Abstract
Objective
This study examined the role of SNP rs2858056 of the MPG gene on the incidence and severity of rheumatoid arthritis (RA).
Methods
This cohort study enrolled 365 RA patients and 375 age- and gender-matched healthy controls, all of whom had Han Chinese ethnicity and were from Taiwan. Gene polymorphism of the SNP rs2858056 of MPG was determined from genomic DNA. Allelic frequencies and genotypes were compared among cases and controls. Quantitation of rs2858056 copy number variation (CNV) was determined. Serum samples from RA patients and controls were analyzed to determine serum levels of MPG. The relationship between rs2858056 polymorphism and clinical manifestations of RA was evaluated.
Results
Our results indicated a statistically significant difference in genotype frequency distributions at rs2858056 for RA patients and controls (p = 0.05) and a significant difference in allelic frequency in patients and controls (p = 0.04). Furthermore, there was a significantly greater level of serum MPG protein in patients than controls (p < 0.001). However, the cases and controls had no significant differences in MPG CNV (p = 0.12). We also did not detect any association of the MPG rs2858056 with rheumatoid factor (RF), extraarticular involvement, or bone erosion in the RA patients.
Conclusion
Our study suggests that RA is associated with a polymorphism in the MPG gene (rs2858056) and increased serum level of the MPG protein.
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease characterized by joint inflammation and numerous peripheral inflammatory manifestations [1]. Patients experience chronic synovitis, destruction of joint tissue (cartilage and bone), and impaired joint function [2]. There are heritable risk factors [3–4], but the genetic basis of RA is not well understood. Previous studies identified nearly 60 genetic loci associated with susceptibility to RA in multiple populations. These include HLA-DRB1 [5], PTPN22 (encoding tyrosin-protein phosphotase non receptor type 22) [6], PADI4 (protein-arginine deiminase type 4) [7], TNFAIP3 (TNF-induced protein 3) [8], and CTLA-4 (cytotoxic T-lymphocyte protein4) [9].
Disruption of DNA repair appears increase the susceptibility to RA. DNA repair enzymes modulate the DNA damage from free-radicals, by repairing DNA mainly through four pathways: mismatch repair (MMR), nucleotide excision repair (NER), direct reversal, and base excision repair (BER). The oxidized base 7,8-dihydroxy-8-oxoguanine (8-oxo-G) is a mutagenic product of oxidative DNA damage and the BER pathway can excise misincorporated 8-oxo-G. In particular, N-methylpurine DNA glycosylase (MPG; MIM 156565) removes diverse damaged bases, including cytotoxic and mutagenic alkylation adducts of purine [10]. Previous studies have examined the role of the MPG gene in breast cancer and lung cancer [11–13]. Recently, Shao et al. reported that the DNA repair system was involved in the pathology of RA [14]. In particular, the T cells of RA patients failed to produce sufficient amounts of the DNA repair kinase, ataxia telangiectasia mutated (ATM).
Our previous study [15] investigated the association of four gene polymorphisms (rs3176364, rs710079, rs2858056, and rs2541632) with susceptibility to RA in 384 Taiwanese individuals (192 RA patients and 192 controls). The results showed that rs710079 and rs2858056 polymorphisms and the GCGC haplotype were associated with increased risk of RA progression. In the present study, we sought to confirm the role of MPG rs2858056 gene polymorphism, copy number variation (CNV), and level of serum MPG protein with the risk of RA. Thus, we compared the genotype and allele frequencies of the MPG polymorphism rs2858056, CNV, and serum MPG levels in RA patients and healthy individuals of Han Chinese ethnicity from central Taiwan. Furthermore, the relationship between rs2858056 polymorphism and clinical manifestations of RA was evaluated.
Materials and Methods
Patient selection
We enrolled 365 patients (285 women and 80 men) diagnosed with RA, based on the 1987 revised criteria of the American College of Rheumatology [16], and 375 unrelated healthy individuals (291 women and 84 men) from the general population were selected from health examination. The cases and controls were matched for age and sex, were of Han Chinese ethnicity, and lived in central Taiwan. The age of the RA patients was 22–78 (mean 51.1 ± 5.9) years. The age of the volunteers was 20–75 (mean 49.3 ± 5.1) years. The duration of disease in RA patients was 0–19 (mean 5.6 ± 1.4) years. The baselines characteristic of 365 patients with RA were shown in Table 1. Nephelometry was used to measure the rheumatoid factor (RF), and values of 20 IU/mL or more were considered positive. The presence or history of extra-articular manifestations ((1) subcutaneous rheumatoid nodules; (2) cutaneous Vasculitis; (3) eosinophilia; (4) lymphadenopathy; (5) pulmonary disease (pleurisy, interstitial fibrosis, nodular lung or pulmonary hypertension); (6) cardiac disease (pericarditis or conduction defect); (7) noncompressive neuropathy; (8) Raynaud’s phenomenon; or (9) sicca syndrome.) was recorded for all RA patients [17]. Radiographs of hands, wrists, and feet of patients were taken, and joint erosion was evaluated by a rheumatologist and a radiologist. We noted only the presence or absence of erosion, with no calculation of radiological score. To assess inflammatory activity we measured C-reactive protein (CRP), and erythrocyte sedimentation rate (ESR). All samples for genomic DNA isolation were collected by venipuncture. This study was approved by the Institutional Review Board (IRB) of China Medical University Hospital (Taichung, Taiwan) prior to patient enrollment. All participants provided written informed consent.
Table 1. Demographics and characteristics of MPG genetic polymorphisms in RA patients.
| MPG-3644 | (rs2858056) | P value | |||
|---|---|---|---|---|---|
| GG(n = 53) | CG(n = 135) | CC(n = 177) | total(n = 365) | ||
| Age. Years (SD)* | 51.2 (6.1) | 49.7 (5.2) | 52.3 (6.5) | 51.1 (5.9) | 0.67 |
| Sex, female, n (%) | 42 (79.2) | 108 (80.0) | 135 (76.3) | 285 (78.1) | 0.71 |
| Disease duration, years, | |||||
| median (IQR) * | 6 (8) | 5 (7) | 6 (8) | 6 (8) | 0.43 |
| ESR**, mm/h (SD) * | 35 (7.6) | 28 (5.3) | 32 (6.4) | 31 (6.2) | 0.72 |
| CRP**, mg/dl (SD) * | 1.7 (0.6) | 1.5 (0.4) | 1.5 (0.4) | 1.5 (0.4) | 0.93 |
| Medications | |||||
| Hydroxylchroloqine, n (%) | 42 (79.2) | 105 (77.8) | 137 (77.4) | 284 (77.8) | 0.96 |
| Sulfasalazin, n (%) | 29 (54.7) | 67 (49.6) | 92 (51.9) | 187 (51.2) | 0.81 |
| Methotrexate, n (%) | 43 (81.1) | 110 (81.5) | 142 (80.2) | 295 (80.8) | 0.96 |
| Leflunomide, n(%) | 21 (39.6) | 62 (45.9) | 70 (39.5) | 153 (41.9) | 0.49 |
| NSAID**, n (%) | 46 (86.8) | 121 (89.6) | 156 (88.1) | 323 (88.5) | 0.85 |
| Steroid, n (%) | 27 (50.9) | 75 (55.6) | 95 (53.7) | 197 (53.9) | 0.84 |
| Anti-TNF**, n (%) | 8 (15.1) | 12 (8.9) | 15 (8.5) | 35 (9.6) | 0.34 |
* Result are expressed as percentages, median (interquartile range, IQR), or mean (SD) as approciate.
** ESR, erythrocyte sedimentation rate; CRP, C-reacive protein; NSAID, non-steroidal anti-inflammatory drug; TNF, tumor necrosis factor
SNP selection
MPG SNPs genotypes information was downloaded in December 2008 from the HapMap CHB + JPT population. HapMap genotypes were analyzed within Haploview and Tag SNPs were selected using the Tagger function by applying the following additional criteria: (i) a threshold minor allele frequency (MAF) in the HapMap CHB + JPT population of 0.05 for”tag SNPs”; and (ii) probe/primers that pass the qualification as recommended by the manufacturer (Applied Biosystems), to ensure a high genotyping success rate.
Genomic DNA extraction and genotyping (polymerase chain reaction and restriction enzyme analysis)
Genomic DNA was prepared from peripheral blood according to the standard protocols of the DNA extraction kit (Genomic DNA kit, Roche, USA). Polymerase chain reaction (PCR) was used to identify the MPG rs2858056 polymorphism. Polymerase chain reaction was carried out in a total volume of 50 μL, containing 50 ng of genomic DNA, 2–6 pmol of each primer, 1× Taq polymerase buffer (1.5 mM MgCl2) and 0.5 units of AmpliTaq DNA polymerase (Perkin Elmer, Foster City, CA, USA). In the study of the EGFR rs2227983 SNP, the primers used were forward-5'-AGAGCTGAGATCACGCCATT-3' and reverse-5'-AGGGCATCCACTAGGAGGTT-3'. PCR amplification was performed in a PCR thermal cycler (GeneAmp PCR System 2400, Perkin Elmer). The PCR cycling conditions were as follows: one cycle at 95°C for 5 min, 35 cycles at 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 45 seconds. One final cycle of extension at 72°C for 7 min, then holding at 25°C. The MPG rs2858056 SNP was analyzed by PCR amplification followed by restriction enzyme analysis with Hae III. Four fragments of 29 bp, 112 bp, 86 bp and 101 bp were present if the product was excised (CC homozygote). The uncut band showed up as three fragments of 29 bp, 169bp and 130 bp length on the gel. The reaction was then incubated for overnight at 65°C, and then 10μl of the products were loaded into a 3% agarose gel containing ethidium bromide for electrophoresis. The MPG rs2858056 SNP was categorized as excisable (CC homozygote), non-excisable (CG homozygote), and (GG heterozygote).
Quantitation of MPG copy number
A pre-designed TaqMan Copy Number assay (Assay ID Hs00453520_cn, Applied Biosystems, USA) was used for quantitation of MPG copy number. PCR reactions contained 2× TaqMan PCR Master Mix (Applied Biosystems, USA), 20× TaqMan Copy Number Assay (Applied Biosystems, USA), 20× TaqMan Copy Number Reference Assay-RNase P-Human (Cat No. 4403328, Applied Biosystems, USA), and 10 ng of genomic DNA. The TaqMan RNase P control reagents kit (Applied Biosystems, USA) was used as an endogenous control. After initial denaturation for 10 min at 95°C, 40 cycles were run, each consisting of denaturation (95°C for 15 s), and annealing (60°C for 60 s). The fluorescence signal was measured using the ABI Prism 7900 Real Time PCR System.
Quantitative determination of MPG in serum
Serum samples were diluted with buffer so they were within the range of the standard curve, and were then quantified by an enzyme-linked immunosorbent assay (ELISA, E97625Hu, Uscn Life Science Inc, Wuhan, China). The serum MPG was bound to monoclonal mouse antibodies against human MPG, which were immobilized on the surface of microtiter plates. After washing, bound MPG was quantified by adding a rabbit anti-human MPG antibody. The bound rabbit antibody was measured by a peroxidase-labeled goat anti-rabbit antibody. The amount of converted substrate was directly proportional to the amount of bound human MPG, and was determined by measurement of OD450 nm.
Statistical analysis
The genotype and allele frequencies of MPG SNP rs2858056 in RA patients and controls were compared using the chi-square test. Allelic frequency was expressed as a percentage of the total number of alleles. Differences between genotypes and allele frequencies and deviations from the Hardy-Weinberg equilibrium were analyzed using the χ2 test and Fisher’s exact test, depending on the minimum expected values. A p-value less than 0.05 was considered statistically significant. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated for allelic frequency of MPG SNP rs2858056 and for CNV. The Mann-Whitney U test or the Kruskal-Wallis test was used for non-parametric comparisons, and Student’s t-test or one-way analysis of variance was used for parametric comparisons. All statistical analysis was performed with SPSS version 11.
Results
Genotype and allele frequency distributions of MPG rs2858056 SNP
Table 2 shows the frequencies of the genotype MPG rs2858056 in the RA and control groups. There was a significant difference in the distribution of the MPG rs2858056 gene polymorphism between patients and controls (OR for GG genotype: 1.79, 95% CI: 1.11–2.89, p = 0.05). In addition, there was a significant difference in allele frequency of MPG rs2858056 between patients and controls (OR for G allele: 1.26, 95% CI: 1.01–1.57, p = 0.04). The statistic power with alpha at 0.05 was more than 89% in this study (89.7%).
Table 2. Genotype and allele frequencies of MPG genetic polymorphisms in RA patients (n = 365) and controls (n = 375).
| Patients, n (%) | Controls, n (%) | OR (95% CI) | P value | |
|---|---|---|---|---|
| MPG-3644 (rs2858056) | ||||
| CC | 177 (48.5) | 197 (52.5) | Ref | 0.05 |
| CG | 135 (37.0) | 145 (38.7) | 1.04 (0.76–1.41) | |
| GG | 53 (14.5) | 33 (8.8) | 1.79 (1.11–2.89) | |
| Allele frequency | ||||
| Allele C | 489 (67.0) | 539 (71.9) | Ref | 0.04 |
| Allele G | 241 (33.0) | 211 (28.1) | 1.26 (1.01–1.57) |
Abnormal MPG CNV in RA patients
Blood leukocyte gDNA samples were available from 235 RA patients (64.4%) and 223 healthy controls (59.5%); for the other subjects, insufficient gDNA was available for quantification of MPG CNV. The presence of RA was not associated with abnormal MPG CNV (Table 3, p = 0.12, Fisher’s test). Most patients and controls had 2 copies of the MPG gene (100% of controls and 98.3% of patients). However, 4 RA patients (1.7%) had 3 copies of the MPG gene, and none of the healthy controls (0%) had 3 copies.
Table 3. Distribution of MPG copy number variation (CNV) in RA patients (n = 235) and controls (n = 223).
| Controls, n (%) | Patients, n (%) | P value(Chi-square test) | P value(Fisher's test) | |
|---|---|---|---|---|
| MPG copies | ||||
| 2 | 223 (100) | 231 (98.3) | 0.05 | 0.12 |
| 3 | 0 (0) | 4 (1.7) |
Increased serum level of MPG in RA patients
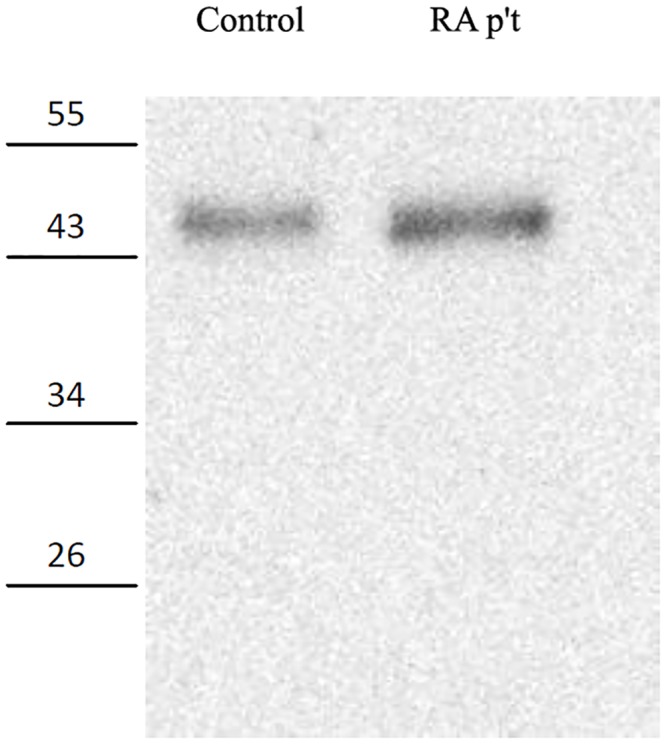
Serum levels of MPG were available from 40 RA patients (10.7%) and 39 healthy controls (10.4%); for the other subjects, insufficient serum was available for quantification of serum MPG (Fig. 1). The results indicate a significantly higher level of MPG in patients than in controls (4.90 ± 2.93 ng/mL vs. 1.96 ± 0.85 ng/mL, p < 0.001). This was confirmed by western blot assays for MPG (Fig. 2).
Fig 1. Serum level of MPG protein in RA patients (n = 40) and controls (n = 39).
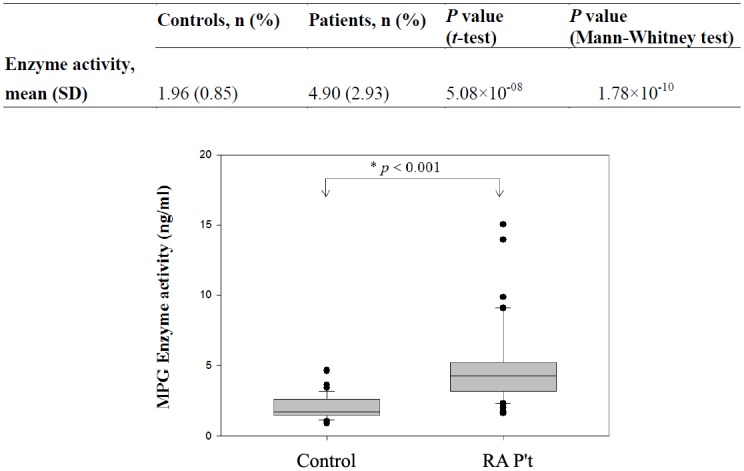
Fig 2. Representative MPG western blot assays of an RA patient and a control patient.
Clinical signs and findings and genotype of MPG polymorphisms in RA patients
Clinical manifestations and laboratory findings of the RA patients are shown in Table 4. The associations of the genotype MPG rs2858056 with particular clinical features of RA were examined in the 365 Chinese patients. We did not detect any association of the MPG rs2858056 genotype with the RF or extra-articular involvement in the RA patients (P = 0.90 and P = 0.18, respectively). We observed increased frequencies of bone erosion among the RA patients with GG genotype (54.7%) when compared with CC genotype (37.9%). However, this difference did not reach statistical significance (P = 0.07, χ2 = 5.33).
Table 4. Relationships between MPG-3644 (rs2858056) genotype and clinical signs and findings in patients with RA.
| GG(n = 53) | CG(n = 135) | CC(n = 177) | total(n = 365) | P value | |
|---|---|---|---|---|---|
| Rheumatoid factor (+) | 38(71.7%) | 101(74.8%) | 130(73.4%) | 269(73.7%) | 0.90 |
| Extra-articular involvement | 21(39.6%) | 65 (48.1%) | 67 (37.9%) | 153(41.9%) | 0.18 |
| Bone erosion | 29(54.7%) | 62 (45.9%) | 67 (37.9%) | 156(42.7%) | 0.07 |
Discussion
Inflammation results from a variety of cellular signals and the formation of reactive oxygen species (ROS), which threaten the integrity of cellular DNA. The oxidized base 7,8-dihydroxy-8-oxoguanine (8-oxo-G) is a mutagenic product of oxidative DNA damage. However, misincorporated 8-oxo-G can be excised by the base excision repair (BER) pathway, and many DNA repair enzymes, including MUTYH, OGG1, MPG, TDG, UNG, EGFR, and SMUG1, are part of, or related to, this pathway [10].
Previous studies have examined the roles of genes related to DNA repair in various cancers, including lung cancer, breast cancer [11–13], gastric cancer [17], head and neck squamous cell carcinomas (HNSCC) [18], and colorectal cancer (CRC) [19]. In addition, there is evidence that ROS-induced DNA damage may contribute to tumor development in a murine model of chronic inflammation [20]. These studies led to our examination of genetic and allelic variations of enzymes that repair ROS-induced DNA damage and the risk of RA.
Our previous research examined the gene and allele frequencies of polymorphisms in DNA repair-related genes (including OGG1, MPG, UNG, and EGFR), and compared the haplotypes of RA patients and healthy controls in a Taiwan population [15, 21–23]. In particular, our 2010 study [15] investigated that the association of four MPG polymorphisms (rs3176364, rs710079, rs2858056, and rs2541632) with RA by examination of 192 patients and 192 healthy controls. The results indicated statistically significant differences in genotype frequency distributions of rs710079 and rs2858056 between RA patients and controls (p = 0.04 and p = 0.029, respectively). In the present study, we increased the case numbers (365 RA patients, 375 healthy controls) to increase the statistical power of the analysis and focused on the association of the MPG rs2858056 polymorphism with the risk of RA. The results showed that there was a statistically significant difference in genotype and allele frequency distributions between RA patients and healthy controls. In particular, individuals with the G genotype at rs2858056 have increased risk for RA. We also used Hardy—Weinberg equilibrium (HWE) testing for data quality control. The results showed that the control group of rs2858056 SNP is under the null hypothesis of no departure from HWE. In the data set, the p value under 0.05 is obtained in RA patients group of rs2858056 SNP, giving the evidence of departure from HWE. It shown that rs2858056 SNP maybe plays an important role in RA development.
We also evaluated the relationship of RA with MPG CNV (Table 2) and MPG expression (Fig. 1). Several large-scale whole-genome studies and focused studies have documented widespread CNV in the human genome [24–27]. This variation accounts for roughly 12% of human genomic DNA, and each variation ranges from about one kilobase to several megabases in size [28]. Gene copy number is often altered in the presence of systemic disease. For instance, the EGFR copy number is elevated in non-small cell lung cancer [29], an elevated copy number of CCL3L1 is associated with lower susceptibility to HIV infection [30], and a low copy number of FCGR3B (the CD16 cell-surface immunoglobulin receptor) increases the susceptibility to systemic lupus erythematosus and similar inflammatory autoimmune disorders [27]. However, although CNV has been proposed to explain the elusive hereditary causes of complex diseases, such as rheumatoid arthritis, the most common CNVs have little or no role in causing disease [31]. Similarly, the results of the present study indicate that RA patients did not have abnormal MPG CNV. Most patients and controls had 2 copies of the MPG gene (100% of controls, 98.3% of patients). However, 4 of the patients (1.7%) had 3 copies, but none of the healthy controls had 3 copies.
We collected blood samples from patients and controls and determined serum MPG levels to verify the functional expression of MPG rs2858056 SNP. The results indicated significantly higher levels of serum MPG protein in patients than controls. In agreement, our research on the role of EGFR gene rs17337023 polymorphism, copy number variation, and serum levels in RA indicated no effect of CNV of this gene (unpublished observation).
The etiology of RA is still incompletely characterized, and this is a very active area of research worldwide. The present investigation examined whether an MPG SNP is associated with RA susceptibility. To our knowledge, from a search of reports of the literature on MEDLINE, an association between the gene polymorphism of MPG rs2858056 and RA has not been demonstrated before. In the present study, the significant differences were observed in the allelic frequencies and genotype of MPG rs2858056 gene polymorphism between RA patients and healthy controls. However, we did not find any relationships between MPG rs2858056 and clinical manifestations and laboratory profiles (RF and extra-articular involvement) of RA in the Chinese patients. Although we observed increased frequencies of bone erosion among RA patients with the GG genotype (54.7%) when compared with CC genotype (37.9%), this difference did not reach statistical significance (P = 0.07). A much larger study group will be required to verify the relationship between disease phenotypes and genotypes.
The generalizability of our results is limited because all of the patients were from single center in Taiwan. Nonetheless, our results strongly suggest a significant role of MPG rs2858056 gene polymorphisms in the risk for RA among subjects from Taiwan. The identification of MPG alterations as a risk factor for RA in Taiwan requires further clinical testing worldwide, especially in populations with different ethnicities.
In conclusion, the present study showed a correlation between increased MPG expression and MPG gene polymorphism with RA.
Acknowledgments
We thank all colleagues at the Genetic Center, Department of Medical Research, China Medical University Hospital for their feedback and technical support.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was supported by the National Science Council in Taiwan (NSC99-2314-B-039-005). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Feldmann M, Brennan FM, Maini RN. Rheumatoid arthritis. Cell. 1996;85:307–310. [DOI] [PubMed] [Google Scholar]
- 2. Harris ED Jr. Rheumatoid arthritis: pathophysiology and implications for therapy. N Engl J Med. 1990; 322:1277–1289. [DOI] [PubMed] [Google Scholar]
- 3. Wordsworth BP, Bell JI. Polygenic susceptibility in rheumatoid arthritis. Ann Rheum Dis 1991; 18:343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gomolka M, Menninger H, Saal JE, Kemmel EM, Albert ED, Niwa O, et al. Immunoprinting: various genes are associated with increased risk to develop rheumatoid arthritis in different groups of adult patients. J Mol Med. 1995; 73:19–29. [DOI] [PubMed] [Google Scholar]
- 5. Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987; 30:1205–1213. [DOI] [PubMed] [Google Scholar]
- 6. Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet; 2004; 75:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003; 34:395–402. [DOI] [PubMed] [Google Scholar]
- 8. Kool M, van Loo G, Waelput W, De Prijck S, Muskens F, Sze M, et al. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity. 2011; 35:82–96. 10.1016/j.immuni.2011.05.013 [DOI] [PubMed] [Google Scholar]
- 9. Plenge RM, Padyukov L, Remmers EF, Purcell S, Lee AT, Karlson EW, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005; 77:1044–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Broderick P, Bagratuni T, Vijayakrishnan J, Lubbe S, Chandler I, Houlston RS. Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer; 2006. 6:243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cerda SR, Turk PW, Thor AD, Weitzman SA. Altered expression of the DNA repair protein, N-methylpurine-DNA glycosylase (MPG) in breast cancer. FEBS Lett. 1998; 431:12–18. [DOI] [PubMed] [Google Scholar]
- 12. Rusin M, Samojedny A, Harris CC, Chorazy M. Novel genetic polymorphisms in DNA repair genes: O(6)-methylguanine-DNA methyltransferase (MGMT) and N-methylpurine-DNA glycosylase (MPG) in lung cancer patients from Poland. Hum Mutat. 1999; 14:269–270. [DOI] [PubMed] [Google Scholar]
- 13. Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006; 27:560–7. [DOI] [PubMed] [Google Scholar]
- 14. Shao L, Fujii H, Colmegna I, Oishi H, Goronzy JJ, Weyand CM. Deficiency of the DNA repair enzyme ATM in rheumatoid arthritis. J Exp Med. 2009; 206:1435–1449. 10.1084/jem.20082251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen SY, Wan L, Huang CM, Huang YC, Sheu JJ, Lin YJ, et al. Genetic polymorphisms of the DNA repair gene MPG may be associated with susceptibility to rheumatoid arthritis. J Appl Genet. 2010; 51:519–521. 10.1007/BF03208883 [DOI] [PubMed] [Google Scholar]
- 16. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1998; 31:315–324. [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y, Liu X, Fan Y, Ding J, Xu A, Zhou X, et al. Germline mutations and polymorphic variants in MMR, E-cadherin and MYH genes associated with familial gastric cancer in Jiangsu of China. Int J Cancer. 2006; 119:2592–2596. [DOI] [PubMed] [Google Scholar]
- 18. Sliwinski T, Markiewicz L, Rusin P, Pietruszewska W, Olszewski J, Morawiec-Sztandera A, et al. Polymorphisms of the DNA base excision repair gene MUTYH in head and neck cancer. Exp Oncol. 2009; 31:57–59. [PubMed] [Google Scholar]
- 19. Chen H, Xu L, Qi Q, Yao Y, Zhu M. A haplotype variation affecting the mitochondrial transportation of hMYH protein could be a risk factor for colorectal cancer in Chinese. BMC Cancer. 2008; 8:269 10.1186/1471-2407-8-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meira LB, Bugni JM, Green SL, Lee CW, Pang B, Borenshtein D, et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice J Clin Invest. 2008; 118:2516–2525. 10.1172/JCI35073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lo SF, Wan L, Huang CM, Lin HC, Chen SY, Liu SC, et al. Genetic polymorphisms of the DNA repair gene UNG are associated with the susceptibility of rheumatoid arthritis. Rheumatol Int. 2012; 32:3723–3727. 10.1007/s00296-011-2185-3 [DOI] [PubMed] [Google Scholar]
- 22. Lo SF, Wan L, Lin HC, Huang CM, Chen SY, Liu SC, et al. Association of rheumatoid arthritis risk with EGFR genetic polymorphisms in Taiwan's Han Chinese population. Rheumatol Int. 2012; 32:2301–2306. 10.1007/s00296-011-1961-4 [DOI] [PubMed] [Google Scholar]
- 23. Chen SY, Wan L, Huang CM, Huang YC, Sheu JJ, Lin YJ, et al. Association of the C-285T and A5954G polymorphisms in the DNA repair gene OGG1 with the susceptibility of rheumatoid arthritis. Rheumatol Int. 2012; 32:1165–1169. 10.1007/s00296-010-1738-1 [DOI] [PubMed] [Google Scholar]
- 24. Sebat J, Lakshmi B, Troge J, Alexander J, Young JL, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004; 305:525–528. [DOI] [PubMed] [Google Scholar]
- 25. Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006; 444:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, et al. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007; 80:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006; 439:851–855. [DOI] [PubMed] [Google Scholar]
- 28. Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010; 61:437–455. 10.1146/annurev-med-100708-204735 [DOI] [PubMed] [Google Scholar]
- 29. Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005; 97:643–655. [DOI] [PubMed] [Google Scholar]
- 30. Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005; 307:1434–1440. [DOI] [PubMed] [Google Scholar]
- 31. Wellcome Trust Case Control Consortium, Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010; 464:713–720. 10.1038/nature08979 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.


