Figure 2. Native intestinal bacterial species conserved across murine and human microbiota are predicted to inhibit C. difficile infection.
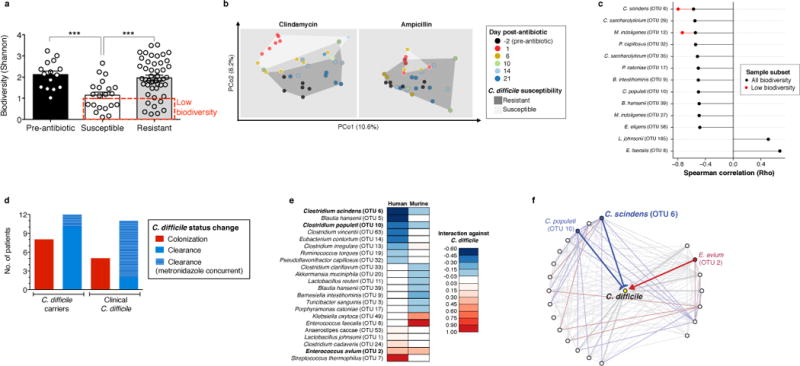
Intestinal microbiota alpha diversity (a) and Beta diversity (weighted UniFrac distances) (b) of antibiotic-naïve (n=15) and antibiotic-exposed animals susceptible (n=21) or resistant (n=47) to C. difficile infection. Correlation of individual bacterial OTUs with susceptibility to C. difficile infection (c). Colonization (C. difficile-negative to -positive) and clearance (C. difficile-positive to –negative) events among C. difficile-diagnosed and carrier patients included in microbiota time-series inference modeling (d). Bacterial species with strong C. difficile interactions in human and murine microbiota models (e) that exist in a conserved subnetwork predicted to inhibit (blue) or positively associate (red) with C. difficile (f). ***P<0.001. In (c), P<0.0005 (“any biodiversity”, n=68) or P<0.05 (“Low biodiversity”, Shannon≤1 (n=16 animals). Center values (mean), error bars (s.e.m.).
