Abstract
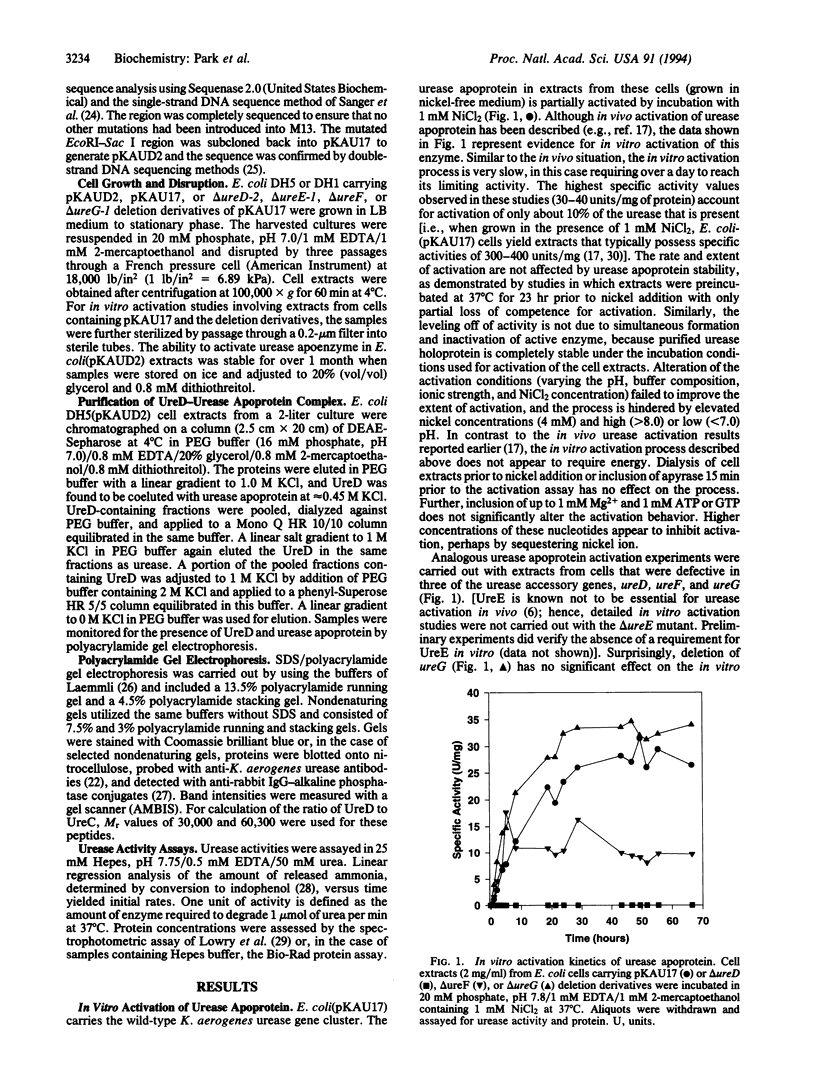
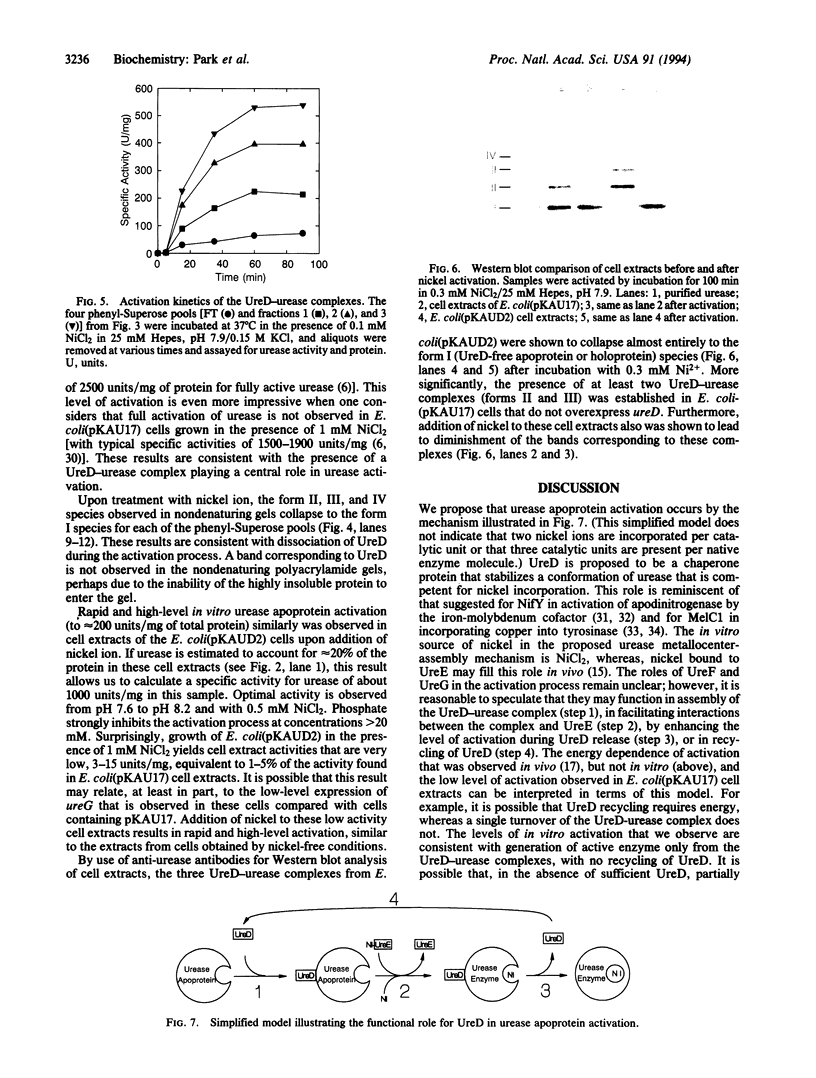
The formation of active urease in Klebsiella aerogenes requires the presence of three structural genes for the apoprotein (ureA, ureB, and ureC), as well as four accessory genes (ureD, ureE, ureF, and ureG) that are involved in functional assembly of the metallocenter in this nickel-containing enzyme. Slow and partial activation of urease apoprotein was observed after addition of nickel ion to extracts of Escherichia coli cells bearing a plasmid containing the K. aerogenes urease gene cluster or derivatives of this plasmid with deletions in ureE, ureF, or ureG. In contrast, extracts of cells containing a ureD deletion derivative failed to generate active urease, thus highlighting a key role for UreD in the metallocenter assembly process. Site-directed mutagenesis methods were used to overexpress ureD in the presence of the other urease genes, and the UreD protein was found to copurify with urease. A molecule of native urease apoprotein is capable of binding 0, 1, 2, or 3 molecules of UreD, consistent with a trimeric structure of urease catalytic units. The UreD-urease apoprotein complexes are competent for activation by nickel, with the level of activity obtained being directly related to the number of UreD molecules bound per urease molecule. Activation of the UreD-urease complexes is rapid and accompanied by UreD dissociation. We propose that UreD is a chaperone protein which stabilizes a urease apoprotein conformation that is competent for nickel incorporation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benson E. W., Howe H. B., Jr Reversion and interallelic complementation at four urease loci in Neurospora crassa. Mol Gen Genet. 1978 Oct 24;165(3):277–282. doi: 10.1007/BF00332527. [DOI] [PubMed] [Google Scholar]
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Chen L. Y., Chen M. Y., Leu W. M., Tsai T. Y., Lee Y. H. Mutational study of Streptomyces tyrosinase trans-activator MelC1. MelC1 is likely a chaperone for apotyrosinase. J Biol Chem. 1993 Sep 5;268(25):18710–18716. [PubMed] [Google Scholar]
- Chen L. Y., Leu W. M., Wang K. T., Lee Y. H. Copper transfer and activation of the Streptomyces apotyrosinase are mediated through a complex formation between apotyrosinase and its trans-activator MelC1. J Biol Chem. 1992 Oct 5;267(28):20100–20107. [PubMed] [Google Scholar]
- Collins C. M., Gutman D. M. Insertional inactivation of an Escherichia coli urease gene by IS3411. J Bacteriol. 1992 Feb;174(3):883–888. doi: 10.1128/jb.174.3.883-888.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cussac V., Ferrero R. L., Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992 Apr;174(8):2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon N. E., Gazzola T. C., blakeley R. L., Zermer B. Letter: Jack bean urease (EC 3.5.1.5). A metalloenzyme. A simple biological role for nickel? J Am Chem Soc. 1975 Jul 9;97(14):4131–4133. doi: 10.1021/ja00847a045. [DOI] [PubMed] [Google Scholar]
- Homer M. J., Paustian T. D., Shah V. K., Roberts G. P. The nifY product of Klebsiella pneumoniae is associated with apodinitrogenase and dissociates upon activation with the iron-molybdenum cofactor. J Bacteriol. 1993 Aug;175(15):4907–4910. doi: 10.1128/jb.175.15.4907-4910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. D., Mobley H. L. Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J Bacteriol. 1989 Dec;171(12):6414–6422. doi: 10.1128/jb.171.12.6414-6422.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee M. H., Mulrooney S. B., Hausinger R. P. Purification, characterization, and in vivo reconstitution of Klebsiella aerogenes urease apoenzyme. J Bacteriol. 1990 Aug;172(8):4427–4431. doi: 10.1128/jb.172.8.4427-4431.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Mulrooney S. B., Renner M. J., Markowicz Y., Hausinger R. P. Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J Bacteriol. 1992 Jul;174(13):4324–4330. doi: 10.1128/jb.174.13.4324-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Pankratz H. S., Wang S., Scott R. A., Finnegan M. G., Johnson M. K., Ippolito J. A., Christianson D. W., Hausinger R. P. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993 Jun;2(6):1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S., Jacobi A., Schlensog V., Böhm R., Sawers G., Böck A. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):123–135. doi: 10.1111/j.1365-2958.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- Mackay E. M., Pateman J. A. Nickel requirement of a urease-deficient mutant in Aspergillus nidulans. J Gen Microbiol. 1980 Jan;116(1):249–251. doi: 10.1099/00221287-116-1-249. [DOI] [PubMed] [Google Scholar]
- Mackay E. M., Pateman J. A. The regulation of urease activity in Aspergillus nidulans. Biochem Genet. 1982 Aug;20(7-8):763–776. doi: 10.1007/BF00483972. [DOI] [PubMed] [Google Scholar]
- Maier T., Jacobi A., Sauter M., Böck A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol. 1993 Feb;175(3):630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Bothling L E, Polacco J C, Cianzio S R. Pleiotropic soybean mutants defective in both urease isozymes. Mol Gen Genet. 1987 Oct;209(3):432–438. doi: 10.1007/BF00331146. [DOI] [PubMed] [Google Scholar]
- Mobley H. L., Hausinger R. P. Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev. 1989 Mar;53(1):85–108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney S. B., Hausinger R. P. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990 Oct;172(10):5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney S. B., Pankratz H. S., Hausinger R. P. Regulation of gene expression and cellular localization of cloned Klebsiella aerogenes (K. pneumoniae) urease. J Gen Microbiol. 1989 Jun;135(6):1769–1776. doi: 10.1099/00221287-135-6-1769. [DOI] [PubMed] [Google Scholar]
- Park I. S., Hausinger R. P. Site-directed mutagenesis of Klebsiella aerogenes urease: identification of histidine residues that appear to function in nickel ligation, substrate binding, and catalysis. Protein Sci. 1993 Jun;2(6):1034–1041. doi: 10.1002/pro.5560020616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990 Nov;15(11):430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Waugh R., Boxer D. H. Pleiotropic hydrogenase mutants of Escherichia coli K12: growth in the presence of nickel can restore hydrogenase activity. Biochimie. 1986 Jan;68(1):157–166. doi: 10.1016/s0300-9084(86)81080-x. [DOI] [PubMed] [Google Scholar]
- White T. C., Harris G. S., Orme-Johnson W. H. Electrophoretic studies on the assembly of the nitrogenase molybdenum-iron protein from the Klebsiella pneumoniae nifD and nifK gene products. J Biol Chem. 1992 Nov 25;267(33):24007–24016. [PubMed] [Google Scholar]
- Wu L. F. Putative nickel-binding sites of microbial proteins. Res Microbiol. 1992 Mar-Apr;143(3):347–351. doi: 10.1016/0923-2508(92)90027-l. [DOI] [PubMed] [Google Scholar]






