Abstract
The biomaterial scaffolds for regenerative medicine need to be rationally designed to achieve the desired cell fate and function. This paper describes the development of hydrogel microstructures for cultivation of primary hepatocytes. Four different micropatterned surfaces are tested: 1) poly(ethyelene glycol) (PEG) microwells patterned on glass, 2) heparin hydrogel microwells patterned on glass, 3) PEG microwells patterned on heparin hydrogel-coated substrates, and 4) heparin hydrogel microwells patterned on heparin hydrogelcoated substrates. The latter surfaces are constructed by a combination of micromolding and microcontact printing techniques to create microwells with both walls and floor composed of heparin hydrogel. Individual microwell dimensions are 200 μm diameter and 20 μm in height. In all cases, the floor of the microwells is modified with collagen I to promote cell adhesion. Cultivation of hepatocytes followed by analysis of hepatic markers (urea production, albumin synthesis, and E-cadherin expression) reveals that the all-heparin gel microwells are most conducive to hepatic phenotype maintenance. For example, ELISA analysis shows 2.3 to 13.1 times higher levels of albumin production in all-heparin gel wells compared with other micropatterned surfaces. Importantly, hepatic phenotype expression can be further enhanced by culturing fibroblasts on the heparin gel walls of the microwells. In the future, multicomponent all-heparin gel microstructures may be employed in designing hepatic niche for liver-specific differentiation of stem cells.
1. Introduction
Cells in vivo are exposed to complex and highly structured microenvironment regulated by multiple niche effectors, including biochemical cues arising from neighboring cells, soluble factors, and extracellular matrix (ECM), and also biophysical cues coming from niche elasticity and geometry.[1–4] Several studies have demonstrated that cell fate is determined by the combination of these niche effectors,[5–7] and it is therefore important to recreate biochemical and mechanical cues for enhanced in vitro culture results.[8] In contrast, the classical cell culture techniques often rely on culturing a single cell type on a two dimensional, rigid substrate and come up short in recreating microenvironment complexity. To overcome these limitations, a number of research groups have been using the microfabrication techniques to enhance complexity of in vitro microenvironment.[9–17] One example of these involves fabricating microwells by photolithography or soft lithography to control the extent of homotypic cellular interactions.[18,19] These microstructures have been mainly fabricated with hydrogels such as poly (ethylene glycol) (PEG) due to their biocompatibility, high water content, and mechanical properties. However, PEG microstructures are biologically inert; they do not promote cell attachment or function. Therefore, researchers have been either functionalizing PEG backbone with cell binding motifs or utilizing natural polymers such as chitosan, gelatin, or denatured collagen.[12,20–22] Other approaches focus on creating heterotypic interactions by co-culturing the cell of primary interest with a secondary or supporting cell type, in an attempt to mimic interactions between epithelium and stroma present in numerous tissues.[20,23,24] Yet another subset of studies focus on defining mechanical properties of the substrate to provide physiologically appropriate mechanical cues to cells.[25,26]
The goal of our paper was to create microstructured hydrogel surfaces where biochemical and biophysical cues as well as co-culture effects could be present simultaneously. We chose to work with heparin hydrogel, as this bioactive material has been shown useful for culturing multiple cell types, including the cell type of our interest–primary hepatocytes.[16,27–31] These cells are notoriously difficult to culture and are commonly used as liver mimics in toxicology/drug screening studies. An important design criterion for us was to develop bioactive microwells where the floor is composed of a soft gel approaching mechanical properties of the healthy liver (E, < 6 kPa).[32–34] As shown in Figure 1, all-heparin hydrogel microstructures were fabricated in two steps. The first heparin gel layer serving as the bottom of microwells was created by photopolymerization. The second layer containing microwells was created by a combination of micromolding and microcontact printing. This combined process allowed to fabricate microwells while immobilizing cell-adhesive collagen layer at the bottom of the wells. Three other types of micropatterned surfaces, those containing PEG microwells on glass, heparin gel microwells on glass, and PEG microwells on heparin gel layer were used as controls. Our experiments revealed that all-heparin hydrogel microwells outperformed other micropatterned surfaces by eliciting highest levels of hepatic function expression. Given that heterotypic interactions are important in the liver, we also tested a co-culture system with hepatocytes seeded inside the microwells and fibroblasts on the hydrogel walls. Inclusion of stromal cells on the microstructured surfaces further enhanced hepatic function. Overall, the multi-layered heparin gel microstructures described here represent a novel tool for engineering the niche for maintaining hepatic function.
Figure 1.
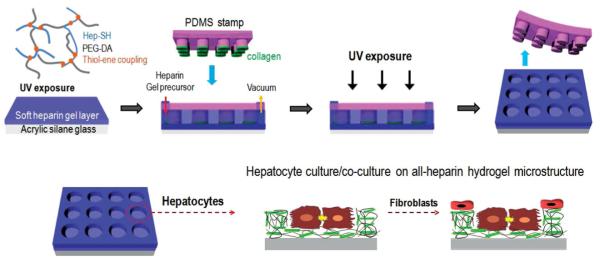
Schematic illustration of the fabrication of multilayered heparin hydrogel microstructure. A combination of micromolding and microcontact printing was used to create microwells where both walls and floor were composed of heparin hydrogel. This protocol allowed to selectively pattern the bottom of the wells with collagen I to promote attachment of hepatocytes. A co-culture could be created by seeding 3T3 fibroblasts on the same surfaces, resulting in stromal cell attachment around the hepatocyte clusters, on the heparin gel walls of the microwells.
2. Results and Discussion
2.1. Fabricating and Characterizing Heparin Gel Microstructures
In this study, heparin-based hydrogels formed via UV-mediated thiol-ene crosslinking reaction were employed to fabricate the multilayered bioactive microstructures for cell cultivation. The selection of heparin hydrogel as a building material was influenced by our previous result demonstrating hepatic phenotype-inductive effects of this biomaterial.[16,27,31] In contrast to previous studies that focused on either defining the cell group size[35,36] or controlling heterotypic interactions via co-cultures[23,24] or providing mechanical cues,[25,26,37] we wanted the ability to create all of the above effects on the same surface. To accomplish this, several design criteria had to be addressed, including: 1) defining hepatocyte attachment sites within the microwells, 2) designing microwells with soft bottom layer approaching mechanical properties of the liver, 3) controlling attachment of secondary cells on the walls around the microwells. Figure 1 describes a two-step process for fabricating microwells composed entirely of heparin gel. First, a bottom heparin gel layer was created by UV photopolymerization of a prepolymer containing diacrylated PEG and thiolated heparin. In our previous study, this formulation of heparin gel was determined to result in a soft substrate (E ≈ 11 kPa) and was found to elicit higher levels of hepatic function when compared with stiffer gel or glass substrates.[27] In the second fabrication step, a polydimethyl siloxane (PDMS) template containing arrays of 200 μm diameter posts was pressed against the gelcoated glass slide, followed by infusion of heparin prepolymer solution into the inlet port of the template. This step was specifically developed to create cell adhesive regions at the bottom of microwells, leaving walls free of hepatocytes and available for subsequent addition of secondary cell type. To accomplish this, we inked PDMS template/stamp in collagen I before pressing it against heparin gel-coated slide. Therefore in this process collagen transfer by microcontact printing was happening concurrently with UV-induced micromolding of the polymer precursor solution. In addition to all-heparin hydrogel microstructured surfaces described in Figure 1 and Figure 2A, we fabricated three other groups of microstructured surfaces serving as controls. Summarized in Figure 2, these surfaces included PEG hydrogel microwells fabricated on glass (Figure 2B), heparin hydrogel microwells on glass (Figure 2C) and PEG microwells on heparin gel-coated glass slides (Figure 2D).
Figure 2.
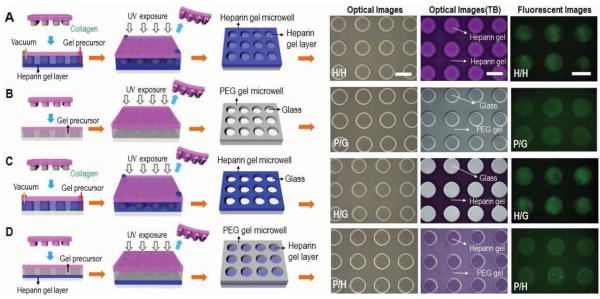
Schematic illustration of four variants of hydrogel microstructures tested in our study. A) Heparin microwells on the heparin gel-coated glass surface (H/H). B) PEG microwells on the glass surface (P/G). C) Heparin gel microwells on the glass surface (H/G). D) PEG microwells on the heparin gel-coated glass surface (P/H). Optical and fluorescent microscopic images of the four kinds of gel microstructures before and after toluidine blue (TB) staining as well as after collagen coating. Scale bar = 200 μm. B) and D) PEG hydrogel microwells were simply fabricated by micromolding technique and then collagen was selectively absorbed on bottom of microwells by immersing in collagen solution due to anti-fouling effect of PEG microwell. A,C) Heparin hydrogel microwells were fabricated by a combination of micromolding and microcontact printing. Collagen transfer by microcontact printing was concurrent to micromolding procedure.
Brightfield images in Figure 2 show that microwells with 200 μm diameter could be successfully fabricated for all four groups of surfaces. To demonstrate presence of heparin, the different types of micropatterned surfaces were stained with toluidine blue O (TB). Optical images of TB staining in Figure 2 demonstrated differences in staining for the different hydrogel micropatterns. Everything is stained for all-heparin gel micropatterns, no staining appears in the case of PEG gel-on-glass micropatterns, only the wells are stained in blue/purple in the case of heparin gel-on-glass micropatterns, and only the bottom is stained for PEG gel-on-heparin gel micropatterns. A set of fluorescence images in Figure 2 shows selective adsorption of fluoresceine-labeled collagen I at the bottom of the microwells. In cases of Figure 2B,D, collagen immobilization was reasonably straightforward because PEG hydrogel walls were non-fouling and protein adsorption was accomplished by simple immersion of surfaces in collagen solution. However, for scenarios involving heparin gel (Figure 2A,C), this simple approach was not sufficient due to possibility of protein adsorbing onto heparin gel, thus compromising our ability to create micropatterned co-cultures later on. For this situation, we developed and deployed a method involving a combination of micromolding and microcontact printing of collagen. As seen in Figure 2A,C, this method resulted in transfer/immobilization of collagen on the underlying substrate. Studies with hepatocytes, described in the following section, supported the effectiveness of micromolding and microcontact printing method in creating surfaces for cell cultivation.
2.2. Cultivating Primary Rat Hepatocytes on Gel Microstructures
As mentioned above, cells respond to multiple signals existing in the niche. Mechanical cues in addition to biochemical cues have garnered a lot of attention recently. While majority of these studies focus on responses of mesenchymal cells to mechanical stimuli,[38,39] there is increasing interest in engineering mechanical microenvironment of epithelial cells such as hepatocytes.[27,37,40]
In our study, primary rat hepatocytes were cultured in four kinds of microwells to investigate the effects of chemical composition and mechanical properties of the microstructured surface on phenotype expression. Figure 3 shows representative images of hepatocytes cultured on gel microstructures at day 1 and day 5. Hepatocytes are known to require adhesive ligands such as collagen I, IV, or laminin for attachment, therefore, these cells selectively attached on collagen I regions at the bottom of the microwells. It is worth noting that minimal attachment of hepatocytes was observed not only on non-fouling PEG gel (Figure 3A,C) but also on heparin gel lacking collagen (Figure 3B,D). Over the culture period, morphology of hepatocytes cultured in PEG gel-on-glass microwells changed from cuboidal/epithelial to elongated/mesenchymal, suggestive of de-differentiation (Figure 3A). In contrast, hepatocytes cultured on heparin gel-containing microstructures appeared to retain typical cuboidal morphology of healthy hepatocytes (Figure 3B–D). More quantitative assessment of hepatic phenotype was carried out based on albumin and urea synthesis. These are the major products of liver metabolism that are frequently used as benchmarks for assessment of hepatic function.
Figure 3.
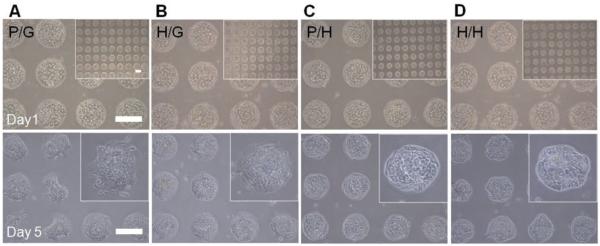
Culturing primary rat hepatocytes on the four kinds of gel microstructures of varying components. A) Bright-field microscopic images of hepatocytes cultured on PEG microwells on the glass surface (P/G). B) Heparin gel microwells on the glass surface (H/G). C) PEG microwells on soft heparin gel layer (P/H). D) Heparin microwells on soft heparin gel layer (H/H) at day 1 and day 5. Scale bar = 200 μm.
The albumin ELISA data presented in Figure 4A shows that heparin gel containing micropatterned surfaces elicited higher level of albumin synthesis from hepatocytes, compared with PEG hydrogel-on-glass micropatterns. For example, compared with PEG hydrogel-on-glass control (0.015 ± 0.001 ng cell−1 d−1) at day 7, the albumin levels were 4.2 times higher for heparin gel-on-glass microwells (0.063 ± 0.026 ng cell−1 d−1), 5.8 times higher for PEG hydrogel-on-heparin gel microwells (0.087 ± 0.005 ng cell−1 d−1), and 13.1 times higher for all-heparin hydrogel microwells (0.196 ± 0.033 ng cell−1 d−1). Interestingly, as seen from Figure 4A, beyond 5 days in culture, the highest levels of albumin synthesis were observed in the case of all-heparin hydrogel microstructures.
Figure 4.
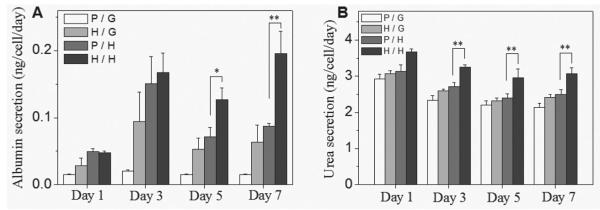
Liver-specific function of primary hepatocytes cultured on the gel microwells of varying components. PEG microwell on pure glass (P/G), heparin gel microwell on pure glass (H/G), PEG microwell on heparin gel layer (P/H), heparin microwell on heparin gel layer (H/H). A) ELISA analysis of albumin secretion and B) urea synthesis by hepatocytes at various time points during 7 day culture. Error bars indicate standard deviation of the mean for n = 3 samples. Significance; * p < 0.05, ** p < 0.001.
Urea arises in the process of converting toxic ammonia.[41] While less pronounced than albumin data, urea analysis results presented in Figure 4B demonstrate that all-heparin hydrogel microstructures were associated with highest levels of urea production. The levels of albumin and urea synthesis in our best case scenario of all-heparin hydrogels are comparable to hepatic function reported for collagen gel sandwich[42,43] or hepatocyte spheroid cultures.[44–46]
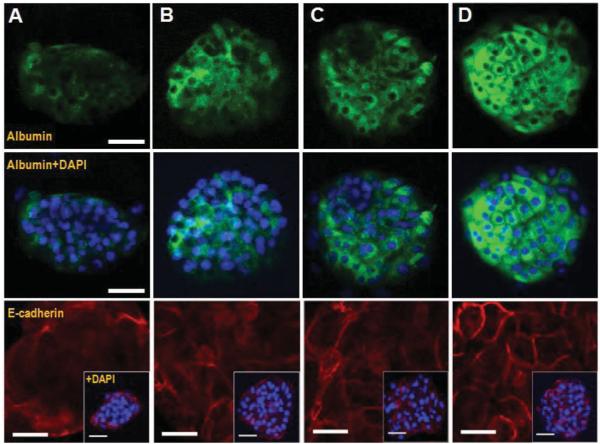
The differences in hepatic phenotype expression were further corroborated by immunofluorescent staining for albumin and E-cadherin–a cell–cell adhesion molecule expressed in epithelial cells such as hepatocytes.[47] As seen from images in Figure 5, after 5 days in culture, hepatocytes in multilayered all-heparin gel microstructures showed much stronger albumin fluorescence signal than the cells on other types of surfaces. These data were in line with albumin ELISA results reported in Figure 4A. Additional evidence of hepatic phenotype expression was obtained by immunostaining for E-cadherin. This is a consensus marker of epithelial hepatic phenotype that is known to be down-regulated during de-differentiation of hepatocytes.[47] As shown in Figure 5, E-cadherin was expressed stronger and was localized at cell–cell junction in hepatocytes cultured on all-heparin hydrogel microstructure as opposed to other micropatterned surfaces. In summary, hepatic function and phenotype analysis clearly showed all-heparin hydrogel microstructures to be superior to other types of micropatterned surfaces tested here. The reasons for this are likely severalfold. 1) Heparin-based hydrogels are bioactive and have been shown by us and others to induce cellular phenotype in the absence of exogenous morphogens.[16,31,48] This likely happens by sequestration of cell-produced (endogenous) growth factors/morphogens expressing heparin-binding domains that are captured in the vicinity of cells and are possibly re-released over time. 2) In addition to bioactivity, low-modulus surfaces employed here may approximate mechanical properties of the native liver and may contribute to phenotype maintenance.
Figure 5.
Intracellular albumin and expression of E-cadherin in hepatocytes cultured on gel microstructures of varying components after 5 day culture. A) PEG microwell on pure glass (P/G), B) heparin gel microwell on pure glass (H/G), C) PEG microwell on heparin gel layer (P/H), D) heparin microwell on heparin gel layer (H/H). Green fluorescence was intracellular albumin and blue fluorescence was DAPI staining of nuclei. Scale bar = 50 μm. Red fluorescence was E-cadherin staining and blue fluorescence was DAPI staining of nuclei. scale bar = 25 μm; E-Cadherin distribution also contains insets of each single microwell. Scale bar = 50 μm.
2.3. Creating Hepatocyte-Fibroblast Co-Cultures in Heparin Gel Microstructures
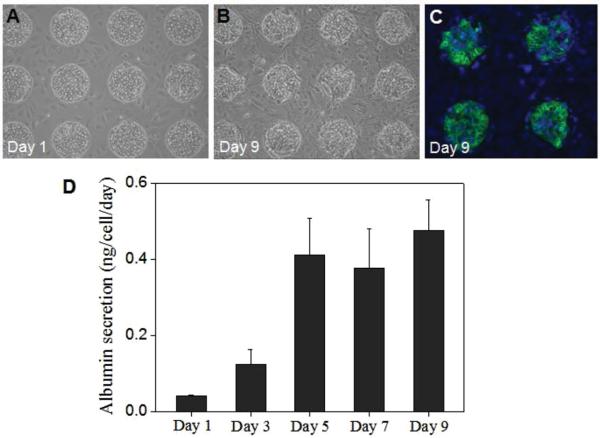
In an attempt to mimic heterotypic cellular interactions present in the liver, hepatocytes have been frequently co-cultured with stromal cells, leading to marked improvement of hepatic phenotype expression and maintenance.[24,49–52] To further enhance complexity of the liver microenvironment in our study, stromal cells were added to hepatocyte micropatterns. As noted earlier in this paper, the micropatterned surfaces were specifically designed to enable co-culture formation. That is, surfaces were micropatterned so as to define sites of hepatocyte attachment, leaving regions of the surface open for the secondary cell type to adhere. The principle of this co-culture formation is similar to the earlier reports of Langer and co-workers.[20] Hepatocyte were seeded first and were given 1 day to form a monolayer inside the microwells. 3T3 fibroblasts were seeded next, adhering on the walls of heparin gel wells but not on the top of the hepatocyte clusters formed within the wells. Fibroblasts are adhesive cells that secrete endogenous ECM proteins to promote attachment. These cells attached well on unmodified heparin gel walls of the microwells. An example of hepatocyte-fibroblast co-cultures formed on microstructured gel surfaces is shown in Figure 6A,B. DAPI in combination with immunofluorescent staining was used to show presence of albumin positive cells (hepatocytes) inside the wells and albumin negative cells (fibroblasts) on the hydrogel wall regions (Figure 6C). Analysis of albumin production by ELISA (Figure 6D) revealed that co-cultures were highly functional up to day 9, after which function began to decay. In comparison to most functional mono-cultures (Figure 4A), co-cultures produced 3.5 fold higher level of albumin. Overall, micropatterned co-cultures of primary hepatocytes and 3T3 fibroblasts showed enhanced hepatic function compared with mono-cultures.
Figure 6.
Application of a multilayered heparin gel microstructure (H/H) for hepatocyte-3T3 cell co-cultures. A) and B) Bright-field microscopic images of co-culture at day 1 and 9. Hepatocyte were cultured on inside microwells and 3T3 fibroblasts were cultured on the wall around microwells. C) Intracellular albumin of hepatocytes co-cultured on all heparin gel microwells (H/H) at day 9. D) ELISA analysis of albumin secretion by hepatocyte co-cultured on H/H during 9 days.
3. Conclusion
In this study, we describe the creation of heparin-based hydrogel microstructures and culturing primary hepatocytes on these microstructured surfaces. Design considerations for these surfaces included 1) controlling the cell group size, 2) creating substrates that mimic mechanical properties of the liver, and 3) introducing heterotypic interactions between hepatocytes and stromal cells. With these goals in mind a fabrication strategy combining photopolymerization, micromolding, and microcontact printing was devised to create arrays of all-heparin hydrogel microwells with collagen-modified attachment sites to promote hepatocyte adhesion. These microstructured surfaces elicited higher levels of hepatic function compared with other micropatterned substrates that were either fabricated on glass or were not entirely made of heparin gel. Importantly, fibroblasts could be added to hepatocyte micropatterns to create micropatterned co-cultures. When compared with mono-cultures, these co-cultures caused a ≈3.5 fold enhancement in production of albumin. Overall, the novel micropatterning and biomaterial microfabrication strategies detailed in this paper provide the flexibility in designing the microenvironment of desired micrometer-scale dimensions, mechanical properties, and cellular composition. We foresee building on these fabrication strategies in the future for improving the microenvironment niche for hepatocyte maintenance or liver-specific differentiation of stem cells.
4. Experimental Section
Chemicals and Materials
Heparin (sodium salt, from porcine intestinal mucosa) was purchased from Celsus Ins. (Cincinnati, IA, USA) and Sigma Aldrich (St. Louis, MO, USA). Thiolated heparin (Hep-SH) was synthesized with the modification of carboxylic groups of heparin using carbodiimide chemistry, as previously reported.[28] Poly(ethylene glycol) diacrylate (PEG-DA, MW 6 kDa, 98% degree of substitution) was purchased from SunBio Inc. (Anyang, Korea). 4-(2-Hydroxyethoxy) phenyl-(2-hydroxy-2-propyl) ketone (Irgacure 2959) was purchased from Ciba Specialty Chemicals Inc. (Basel, Switzerland). Sulfuric acid, ethanol, bovine serum albumin (BSA), and toluidine blue O were purchased from Sigma Aldrich (St. Louis, MO, USA). Glucagon and recombinant human insulin were obtained from Eli Lilly (Indianapolis, IN, USA), and hydrocortisone sodium succinate was obtained from Pfizer Inc. (Ann Arbor, MI, USA). Phosphate-buffered saline (PBS) was purchased from Gibco (Grand Island, NY, USA). Dulbecco’s modified Eagle’s medium (DMEM), sodium pyruvate, fetal bovine serum (FBS), penicillin/streptomycin were purchased from Invitrogen (Carlsbad, CA, USA). Rat albumin ELISA kit was obtained from Bethyl Laboratories (Montgomery, TX, USA) and urea analysis kit was purchased from Bioassay Systems (Hayward, CA, USA). Paraformaldehyde was purchased from Election Microscopy Sciences (Hatfield, PA, USA). Sheep anti-albumin and FITC-anti-sheep IgG were obtained from Bethyl Labs and Santa Cruz Biotechnologies, Inc. (Santa Cruz, CA, USA). Mouse anti-E-cadherin and Alexafluor 546 anti-mouse were purchased from BD Science and Invitrogen. Mounting medium with DAPI was purchased from Vector Laboratories, Inc. (Burlingame, CA, USA).
Fabricating Gel Microstructures on Glass
In this study, we prepared and tested four types of micropatterned surfaces. PEG hydrogel microwells as shown in Figure 2B,D were simply fabricated in a micromold and heparin hydrogel microwells as shown in Figure 2A,C were constructed using a combination of micromolding and microcontact printing. Specifics of fabricating each surface are discussed later in this section. The common features for all micropatterned surfaces included silanization of glass and fabrication of PDMS molds. To promote attachment of hydrogels, the glass substrates were modified with 3-acryloxypropyl trichlorosilane (Gelest, Morrisville, PA) according to the protocol reported previously.[16] All processes relied on PDMS templates that were fabricated using traditional soft lithography approaches[53,54] to contain arrays of posts 200 μm in diameter and 20 μm in height. After peeling off PDMS template the surfaces contained arrays of 200 μm diameter wells with depth of ≈20 μm. Presented below are protocols used to fabricate four different surfaces used in our studies.
1) Heparin gel microwells on heparin gel (Figure 2A). In this case, the prepolymer solution was composed of thiolated Hep-SH and 6 kDa PEG-DA (1:1 molar ratio of thiol to acrylate group). These two components were mixed in PBS containing 1% w/v photo-initiator (Irgacure 2959), which was dissolved in 70% v/v ethanol, to achieve 5% w/v of gel precursor solution. To make multi-layered all-heparin microwells in which both the walls and the bottom of microwells were composed of heparin gel, the substrates were first coated with heparin gel precursor solution based on photopolymerization. 6 μL of prepolymer solution was dispensed onto a 12 mm × 12 mm glass slide, covered with a cover slip (25 mm × 25 mm × 0.13 mm) to create a uniform liquid layer and exposed to UV (365 nm, 18 W cm−2, OmniCure series 1000 light source, EXFO, Vanier, Quebec, Canada) for 5 s. After removing the cover slip, the fabrication of heparin gel microwells was performed by a combination of micromolding and microcontact printing. The PDMS template for this combined micromolding procedure contained two ports, one for injection of liquid prepolymer, another for application of negative pressure. The PDMS template was first gently pressed against the substrate, then the liquid prepolymer was loaded into the inlet port and vacuum was applied to the outlet port to pull prepolymer solution. These constructs were subsequently exposed to UV light for 10 s (365 nm, 18 W cm−2) to cross-link the prepolymer. Inclusion of heparin diminished non-fouling properties of the hydrogel walls, making it difficult to selective adsorb collagen at the bottom of wells as described for construction of PEG microwells in scenario 2. To resolve this challenge, we developed a combined approach of micromolding and microcontact printing where a PDMS template was inked in collagen I prior to micromolding process. For collagen inking protocol, 0.1 mg mL−1 collagen solution was spin-coated onto a glass slide at 1000 rpm to create a thin liquid layer on the surface. That way, when the array of PDMS pillars was brought in contact with the protein layer, only the tips of the pillars became functionalized with protein. Subsequently, when the PDMS template was pressed against the substrate for micromolding, collagen was transferred into the regions of heparin gel layer that become the floor of the wells during micromolding procedure. Therefore, in this method, formation of microwells and functionalization of wells with collagen was happening concurrently.
2) PEG hydrogel microwells on glass (Figure 2B). 6 kDa PEG-DA was dissolved in PBS to achieve 10% w/v of gel precursor solution. Using this prepolymer, PEG microwells were simply constructed via a micromolding. The PEG precursor solution (10 μL) was dropped onto silane-modified glass substrate (12 × 12 mm) and then PDMS mold was brought into contact with the solution and gently pressed. It was exposed to UV irradiation for 10 s and then the PDMS mold was peeled from the surface. After fabricating microwells, the substrates were immersed in 0.1 mg mL−1 solution of collagen I for 1 h. Because the microwells were composed of non-fouling PEG, the protein became selectively adsorbed on the glass regions inside the microwells.
3) Heparin gel microwells on glass (Figure 2C). These microwells were also fabricated by a combination of micromolding and microcontact printing as described in case 1 but with the exception that the underlying glass substrate was not coated with heparin gel. The combined technique of micromolding and microcontact printing developed by us allowed for formation of the microwells and deposition of collagen at the bottom of the wells to happen concurrently.
4) PEG gel microwells on heparin gel (Figure 2D). To make this type of microwells, the substrates were first coated with heparin gel precursor solution based on photopolymerization as described in case 1. After removing the cover slip, PEG gel microwells were simply constructed by the micromolding process described above in case 2. Because these substrates contained non-fouling PEG wells on the top of heparin gel, the bottom of the wells could be made cell-adhesive by incubating in 0.1 mg mL−1 solution of collagen I.
Toluidine blue O staining was used to identify presence and location of heparin in microstructures, as reported previously.[55,56] Staining of negatively charged heparin molecules by this dye (purple color) was visualized using optical microscopy (Zeiss Axiovert 40, Carl Zeiss, NJ, USA).
Cell Culture on Gel Microstructures
All four types of gel microstructures had similar microwell dimensions −200 μm in diameter and 20 μm in height. These micropatterned surfaces were used to create mono- and co-cultures of hepatocytes. Primary hepatocytes were isolated from adult female Lewis rats (Charles River Laboratories, Boston, MA, USA) weighing 125–200 g, using a two-step collagenase perfusion procedure as described previously.[41] Primary hepatocytes were maintained in DMEM supplemented with 10% FBS, 200 U mL−1 penicillin, 200 mg mL−1 streptomycin, 7.5 mg mL−1 hydrocortisone sodium succinate, 20 ng mL−1 EGF, 14 ng mL−1 glucagon, and 0.5 U mL−1 recombinant human insulin at 37 °C in a humidified 5% CO2 atmosphere. A glass piece containing gel microstructures was placed into a well of a 12-well plate and then immersed in 2 mL solution containing 1 × 106 hepatocytes. After 1 h of incubation at 37 °C, the samples were washed twice in PBS to remove unattached hepatocytes and fresh medium was added to the sample wells.
For co-culture experiments, murine 3T3 fibroblasts were maintained in the DMEM supplemented with 10% FBS, 200 U mL−1 penicillin and 200 mg mL−1 streptomycin. The cells were growth arrested by treatment with 10 μg mL−1 of mitomycin C (Sigma-Aldrich) for 2 h prior to cell seeding. Hepatocytes were cultured on micropatterned surfaces for 1 d prior to introduction of fibroblasts. To create co-cultures, fibroblasts were added at a concentration of 1.2 × 10 6 cells mL−1 into the wells already containing hepatocyte micropatterns. After 2 h of incubation at 37 °C, unattached fibroblasts were washed away and then co-culture of hepatocytes and fibroblasts was maintained in hepatocyte culture medium at 37 °C in a humidified 5% CO2 atmosphere.
Analysis of Hepatic Function and Phenotype
The number of hepatocytes attached on each microwell was counted with six microscopic images of 10 × magnification fields per condition 1 d after cell seeding. For analysis of hepatic function, culture medium was collected and analyzed using a urea kit and rat albumin ELISA kit. To detect intracellular albumin and E-cadherin, gel microstructures were fixed in 4% paraformaldehyde and were washed in PBS solution and then incubated with primary antibodies for albumin (1:100 dilution in PBS) and for E-cadherin (1:50 dilution in PBS). After overnight incubation at 4 °C, gel microstructures were washed in PBS solution and then stained with secondary antibodies for albumin (1:200 dilution in PBS) and for E-cadherin (1:750 dilution in PBS). After 1 h of incubation at room temperature, the gel microstructures were washed in PBS and were mounted using a mounting medium containing DAPI to determine the location of nuclei. Stained cells were visualized and imaged using a laser scanning confocal microscope (LSM700, Carl Zeiss, Jena, Germany).
Statistical Analysis
The results were expressed as the mean ± standard deviation. Student t-test analysis was used for statistical analysis. Differences were considered to be statistically significant at p < 0.05.
Acknowledgements
The authors thank Prof. Tae for providing heparin thiol and also Profs. Marcu and Louie for use of their microscopy facilities. Financial support for this work was provided by the NIH grants DK073901.
References
- [1].Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Nat. Meth. 2006;3:369. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- [2].Gobaa S, Hoehnel S, Roccio M, Negro A, Kobel S, Lutolf MP. Nat. Meth. 2011;8:949. doi: 10.1038/nmeth.1732. [DOI] [PubMed] [Google Scholar]
- [3].Guillame-Gentil O, Semenov O, Roca AS, Groth T, Zahn R, Voros J, Zenobi-Wong M. Adv. Mater. 2010;22:5443. doi: 10.1002/adma.201001747. [DOI] [PubMed] [Google Scholar]
- [4].Zorlutuna P, Annabi N, Camci-Unal G, Nikkhah M, Cha JM, Nichol JW, Manbachi A, Bae HJ, Chen SC, Khademhosseini A. Adv. Mater. 2012;24:1782. doi: 10.1002/adma.201104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- [6].Morrison SJ, Spradling AC. Cell. 2008;132:598. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1673. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Thery M. J. Cell Sci. 2010;123:4201. doi: 10.1242/jcs.075150. [DOI] [PubMed] [Google Scholar]
- [9].Suh KY, Khademhosseini A, Yang JM, Eng G, Langer R. Adv. Mater. 2004;16:584. [Google Scholar]
- [10].Revzin A, Russell RJ, Yadavalli VK, Koh WG, Deister C, Hile DD, Mellott MB, Pishko MV. Langmuir. 2001;17:5440. doi: 10.1021/la010075w. [DOI] [PubMed] [Google Scholar]
- [11].Albrecht DR, Tsang VL, Sah RL, Bhatia SN. Lab Chip. 2005;5:111. doi: 10.1039/b406953f. [DOI] [PubMed] [Google Scholar]
- [12].Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bauwens CL, Peerani R, Niebruegge S, Woodhouse KA, Kumacheva E, Husain M, Zandstra PW. Stem Cells. 2008;26:2300. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- [14].Dusseiller MR, Schlaepfer D, Koch M, Kroschewski R, Textor M. Biomaterials. 2005;26:5917. doi: 10.1016/j.biomaterials.2005.02.032. [DOI] [PubMed] [Google Scholar]
- [15].Wang WJ, Itaka K, Ohba S, Nishiyama N, Chung UI, Yamasaki Y, Kataoka K. Biomaterials. 2009;30:2705. doi: 10.1016/j.biomaterials.2009.01.030. [DOI] [PubMed] [Google Scholar]
- [16].Shah SS, Kim M, Cahill-Thompson K, Tae G, Revzin A. Soft Matter. 2011;7:3133. [Google Scholar]
- [17].Tuleuova N, Lee JY, Lee J, Ramanculov E, Zern MA. A. Revzin, Biomaterials. 2010;31:9221. doi: 10.1016/j.biomaterials.2010.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Moeller HC, Mian MK, Shrivastava S, Chung BG, Khademhosseini A. Biomaterials. 2008;29:752. doi: 10.1016/j.biomaterials.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Charnley M, Textor M, Khademhosseini A, Lutolf MP. Integrative Bio. 2009;1:625. doi: 10.1039/b918172p. [DOI] [PubMed] [Google Scholar]
- [20].Fukuda J, Khademhosseini A, Yeo Y, Yang XY, Yeh J, Eng G, Blumling J, Wang CF, Kohane DS, Langer R. Biomaterials. 2006;27:5259. doi: 10.1016/j.biomaterials.2006.05.044. [DOI] [PubMed] [Google Scholar]
- [21].Lee HJ, Lee JS, Chansakul T, Yu C, Elisseeff JH, Yu SM. Biomaterials. 2006;27:5268. doi: 10.1016/j.biomaterials.2006.06.001. [DOI] [PubMed] [Google Scholar]
- [22].Tsang VL, Chen AA, Cho LM, Jadin KD, Sah RL, DeLong S, West JL, Bhatia SN. FASEB J. 2007;21:790. doi: 10.1096/fj.06-7117com. [DOI] [PubMed] [Google Scholar]
- [23].Lee JY, Tuleuova N, Jones CN, Ramanculov E, Zern MA, Revzin A. Integ. Biol. 2009;1:460. doi: 10.1039/b905757a. [DOI] [PubMed] [Google Scholar]
- [24].Bhatia SN, Balis UJ, Yarmush ML, Toner M. J. Biomater. Sci., Polym. Ed. 1998;9:1137. doi: 10.1163/156856298x00695. [DOI] [PubMed] [Google Scholar]
- [25].Kolind K, Leong KW, Besenbacher F, Foss M. Biomaterials. 2012;33:6626. doi: 10.1016/j.biomaterials.2012.05.070. [DOI] [PubMed] [Google Scholar]
- [26].Wells RG. Hepatology. 2008;47:1394. doi: 10.1002/hep.22193. [DOI] [PubMed] [Google Scholar]
- [27].You J, Park S-A, Shin D-S, Patel D, Raghunathan VK, Kim M, Murphy CJ, Tae G, Revzin A. Tissue Eng. doi: 10.1089/ten.tea.2012.0681. Part A DOI: 10.1089/ten.TEA.20120681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tae G, Kim YJ, Choi WI, Kim M, Stayton PS, Hoffman AS. Biomacromolecules. 2007;8:1979. doi: 10.1021/bm0701189. [DOI] [PubMed] [Google Scholar]
- [29].Tae G, Kornfield JA, Hubbell JA. Biomaterials. 2005;26:5259. doi: 10.1016/j.biomaterials.2005.01.042. [DOI] [PubMed] [Google Scholar]
- [30].Tae G, Scatena M, Stayton PS, Hoffman AS. J. Biomater. Sci., Polym. Ed. 2006;17:187. doi: 10.1163/156856206774879090. [DOI] [PubMed] [Google Scholar]
- [31].Kim M, Lee JY, Jones CN, Revzin A, Tae G. Biomaterials. 2010;31:3596. doi: 10.1016/j.biomaterials.2010.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mederacke I, Wursthorn K, Kirschner J, Rifai K, Manns MP, Wedemeyer H, Bahr MJ. Liver Int. 2009;29:1500. doi: 10.1111/j.1478-3231.2009.02100.x. [DOI] [PubMed] [Google Scholar]
- [33].Georges PC, Hui JJ, Gombos Z, McCormick ME, Wang AY, Uemura M, Mick R, Janmey PA, Furth EE, Wells RG. Am. J. Physiology Gastrointestinal Liver Physiology. 2007;293:G1147. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- [34].Bastard C, Bosisio MR, Chabert M, Kalopissis AD, Mahrouf-Yorgov M, Gilgenkrantz H, Mueller S, Sandrin L. World J. Gastroenterology. 2011;17:968. doi: 10.3748/wjg.v17.i8.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mori R, Sakai Y, Nakazawa K. J. Biosci. Bioeng. 2008;106:237. doi: 10.1263/jbb.106.237. [DOI] [PubMed] [Google Scholar]
- [36].Wong SF, No DY, Choi YY, Kim DS, Chung BG, Lee SH. Biomaterials. 2011;32:8087. doi: 10.1016/j.biomaterials.2011.07.028. [DOI] [PubMed] [Google Scholar]
- [37].Chen AA, Khetani SR, Lee S, Bhatia SN, Van Vliet KJ. Biomaterials. 2009;30:1113. doi: 10.1016/j.biomaterials.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Park JS, Chu JS, Tsou AD, Diop R, Tang Z, Wang A, Li S. Biomaterials. 2011;32:3921. doi: 10.1016/j.biomaterials.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Guvendiren M, Burdick JA. Nat. Commun. 2012;3:1269. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- [40].Semler EJ, Lancin PA, Dasgupta A, Moghe PV. Biotechnol. Bioeng. 2005;89:296. doi: 10.1002/bit.20328. [DOI] [PubMed] [Google Scholar]
- [41].Kane BJ, Zinner MJ, Yarmush ML, Toner M. Anal. Chem. 2006;78:4291. doi: 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- [42].Dunn JCY, Yarmush ML, Koebe HG, Tompkins RG. FASEB J. 1989;3:174. doi: 10.1096/fasebj.3.2.2914628. [DOI] [PubMed] [Google Scholar]
- [43].Dunn JC, Tompkins RG, Yarmush ML. Biotechnol. Prog. 1991;7:237. doi: 10.1021/bp00009a007. [DOI] [PubMed] [Google Scholar]
- [44].Xia L, Sakban RB, Qu YH, Hong X, Zhang WX, Nugraha B, Tong WH, Ananthanarayanan A, Zheng BX, Chau IYY, Jia RR, McMillian M, Silva J, Dallas S, Yu H. Biomaterials. 2012;33:2165. doi: 10.1016/j.biomaterials.2011.12.006. [DOI] [PubMed] [Google Scholar]
- [45].Otsuka H, Hirano A, Nagasaki Y, Okano T, Horiike Y, Kataoka K. ChemBioChem. 2004;5:850. doi: 10.1002/cbic.200300822. [DOI] [PubMed] [Google Scholar]
- [46].Fukuda J, Sakai Y, Nakazawa K. Biomaterials. 2006;27:1061. doi: 10.1016/j.biomaterials.2005.07.031. [DOI] [PubMed] [Google Scholar]
- [47].Godoy P, Hengstler JG, Ilkavets I, Meyer C, Bachmann A, Muller A, Tuschl G, Mueller SO, Dooley S. Hepatology. 2009;49:2031. doi: 10.1002/hep.22880. [DOI] [PubMed] [Google Scholar]
- [48].Beamish JA, Geyer LC, Haq-Siddiqi NA, Kottke-Marchant K, Marchant RE. Biomaterials. 2009;30:6286. doi: 10.1016/j.biomaterials.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lee JY, Shah SS, Yan J, Howland MC, Parikh AN, Pan TR, Revzin A. Langmuir. 2009;25:3880. doi: 10.1021/la803635r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Revzin A, Rajagopalan P, Tilles AW, Berthiaume FO, Yarmush ML, Toner M. Langmuir. 2004;20:2999. doi: 10.1021/la035827w. [DOI] [PubMed] [Google Scholar]
- [51].Geerts A. Semin. Liver Dise. 2001;21:311. doi: 10.1055/s-2001-17550. [DOI] [PubMed] [Google Scholar]
- [52].Wisse E. Med. Chir. Dig. 1977;6:409. [PubMed] [Google Scholar]
- [53].Qin D, Xia YN, Whitesides GM. Nat. Protocols. 2010;5:491. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- [54].Tang MD, Golden AP, Tien J. J. Am. Chem. Soc. 2003;125:1–2988. doi: 10.1021/ja037677h. [DOI] [PubMed] [Google Scholar]
- [55].Gosey LL, Howard RM, Witebsky FG, Ognibene FP, Wu TC, Gill VJ, MacLowry JD. J. Clin. Microbiol. 1985;22:803. doi: 10.1128/jcm.22.5.803-807.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yamaguchi N, Kiick KL. Biomacromolecules. 2005;6:1921. doi: 10.1021/bm050003+. [DOI] [PMC free article] [PubMed] [Google Scholar]