Abstract
Cleft lip with or without cleft palate (CL/P) is one of the most common birth defects; it is a multifactorial disease affecting > 1/1,000 live births in Europe, and its etiology is largely unknown, although it is very likely genetic and environmental factors contribute to this malformation. Orofacial development is a complex process involving many genes and signaling pathways. Mutations in the gene for the interferon regulatory factor 6 (IRF6) cause a hereditary dominant malformation syndrome including CL/P, and polymorphisms are associated with non-syndromic CL/P (MIM 119530). Five SNPs at the locus with high heterozygosity in Caucasian populations were chosen for the present research due to their very strong association with CL/P. A case–parent trio study was performed using 292 samples from Mexico. Association with the rs1319435-C/C genotype (P = 0.02) was found in patients (73) as compared to pseudocontrols (219), while the genotype rs1319435-T/C was related with protection (P = 0.041) in the triad design. Significant over-transmission of the G allele for marker rs2235375 (P = 0.049) was found. Only the TACGT haplotype was diminished in the affected child, either in single (P = 0.0208) or double (P = 0.0208) dose. The pairwise analysis showed rs2235543 and rs2235371 were in strong linkage disequilibrium. These results point to a substantial contribution of IRF6 in the etiology of non-syndromic CL/P in a sample of the Mexican population.
Keywords: Birth defects, Congenital abnormalities, Cleft lip, Cleft palate, Craniofacial development, IRF6
Highlights
-
•
Cleft lip with or without cleft palate (CL/P) is one of the most common birth defects.
-
•
Association with the rs1319435-C/C genotype was found in patients as compared to pseudocontrols.
-
•
In the triad design, the genotype rs1319435-T/C was related with protection.
-
•
The IRF6 contributes to the etiology of non-syndromic CL/P in a sample of the Mexican population.
Introduction
Cleft lip with or without cleft palate (CL/P) is a common congenital structural anomaly notable for significant lifelong morbidity and its complex etiology. The prevalence of orofacial clefts varies from 1/500 to 1/2,500 births depending on geographic origin [Vanderas, 1987], racial and ethnic backgrounds [Croen et al., 1998, Tolarova and Cervenka, 1998], and socioeconomic status [Murray et al., 1997]. Non-syndromic CL/P (MIM 119530) is a complex multifactorial disease affecting 1/1,000 live births in Europe [Carinci et al., 2003]. The causes of orofacial clefts involve genetic and environmental factors [Schutte and Murray, 1999]. Epidemiological data in different populations have shown the prevalence of CP is generally lower than that of CL/P and families at high risk for one type of cleft are not at increased risk for the other type. A further subdivision of orofacial clefts into syndromic vs. isolated forms depends on whether additional structural and/or developmental anomalies occur with the cleft [Jugessur and Murray, 2005]. Orofacial development is a complex process involving many genes and signaling pathways [Murray and Schutte, 2004]. Alterations in one or more genes could cause CL/P.
Interferon regulatory factor 6 (IRF6) on 1q32.2 is consistent in its contribution to orofacial clefting among the large number of candidate genes and mutations in this gene cause a dominant malformations syndrome including CL/P and polymorphic markers strongly associated with CL/P [Jugessur et al., 2008, Rahimov et al., 2008].
IRF6 belongs to a family of nine transcription factors that share a high conserved winged helix DNA binding domain and a less conserved protein-binding domain [Kondo et al., 2002]. IRF6 mutations may produce a non-functional protein leading to haplo-insufficiency, affecting the DNA binding domain and cause a dominant negative effect, resulting in severe phenotypes [Houdayer et al., 2001]. Also mutations in the IRF6 transcriptional activation domain may inhibit transcriptional activation, conferring different effects on the function of IRF6 [Little et al., 2009].
A common polymorphic variant, in which isoleucine is substituted for valine at amino acid position 274 (V274I) in the protein-binding domain of IRF6, was identified in an analysis of IRF6 mutations [Kondo et al., 2002]. A family study showed highly significant transmission disequilibrium for this V274I variant with CL/P [Zucchero et al., 2004]. In an Italian population, four different SNPs were analyzed and revealed strong evidence of linkage disequilibrium between CL/P and the markers rs2013162 and rs2235375 [Scapoli et al., 2005].
In the present study, we analyzed five IRF6 SNPs (rs1319435, rs2013162, rs2235375, rs2235371, rs2235543) in 68 Mexican families to test for association with non-syndromic CL/P.
Materials and methods
Sample study
Our study sample consisted of 73 families including father, mother, and two brothers or sisters (without CL/P) of the affected child, corresponding to 292 subjects. All participants were recruited between 2009 and 2011, and all patients were screened for the presence of associated anomalies or syndromes by expert geneticists, and only those determined to have isolated cleft lip with or without cleft palate were included. This study was approved by the local Ethics and Research Committees of the Hospital General “Dr. Manuel Gea González”. Written informed consent was obtained from each person or legal representative of children. DNA was extracted from peripheral blood [Sambrook et al., 2001]. Polymerase chain reaction (PCR) of the IRF6 gene was performed using the primers described by Zucchero et al., 2004 and Scapoli et al., 2005. PCR was performed following manufacturer indications (Epicentre Biotechnologies, USA), and alleles were determined by dot-blot and specific digoxigenin-11-ddUTP-labeled oligonucleotide probes and visualized by chemiluminescence [Bignon and Fernandez-Vina, 1995]. Hybridization results were interpreted by direct observation, considering positive as a strong black dot [Jimenez-Gonzalez et al., 2012].
Statistical analysis
Two approaches were performed: cases and pseudocontrols, according to the Falk and Rubistein, 1987, and Spielman et al., 1993 description, “two parental genes not transmitted to their diseased offspring could be used as the control sample.” This would assure that both samples of the case's genes and its matched control genes (i.e., nontransmitted parental genes) would come from the same genetic population. The second one consisted in a triad analysis considering child, mother, and father.
The data were analyzed by bivariate and multivariate statistics. Allele frequencies (AF) and genotype frequencies (GF) were calculated by direct counting of alleles and genotypes and were compared between the patients and the pseudocontrols using a Chi-squared test or a two-tailed Fisher's exact test when the expected frequency in at least one cell was less than 5. Relative risk for alleles, genotypes, and haplotypes were calculated as an odds ratio (OR) according to Woolf formula [Woolf, 1955], using a 2 × 2 contingence table for each instance. Ninety-five percent confidence intervals (95%CI) were obtained by using Cornfield's approximation. A log-linear method to test for asymmetric distribution of a particular variant allele among affected offspring and their biologic parents assuming Mendelian inheritance with or without Hardy–Weinberg equilibrium (HWE) was performed [Weinberg et al., 1998, Wilcox et al., 1998, Weinberg, 1999], using the program HAPLIN (version 5.3) [Gjessing and Lie, 2006]. This program was chosen because it was specifically designed to analyze genetic risk factors in offspring-parent triads and case–control collections, based on log-linear modeling, implementing a full maximum-likelihood model for estimation. It computes explicit estimates of relative risks with asymptotic standard errors and confidence intervals. It uses the expectation-maximization (EM) algorithm to impute genotypes that are missing [Jugessur et al., 2012]. The most frequent haplotype was use as reference for estimating the effect in a multiplicative model.
Linkage disequilibrium (LD) was determined using Haploview (version 4.2) to reconstruct haplotypes and to estimate the relative risk associated with a single or double dose of each haplotype among the mother–father–child triads. Association analyses were performed under additive or genotype-wise models using family-based conditional logistic regression analysis [Cordell et al., 2004, Barrett et al., 2005, Gjessing and Lie, 2006].
To establish the most informative model of Mendelian association (dominant, co-dominant, recessive, overdominant, or log additive), the online SNPStats program was used in case–pseudocontrols and triads (Solé et al., 2006).
Results
Allelic and genotypic frequencies of all IRF6 polymorphisms, showing P, OR, and 95% CI, obtained by the bivariate analysis in patients and pseudocontrols, are summarized in Table 1. The most frequent alleles were rs1319435-T, rs2013162-A, rs2235375-G, rs2235371-A, and rs2235543-T, while the more frequent genotypes were the heterozygous ones of each SNP. No association was found with any allele when considered alone. However, the genotype rs1319435-C/C showed increased risk (P = 0.02, OR (95 % IC) = 3.84 (1.12–12.78), while the C/C genotype at rs2235375 showed a marginal association with a protective effect P = 0.09, 0.52 (0.24–1.12). Allele and genotype frequencies in these triads (mother–father case) showed no significant differences with any individual allele, but the genotype rs1319435-T/C was significantly increased among parents (Table 2). Table 3 shows overtransmitted alleles in the child obtained with Haploview v4.2, being allele rs2235375-G significant (P = 0.049).
Table 1.
IRF6 allele and genotype frequencies in CL/P patients and pseudocontrols analyzed by bivariate statistic.
| Alleles and genotypes | Affected (n = 73; %) | Non-affected (n = 219; %) | P | OR(95%IC)* |
|---|---|---|---|---|
| rs1319435 | ||||
| T | 60 | 61 | 0.89 | 0.97 (0.66–1.43) |
| C | 40 | 39 | 0.89 | 1.02 (0.69–1.50) |
| T/T | 29 | 24 | 0.43 | 1.27 (0.70–2.30) |
| T/C | 63 | 74 | 0.08 | 0.61 (0.35–1.07) |
| C/C | 8 | 2 | 0.02 | 3.84 (1.12–12.78) |
| rs2013162 | ||||
| A | 62 | 63 | 0.81 | 0.95 (0.65–1.40) |
| C | 38 | 37 | 0.81 | 1.04 (0.71–1.54) |
| A/A | 23 | 25 | 0.71 | 0.89 (0.48–1.66) |
| A/C | 77 | 75 | 0.71 | 1.13 (0.60–2.10) |
| rs2235375 | ||||
| C | 45 | 50 | 0.25 | 0.80 (0.55–1.17) |
| G | 55 | 50 | 0.25 | 1.25 (0.85–1.82) |
| C/C | 12 | 21 | 0.09 | 0.52 (0.24–1.12) |
| C/G | 64 | 58 | 0.30 | 1.34 (0.77–2.32) |
| G/G | 23 | 21 | 0.72 | 1.12 (0.60–2.11) |
| rs2235371 (V274I) | ||||
| A | 56 | 56 | 0.89 | 1.03 (0.70–1.50) |
| G | 44 | 44 | 0.89 | 0.98 (0.67–1.42) |
| A/A | 12 | 13 | 0.97 | 0.98 (0.44–2.20) |
| A/G | 88 | 86 | 0.74 | 1.15 (0.52–2.55) |
| G/G | 0 | 1 | 0.64 | 0.49 (0.02–9.89) |
| rs2235543 | ||||
| T | 52 | 51 | 0.81 | 1.04 (0.72–1.52) |
| C | 48 | 49 | 0.81 | 0.96 (0.66–1.39) |
| T/T | 4 | 3 | 0.57 | 1.50 (0.37–6.16) |
| T/C | 96 | 96 | 0.88 | 0.90 (0.23–3.48) |
| C/C | 0 | 1 | 0.85 | 0.74 (0.03–16.60) |
A character in bold and italics indicates association.
Table 2.
IRF6 allele and genotype frequencies of CL/P triads mother–father case analyzed by bivariate statistic.
| Allele/genotype | Cases (n = 68; %) | Parents (n = 136; %) | P | OR(95%IC)* |
|---|---|---|---|---|
| rs1319435 | ||||
| T | 61 | 60 | 0.83 | 1.05 (0.69–1.60) |
| C | 39 | 40 | 0.83 | 0.95 (0.62–1.46) |
| T/T | 31 | 23 | 0.21 | 1.51 (0.78–2.90) |
| T/C | 60 | 74 | 0.04 | 0.53 (0.28–0.98) |
| C/C | 9 | 3 | 0.07 | 3.19 (0.87–11.73) |
| rs2013162 | ||||
| A | 62 | 62 | 0.66 | 0.91 (0.59–1.39) |
| C | 38 | 38 | 0.66 | 1.10 (0.72–1.68) |
| A/A | 24 | 25 | 0.49 | 0.78 (0.39–1.57) |
| A/C | 76 | 75 | 0.49 | 1.27 (0.64–2.55) |
| rs2235375 | ||||
| C | 43 | 50 | 0.13 | 0.72 (0.48–1.10) |
| G | 57 | 50 | 0.13 | 1.39 (0.91–2.10) |
| C/C | 12 | 19 | 0.10 | 0.50 (0.21–1.16) |
| C/G | 63 | 62 | 0.71 | 1.12 (0.62–2.03) |
| G/G | 25 | 19 | 0.29 | 1.44 (0.73–2.83) |
| rs2235371 (V274I) | ||||
| A | 56 | 57 | 0.93 | 1.02 (0.67–1.54) |
| G | 44 | 43 | 0.93 | 0.98 (0.65–1.49) |
| A/A | 12 | 14 | 0.85 | 0.92 (0.39–2.17) |
| A/G | 88 | 85 | 0.95 | 0.97 (0.44–2.17) |
| G/G | 0 | 1 | 0.53 | 1.88 (0.26–13.67) |
| rs2235543 | ||||
| T | 52 | 51 | 0.84 | 1.04 (0.69–1.58) |
| C | 48 | 49 | 0.84 | 0.96 (0.63–1.45) |
| T/T | 4 | 2 | 0.40 | 1.98 (0.39–10.11) |
| T/C | 96 | 98 | 0.40 | 0.50 (0.10–2.57) |
A character in bold and italics indicates association.
Table 3.
Transmission disequilibrium test (TDT) results.
| SNP | Overtransmitted | T:Ua | Chi-square | P value | |
|---|---|---|---|---|---|
| 1 | rs2235543 | T | 68:65 | 0.07 | 0.794 |
| 2 | rs2235371 | G | 60:56 | 0.14 | 0.710 |
| 3 | rs2235375 | G | 51:33 | 3.86 | 0.049 |
| 4 | rs2013162 | C | 52:50 | 0.04 | 0.843 |
| 5 | rs1319435 | T | 52:49 | 0.09 | 0.765 |
Ratio of transmissions to non-transmissions of the overtransmitted allele. A character in bold and italics indicates association.
The haplotype frequencies of the child and the mother in a single dose (one copy) or double dose (two copies) were obtained with HAPLIN v5.3 and are shown in Fig. 1, where OR are presented as relative risk (RR), and 15 haplotypes were dropped from the analysis because their frequency was less than 1%. Seventeen 5 SNP haplotypes were found for both mother and child, using CCGGT as the reference haplotype. TACGT haplotype was underrepresented in the affected child, either in single (P = 0.0208 RR (95% CI) = 0.157 (0.032–0.762)) or double (P = 0.0208; RR (95% CI) = 0.0247 (0.00102–0.58)) dose. Heterozygotes for the haplotype TAGGT showed a slight excess (P = 0.0674; RR (95% CI) = 6.95 (0.856–57.3)).
Fig. 1.
Estimated relative risks of cleft lip with or without cleft palate (CL/P) in 68 Mexican children carrying one or two copies of the each haplotype. Upper half estimates based on haplotypes in the child; lower half estimates based on haplotypes in the mother. Vertical bars, 95% confidence intervals are on a logarithmic scale.
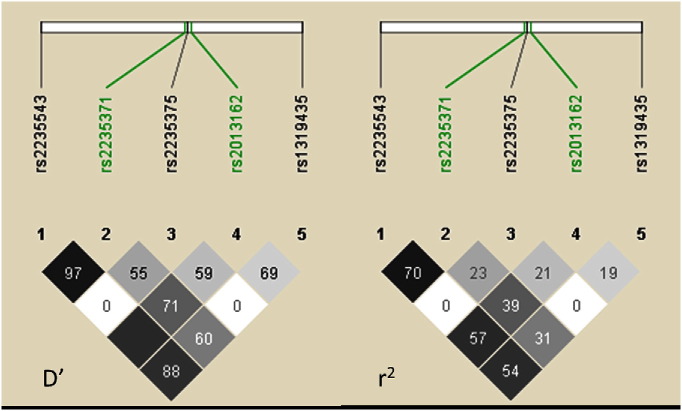
The pairwise LD (D′ and r2) for all 5 SNPs is presented in Fig. 2. Two SNPs were in coding regions: rs2235371 in exon 7 producing a change of the amino acid valine to isoleucine at codon 274 (V274I) and rs2013162 in exon 5 is a silent variant (Ser153Ser), which are noted in green in Fig. 2. SNP rs2235543 is also intragenic, located in intron 6, rs1319435 is 70 kb 5' of IRF6 and rs2235543 is 100 kb 3' of IRF6 [Scapoli et al., 2005]. The LD plot in Fig. 2 indicates alleles at rs2235543 and rs2235371 are in strong LD among parents in these trios, while rs2235543, rs2235375, and rs1319435 seem be almost independent of one another. However, rs2235543 with rs2013162 and rs2235543 with rs1319435 also showed strong LD.
Fig. 2.
Linkage disequilibrium (LD) between the IRF6 genotyped SNPs is shown for which the pairwise LD plot was created by HAPLOVIEW 4.2. Within each diamond the pairwise correlation coefficient (r2) or the standardized coefficient of LD (D′) are presented. Standard color coding was used for the Haploview LD plots using the confidence limits as a color scheme, for D′ LD plots: white = strong evidence of recombination; light gray = uninformative; dark gray = strong evidence of LD; for r2 LD plots: white r2 = 0, shades of gray 0 < r2 < 1, black r2 = 1. White squares indicate absence of LD r2 = 0 or D′ = 0. Intragenic SNPs rs2013162and rs2235371 are shown in green. A marker pair shows moderate or usable LD if D′ is between 0.33 and 0.5, and strong LD if D′ is 0.5 or above (i.e., at least half the maximum value).
Genetic models were fitted for each SNP; odd ratios (OR) and 95% confidence intervals (CI) were calculate to check the relative risk for association with the online SNPStats program (Solé et al., 2006). The results are showed in the Table 4. No inheritance model was found for rs2013162 in the case–pseudocontrol approach, as well as for rs2013162 and rs2235543 in triads.
Table 4.
SNP model inheritance for case–pseudocontrols and triads.
| SNP | Model | Genotype | OR (CI 95%) | P value |
|---|---|---|---|---|
| Case–pseudocontrols | ||||
| rs2235543 | Log additive | − | .1.76 (0.48–6.39) | 0.40 |
| rs2235371 | Co-dominant | G/A | 0.97 (0.43–2.17) | 0.41 |
| rs2235375 | Recessive | C/C | 1.92 (0.89–4.16) | 0.08 |
| rs1319435 | Recessive | C/C | 0.26 (0.08–0.89) | 0.035 |
| Triads | ||||
| rs2235371 | Co-dominant | G/A | 0.81 (0.34–1.97) | 0.60 |
| rs2235375 | Recessive | C/C | 1.77 (0.76–4.16) | 0.17 |
| rs1319435 | Overdominant | C/T | 1.90 (1.02–3.53) | 0.043 |
A character in bold and italics indicates association.
Discussion
The objective in this study was to test for evidence of association and linkage between five SNPs in the IRF6 gene and to calculate the risk of cleft lip with or without cleft palate. We used a population based case–pseudocontrol and case triad study of CL/P in a Mexican sample. Two studies that analyzed Asian and South American populations [Zucchero et al., 2004, Srichomthong et al., 2005] showed a significant association between CL/P and the rs2235371 (V274I) polymorphism in IRF6; in the Zucchero et al. (2004) study, no association with IRF6 was found between cleft palate alone and the V allele in the South American population, but in the South American and Asian groups, the results were highly significant (P < 0.001) for the association between cleft lip or palate and the V allele and also between cleft lip alone and the V allele. In the bivariate analysis of our study, this polymorphism alone did not show association with CL/P. However, an association with increased risk (P = 0.02; OR [95%CI] = 3.84 [1.12–12.78]) was seen with the homozygous genotype rs1319435-C/C in the case–pseudocontrol comparison, while the rs1319435-T/C genotype in the triad analysis was underrepresented among cases (P = 0.04; OR [95%CI] = 0.53 [0.28–0.98]).
Vieira et al. (2002) describe that Amerindian-specific haplotype D of mtDNA had a high frequency. Afterward, Vieira et al. (2007) investigated the specific maternal origin association with IRF6. Individuals with mtDNA haplotype D did not show association with the IRF6 alleles (P = 0.259), but individuals with mtDNA haplotypes other than haplotype D showed a trend for association with IRF6 (P = 0.08). Nevertheless, when they stratified by mitochondrial haplotype and for CL/P, the results indicated an association of IRF6 and risk of NSOFC among individuals with mitochondrial DNA haplotype other than haplotype D (P = 0.023).
On the other hand, admixture analysis of Mexican mestizos from different regions of the country has been carried out with different genetic systems. Few analyses have described the maternal lineages variability (mtDNA) throughout the Mexican territory. Martínez-Cortés et al. (2013) studied the mtDNA variation in 10 populations from different regions of Mexico. The matrilineal diversity estimated in the 742 Mexican mestizos studied was defined by nine haplogroups and paragroups. In the whole population sample Native American haplogroups A, B, C, and D were prevalent (92.9%), with frequencies of 47, 23.7, 15.9, and 6.2, respectively. In the Martínez-Cortés et al., 2013 work, the haplogroup D had the lowest total frecuency (6.2%) without a clear geographical pattern. In our study, mtDNA was not analyzed, but if we considered the results of Vieira et al. (2007) and Martínez-Cortés et al. (2013) , one would think that similar results could be found in our C/LP population. This kind of analysis would be important to perform in the next future, since in this moment it is not possible.
Since we had families with only one affected child, no model free linkage approach could be done because it is necessary the inclusion of multicase families for the calculation of identical-by-descent allele-sharing-based method.
The fitted models obtained by the SNPStats program were the recessive model for rs1319435 in case–pseudocontrols, which was associated with a protective effect (P = 0.035 OR (95% CI) = 0.26(0.08–0.89)), and the overdominant model for rs1319435 in triads, that was significantly increased in affected child (P = 0.043 OR (95% CI) = 1.90 (1.02–3.53)).
Linkage disequilibrium between markers rs2235543, rs2235375, and rs1319435 was low in this population. TDT analysis showed a significant under-transmission of the C common allele and significant over-transmission of the G allele at marker rs2235375 (P = 0.0495) similar to the data of Huang et al., 2009]. SNP rs2235375 showed the strongest association with over-transmission of the G allele with CL/P in our population, as reported in European-American, Taiwanese, Singaporean, Korean, and Western Chinese case–parent triads in a genome wide TDT analysis; however, the SNP rs2013162 was also significant in the same populations, except for Western Chinese [Huang et al., 2009, Park et al., 2007] and Mexican (this study).
IRF6 genotypes influence the risk of CL/P during development, and environment and ethnic factors should be considered as triggers since they could interact with the IRF6 pathway, altering fetal cell differentiation during palate formation. Our results agree with Park et al. (2007), who pointed that “significant results observed from SNPs other than rs2235371 (p.V274I) suggest that rs2235371 itself is not causal, but rather in LD with some causal mutation in IRF6.” Additionally, the lack of association of the rs2235371 SNP and CL/P may be influenced by the relatively small sample size analyzed in this work, although we calculated the statistical power for the case–pseudocontrol and triads association, being 0.62 and 0.53, respectively, and these values are under the rejection level of the null hypothesis, it is important to describe the behavior of these polymorphisms in a sample of the Mexican population, to increase the knowledge of CL/P for further extensive analysis.
Acknowledgments
The authors wish to thank Rocío Jimenez-Lucio and David Sierra-Barrera for technical support.
References
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bignon J.D., Fernandez-Vina F. HLA et medecine. In: Charron D., Fauchet R., editors. XIIth International Histocompatibility Workshop Technical Handbook. EDK Publishers; Paris: 1995. [Google Scholar]
- Carinci F., Pezzetti F., Scapoli L., Martinelli M., Avantaggiato A., Carinci P. Recent developments in orofacial cleft genetics. J. Craniofac. Surg. 2003;14:130–143. doi: 10.1097/00001665-200303000-00002. [DOI] [PubMed] [Google Scholar]
- Cordell H.J., Barratt B.J., Clayton D.G. Case/pseudocontrol analysis in genetic association studies: a unified framework for detection of genotype and haplotype associations, gene–gene and gene–environment interactions, and parent-of-origin effects. Genet. Epidemiol. 2004;26:167–185. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- Croen L.A., Shaw G.M., Wasserman C.R., Tolarova M.M. Racial and ethnic variations in the prevalence of orofacial clefts in California, 1983–1992. Am. J. Med. Genet. 1998;79:42–47. doi: 10.1002/(sici)1096-8628(19980827)79:1<42::aid-ajmg11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Falk C.T., Rubistein P. Haplotype relative risk: an easy reliable way to construct a proper control sample for risk calculations. Ann. Hum. Genet. 1987;51:227–233. doi: 10.1111/j.1469-1809.1987.tb00875.x. [DOI] [PubMed] [Google Scholar]
- Gjessing H.K., Lie R.T. Case–parent triads: estimating single- and double-dose effects of fetal and maternal disease gene haplotypes. Ann. Hum. Genet. 2006;70:382–396. doi: 10.1111/j.1529-8817.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- Houdayer C., Bonaïti-Pellié C., Erguy C., Soupre V., Dondon M.G., Bürglen L. Possible relationship between the van der Woude syndrome (vWS) locus and nonsyndromic cleft lip with or without cleft palate (NSCL/P) Am. J. Med. Genet. 2001;104:86–92. doi: 10.1002/1096-8628(20011115)104:1<86::aid-ajmg10053>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Huang Y., Wu J., Ma J., Beaty T.H., Sull J.W., Zhu L. Association between IRF6 SNPs and oral clefts in West China. J. Dent. Res. 2009;88:715–718. doi: 10.1177/0022034509341040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Gonzalez D.E., Martinez-Flores W.A., Reyes-Gordillo J., Ramirez-Miranda M.E., Arroyo-Escalante S., Romero-Valdovinos M. Blastocystis infection is associated with irritable bowel syndrome in a Mexican patient population. Parasitol. Res. 2012;110:1269–1275. doi: 10.1007/s00436-011-2626-7. [DOI] [PubMed] [Google Scholar]
- Jugessur A., Murray J.C. Orofacial clefting: recent insights into a complex trait. Curr. Opin. Genet. Dev. 2005;15:270–278. doi: 10.1016/j.gde.2005.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A., Rahimov F., Lie R.T., Wilcox A.J., Gjessing H.K., Nilsen R.M. Genetic variants in IRF6 and the risk of facial clefts: single-marker and haplotype-based analyses in a population-based case–control study of facial clefts in Norway. Genet. Epidemiol. 2008;32:413–424. doi: 10.1002/gepi.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A., Skare Ø., Lie R.T., Wilcox A.J., Christensen K., Christiansen L. X-linked genes and risk of orofacial clefts: evidence from two population-based studies in Scandinavia. PLoS ONE. 2012;7(6):e39240. doi: 10.1371/journal.pone.0039240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Schutte B.C., Richardson R.J., Bjork B.C., Knight A.S., Watanabe Y. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little H.J., Rorick N.K., Su L.I., Baldock C., Malhotra S., Jowitt T. Missense mutations that cause Van der Woude syndrome and popliteal pterygium syndrome affect the DNA-binding and transcriptional activation functions of IRF6. Hum. Mol. Genet. 2009;18:535–545. doi: 10.1093/hmg/ddn381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cortés G., Salazar-Flores J., Haro-Guerrero J., Rubi-Castellanos R., Velarde-Félix J.S., Muñoz-Valle J.F. Maternal admixture and population structure in Mexican-Mestizos based on mtDNA haplogroups. Am. J. Phys. Anthropol. 2013;151:526–537. doi: 10.1002/ajpa.22293. [DOI] [PubMed] [Google Scholar]
- Murray J.C., Schutte B.C. Cleft palate: players, pathways, and persuits. J. Clin. Invest. 2004;113:1676–1678. doi: 10.1172/JCI22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.C., Daack-Hirsch S., Buetow K.H., Munger R., Espina L., Paglinawan N. Clinical and epidemiologic studies of cleft lip and palate in the Philippines. Cleft Palate Craniofac. J. 1997;34:7–10. doi: 10.1597/1545-1569_1997_034_0007_caesoc_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- Park J.W., McIntosh I., Hetmanski J.B., Jabs E.W., Vander Kolk C.A., Wu-Chou Y.H. Association between IRF6 and nonsyndromic cleft lip with or without cleft palate in four populations. Genet. Med. 2007;9:219–227. doi: 10.1097/GIM.0b013e3180423cca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F., Marazita M.L., Visel A., Cooper M.E., Hitchler M.J., Rubini M. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat. Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fitsch E.F., Maniatis T. Cold Spring Harbor. 3rd ed. Cold Spring Harbor Laboratory Press; 2001. Molecular cloning: a laboratory manual. [Google Scholar]
- Scapoli L., Palmieri A., Martinelli M., Pezzetti F., Carinci P., Tognon M. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am. J. Hum. Genet. 2005;76:180–183. doi: 10.1086/427344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte B.C., Murray J.C. The many faces and factors of orofacial clefts. Hum. Mol. Genet. 1999;8:1853–1859. doi: 10.1093/hmg/8.10.1853. [DOI] [PubMed] [Google Scholar]
- Solé X., Guino E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- Spielman R.S., McGinnis R.E., Ewens W.J. Transmission test for linkage disequilibrium: the insulin gene region an insulin-dependent diabetes mellitus (IDDM) Am. J. Hum. Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Srichomthong C., Siriwan P., Shotelersuk V. Significant association between IRF6 820G- > A and non-syndromic cleft lip with or without cleft palate in the Thai population. J. Med. Genet. 2005;42:e46. doi: 10.1136/jmg.2005.032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolarova M.M., Cervenka J. Classification and birth prevalence of orofacial clefts. Am. J. Med. Genet. 1998;75:126–137. [PubMed] [Google Scholar]
- Vanderas A.P. Incidence of cleft lip, cleft palate, and cleft lip and palate among races: a review. Cleft Palate J. 1987;24:216–225. [PubMed] [Google Scholar]
- Vieira A.R., Karras J.C., Orioli I.M., Castilla E.E., Murray J.C. Genetic origins in a South American clefting population. Clin. Genet. 2002;62:458–463. doi: 10.1034/j.1399-0004.2002.620606.x. [DOI] [PubMed] [Google Scholar]
- Vieira A.R., Cooper M.E., Marazita M.L., Orioli I.M., Castilla E.E. Interferon regulatory factor 6 (IRF6) is associated with oral-facial cleft in individuals that originate in South America. Am. J. Med. Genet. A. 2007;143A:2075–2078. doi: 10.1002/ajmg.a.31884. [DOI] [PubMed] [Google Scholar]
- Weinberg C.R. Methods for detection of parent-of-origin effects in genetic studies of case–parents triads. Am. J. Hum. Genet. 1999;65:229–235. doi: 10.1086/302466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg C.R., Wilcox A.J., Lie R.T. A log-linear approach to case–parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am. J. Hum. Genet. 1998;62:969–978. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox A.J., Weinberg C.R., Lie R.T. Distinguishing the effects of maternal and offspring genes through studies of “case–parent triads”. Am. J. Epidemiol. 1998;148:893–901. doi: 10.1093/oxfordjournals.aje.a009715. [DOI] [PubMed] [Google Scholar]
- Woolf B. On estimating the relation between blood group and disease. Ann. Hum. Genet. 1955;19:251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Zucchero T.M., Cooper M.E., Maher B.S., Daack-Hirsch S., Nepomuceno B., Ribeiro L. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N. Engl. J. Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]