular F-actin levels as a marker for cellular transformation: correlation with bladder cancer risk. Cancer Res. 1991;51:2762–2767. [PubMed] [Google Scholar]
- 60.Rao J, Li N. Microfilament actin remodeling as a potential target for cancer drug development. Curr Cancer Drug Targets. 2004;4:345–354. doi: 10.2174/1568009043332998. [DOI] [PubMed] [Google Scholar]
- 61.Ochalek T, Nordt FJ, Tullberg K, Burger MM. Correlation between cell deformability and metastatic potential in B16-F1 melanoma cell variants. Cancer Research. 1998;48:5124–5128. [PubMed] [Google Scholar]
- 62.Xu WW, Mezencev R, Kim B, Wang LJ, McDonald J, Sulchek T. Cell Stiffness Is a Biomarker of the Metastatic Potential of Ovarian Cancer Cells. Plos One. 2012;7 doi: 10.1371/journal.pone.0046609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hawkins RJ, Piel M, Faure-Andre G, Lennon-Dumenil AM, Joanny JF, Prost J, Voituriez R. Pushing off the walls: a mechanism of cell motility in confinement. Phys Rev Lett. 2009;102:058103. doi: 10.1103/PhysRevLett.102.058103. [DOI] [PubMed] [Google Scholar]
- 64.Papadopoulos MC, Saadoun S, Verkman AS. Aquaporins and cell migration. Pflugers Arch. 2008;456:693–700. doi: 10.1007/s00424-007-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwab A, Fabian A, Hanley PJ, Stock C. Role of Ion Channels and Transporters in Cell Migration. Physiological Reviews. 2012;92:1865–1913. doi: 10.1152/physrev.00018.2011. [DOI] [PubMed] [Google Scholar]
- 66.Schwab A, Nechyporuk-Zloy V, Fabian A, Stock C. Cells move when ions and water flow. Pflugers Archiv-European Journal of Physiology. 2007;453:421–432. doi: 10.1007/s00424-006-0138-6. [DOI] [PubMed] [Google Scholar]
- 67.Goss GG, Woodside M, Wakabayashi S, Pouyssegur J, Waddell T, Downey GP, Grinstein S. ATP dependence of NHE-1, the ubiquitous isoform of the Na+/H+ antiporter. Analysis of phosphorylation and subcellular localization. J Biol Chem. 1994;269:8741–8748. [PubMed] [Google Scholar]
- 68.Grinstein S, Woodside M, Waddell TK, Downey GP, Orlowski J, Pouyssegur J, Wong DC, Foskett JK. Focal localization of the NHE-1 isoform of the Na+/H+ antiport: assessment of effects on intracellular pH. EMBO J. 1993;12:5209–5218. doi: 10.1002/j.1460-2075.1993.tb06216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mazzochi C, Bubien JK, Smith PR, Benos DJ. The carboxyl terminus of the alpha-subunit of the amiloride-sensitive epithelial sodium channel binds to F-actin. J Biol Chem. 2006;281:6528–6538. doi: 10.1074/jbc.M509386200. [DOI] [PubMed] [Google Scholar]
- 70.Klein M, Seeger P, Schuricht B, Alper SL, Schwab A. Polarization of Na+/H+ and Cl-/HCO3- exchangers in migrating renal epithelial cells. Journal of General Physiology. 2000;115:599–607. doi: 10.1085/jgp.115.5.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ritter M, Schratzberger P, Rossmann H, Woll E, Seiler K, Seidler U, Reinisch N, Kahler CM, Zwierzina H, Lang HJ, et al. Effect of inhibitors of Na+/H+-exchange and gastric H+/K+ ATPase on cell volume, intracellular pH and migration of human polymorphonuclear leucocytes. British Journal of Pharmacology. 1998;124:627–638. doi: 10.1038/sj.bjp.0701864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chae YK, Woo J, Kim MJ, Kang SK, Kim MS, Lee J, Lee SK, Gong G, Kim YH, Soria JC, et al. Expression of Aquaporin 5 (AQP5) Promotes Tumor Invasion in Human Non Small Cell Lung Cancer. Plos One. 2008;3 doi: 10.1371/journal.pone.0002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jung HJ, Park JY, Jeon HS, Kwon TH. Aquaporin-5: A Marker Protein for Proliferation and Migration of Human Breast Cancer Cells. Plos One. 2011;6 doi: 10.1371/journal.pone.0028492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sidhaye VK, Chau E, Srivastava V, Sirimalle S, Balabhadrapatruni C, Aggarwal NR, D'Alessio FR, Robinson DN, King LS. A Novel Role for Aquaporin-5 in Enhancing Microtubule Organization and Stability. Plos One. 2012;7 doi: 10.1371/journal.pone.0038717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Y, Rice W, Gu Z, Li J, Huang J, Brenner MB, Van Hoek A, Xiong J, Gundersen GG, Norman JC, et al. Aquaporin 2 promotes cell migration and epithelial morphogenesis. J Am Soc Nephrol. 2012;23:1506–1517. doi: 10.1681/ASN.2012010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tamma G, Lasorsa D, Ranieri M, Mastrofrancesco L, Valenti G, Svelto M. Integrin signaling modulates AQP2 trafficking via Arg-Gly-Asp (RGD) motif. Cell Physiol Biochem. 2011;27:739–748. doi: 10.1159/000330082. [DOI] [PubMed] [Google Scholar]
- 77.Tham DK, Moukhles H. Regulation of Kir4.1 and AQP4 expression and stability at the basolateral domain of epithelial MDCK cells by the extracellular matrix. Am J Physiol Renal Physiol. 2011;301:F396–F409. doi: 10.1152/ajprenal.00315.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jiang HY, Sun SX. Cellular Pressure and Volume Regulation and Implications for Cell Mechanics. Biophysical Journal. 2013;105:609–619. doi: 10.1016/j.bpj.2013.06.021. Describes a mathematical model of cellular volume and pressure regulation and impacts on cell mechanics when cells are exposed to an externally applied load and demonstrates the importance of water flow in cells
- 79.Stroka KM, Jiang H, Chen S-H, Tong Z, Wirtz D, Sun SX, Konstantopoulos K. Water permeation drives tumor cell migration in confined microenvironments. Cell. 2014 doi: 10.1016/j.cell.2014.02.052. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison TJ. Non-equilibration of hydrostatic pressure in blebbing cells. Nature. 2005;435:365–369. doi: 10.1038/nature03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iwasaki T, Wang YI. Cytoplasmic force gradient in migrating adhesive cells. Biophysical Journal. 2008;94:L35–L37. doi: 10.1529/biophysj.107.124479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jaeger M, Carin M, Medale M, Tryggvason G. The osmotic migration of cells in a solute gradient. Biophysical Journal. 1999;77:1257–1267. doi: 10.1016/S0006-3495(99)76977-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keren K, Yam PT, Kinkhabwala A, Mogilner A, Theriot JA. Intracellular fluid flow in rapidly moving cells. Nature Cell Biology. 2009;11 doi: 10.1038/ncb1965. 1219-U1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mitchison TJ, Charras GT, Mahadevan L. Implications of a poroelastic cytoplasm for the dynamics of animal cell shape. Seminars in Cell & Developmental Biology. 2008;19:215–223. doi: 10.1016/j.semcdb.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oster GF, Perelson AS. The physics of cell motility. J Cell Sci Suppl. 1987;8:35–54. doi: 10.1242/jcs.1987.supplement_8.3. [DOI] [PubMed] [Google Scholar]
- 86.Peskin CS, Odell GM, Oster GF. Cellular motions thermal fluctuations the Brownian ratchet. Biophys J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Spagnolie SE, Lauga E. Jet propulsion without inertia. Physics of Fluids. 2010;22 [Google Scholar]
- 88.Taber LA, Shi YF, Yang L, Bayly PV. A Poroelastic Model for Cell Crawling Including Mechanical Coupling between Cytoskeletal Contraction and Actin Polymerization. Journal of Mechanics of Materials and Structures. 2011;6:569–589. doi: 10.2140/jomms.2011.6.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Charras GT, Mitchison TJ, Mahadevan L. Animal cell hydraulics. J Cell Sci. 2009;122:3233–3241. doi: 10.1242/jcs.049262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Machida Y, Ueda Y, Shimasaki M, Sato K, Sagawa M, Katsuda S, Sakuma T. Relationship of aquaporin 1, 3, and 5 expression in lung cancer cells to cellular differentiation, invasive growth, and metastasis potential. Human Pathology. 2011;42:669–678. doi: 10.1016/j.humpath.2010.07.022. [DOI] [PubMed] [Google Scholar]
- 91.Tominaga T, Ishizaki T, Narumiya S, Barber DL. p160ROCK mediates RhoA activation of Na-H exchange. EMBO J. 1998;17:4712–4722. doi: 10.1093/emboj/17.16.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Magalhaes MA, Larson DR, Mader CC, Bravo-Cordero JJ, Gil-Henn H, Oser M, Chen X, Koleske AJ, Condeelis J. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J Cell Biol. 2011;195:903–920. doi: 10.1083/jcb.201103045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schwartz MA. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol. 2010;2:a005066. doi: 10.1101/cshperspect.a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang P, Zhu F, Lee NH, Konstantopoulos K. Shear-induced interleukin-6 synthesis in chondrocytes: roles of E prostanoid (EP) 2 and EP3 in cAMP/protein kinase A- and PI3-K/Akt-dependent NF-kappaB activation. J Biol Chem. 2010;285:24793–24804. doi: 10.1074/jbc.M110.110320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rape A, Guo WH, Wang YL. Microtubule depolymerization induces traction force increase through two distinct pathways. Journal of Cell Science. 2011;124:4233–4240. doi: 10.1242/jcs.090563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang P, Zhu F, Konstantopoulos K. The antagonistic actions of endogenous interleukin-1beta and 15-deoxy-Delta12,14-prostaglandin J2 regulate the temporal synthesis of matrix metalloproteinase-9 in sheared chondrocytes. J Biol Chem. 2012;287:31877–31893. doi: 10.1074/jbc.M112.362731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patsialou A, Wang Y, Lin J, Whitney K, Goswami S, Kenny PA, Condeelis JS. Selective gene-expression profiling of migratory tumor cells in vivo predicts clinical outcome in breast cancer patients. Breast Cancer Res. 2012;14:R139. doi: 10.1186/bcr3344. [DOI] [PMC free article] [PubMed] [Google Scholar]