Abstract
Tuberous sclerosis complex (TSC) is a genetic disorder characterized by seizures and tumor formation in multiple organs, mainly in the brain, skin, kidney, lung and heart. Renal cell carcinoma (RCC) occurs in ∼3% of TSC patients, and typically develops at age <50. Here we describe genetic findings in two TSC patients with multiple renal tumors, each of whom had the germline mutation TSC2 p.R905Q. The first (female) TSC patient had a left followed by a right nephrectomy at ages 24 and 27. Both kidneys showed multifocal TSC-associated papillary RCC (PRCC). Targeted, next-generation sequencing (NGS) analysis of TSC2 in five tumors (four from the left kidney, one from the right) showed loss of heterozygosity in one tumor, and four different TSC2 point mutations (p.E1351*, p.R1032*, p.R1713H, c.4178_4179delCT) in the other four samples. Only one of the 11 other tumors available from this patient had one of the TSC2 second hit mutations identified. Whole-exome analysis of the five tumors identified a very small number of additional mutated genes, with an average of 3.4 nonsilent coding, somatic mutations per tumor, none of which were seen in >1 tumor. The second (male) TSC patient had bilateral partial nephrectomies (both at age 36), with similar findings of multifocal PRCC. NGS analysis of TSC2 in two of these tumors identified a second hit mutation c.2355+1G>T in one sample that was not seen in other tumors. In conclusion, we report the first detailed genetic analysis of RCCs in TSC patients. Molecular studies indicate that tumors developed independently due to various second hit events, suggesting that these patients experienced a ‘shower’ of second hit mutations in TSC2 during kidney development leading to this severe phenotype.
Introduction
Tuberous sclerosis complex (TSC) is an autosomal dominant genetic disorder characterized by tumor development in multiple organs including the kidney, due to inactivating mutations in either TSC1 or TSC2 (1). Renal disease is very common in TSC, and occurs in three major forms, angiomyolipoma, cystic disease and renal cell carcinoma (RCC), that are often seen simultaneously in the same patient (2,3). Renal angiomyolipoma is seen in ∼80% of TSC subjects and develops progressively with age (4,5). Angiomyolipoma is commonly bilateral and multifocal. Renal cysts are also often seen in TSC, and vary from early childhood onset relatively severe polycystic disease, to late-onset milder polycystic kidney disease, to singleton cysts (3,6).
Although much rarer than angiomyolipoma, TSC RCC were first described decades ago (7). However, the possibility of confusion of epithelioid angiomyolipoma with RCC has contributed to uncertainty in regards to the frequency of renal carcinoma in TSC (8). More recent studies have delineated multiple types of RCC occurring in TSC, including our recent report using modern immunohistochemistry antibody sets to identify three distinct groups of RCC in 19 TSC patients (9). The most common type of RCC we identified (24 of 46 tumors, 52%) had a unique morphology distinct from all of the conventional forms of RCC. Hallmark features of this type of TSC-associated RCC were prominent papillary architecture and a uniform lack of expression of succinate dehydrogenase (SDH) subunit B, prompting the term ‘TSC-associated papillary RCC’ (9). The second most common group of TSC RCC (15 of 46, 33%) was morphologically similar to a hybrid oncocytic/chromophobe tumor (9). The third group of TSC renal epithelial neoplasms (7 of 46, 15%) had features distinct from the first two groups as well as classic types of RCC, and could not be classified further (9).
Multifocality of RCC in TSC has also been a consistent observation going back to the earliest reports (10–13). Here we report the first detailed genetic analysis of multifocal RCC in two TSC patients.
Results
First TSC patient with multifocal RCC
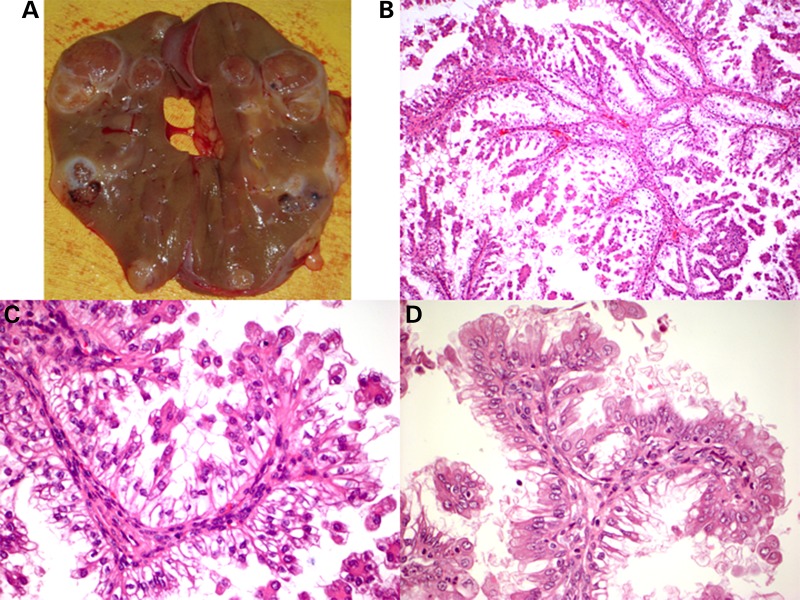
Patient 1 was diagnosed with TSC at age 14 when she presented with seizures, hypomelanotic macules and mild facial angiofibroma. She did not have intellectual disability or behavior disorders, and her epilepsy was easily managed. Blood DNA analysis identified a well-known TSC2 missense mutation c.2714G>A, p.(R905Q), associated with a mild phenotype (14). Renal masses were identified by ultrasound when she was 16. At age 24 she developed intermittent fevers and left sided abdominal pain. CT scan showed a 7 cm tumor in her left kidney which had doubled in size in the preceding 6 months, and several smaller lesions. Left nephrectomy was performed, with findings of multiple solid lesions without fat component (Fig. 1A). Pathological analysis showed that there were 12 apparently separate tumor nodules, all with a distinct papillary morphology as described (9) (Fig. 1B and C). During follow-up after her surgery, renal lesions in the remaining right kidney were observed to grow progressively, and right nephrectomy was performed at age 27. Pathological evaluation showed the presence of three apparently separate tumor nodules, and histologic analysis showed a similar papillary morphology (Fig. 1D).
Figure 1.
Pathology of papillary tumors seen in the first TSC patient. (A) Gross photograph of left nephrectomy from the first patient. (B–D) Microscopic findings from two different RCC tumors (C, D) from the first patient. (B) 100×; (C) 1000×; (D) 1000×.
Molecular studies on patient 1
Fresh frozen tissue was available from five tumors, four from the left kidney and one from the right. Loss of heterozygosity (LOH) analysis using both sequencing across the germline c.2714G>A, p.(R905Q) mutation and microsatellite markers near TSC2 showed LOH in one tumor sample, but none of the others (data not shown). The four tumor samples without LOH were subject to targeted next-generation sequencing (NGS) at high read depth for analysis of mutations in TSC2 (Table 1). This confirmed that none of the four tumors studied showed LOH at c.2714G>A, but instead each had a distinct second mutation in TSC2, comprising two nonsense mutations, a 2 nt deletion mutation, and a missense mutation, thought to be inactivating (http://chromium.liacs.nl/LOVD2/TSC). All four sites of mutation were confirmed by Sanger sequencing in the respective samples. The four mutations were also evaluated by Sanger sequencing on the additional 10 tumor specimens that had DNA available from formalin-fixed paraffin-embedded (FFPE) tissues. An FFPE sample, from the ipsilateral kidney, had one of the TSC2 nonsense mutations. It is possible that this second tumor arose from the same tumor clone as the first; however, it is also possible that this second hit mutation had occurred independently in two separate tumors. None of the other mutations were seen recurrently. None of the other 10 tumor samples showed evidence of LOH by either Sanger sequencing at the germline mutation site, or using microsatellite markers near TSC2.
Table 1.
Genetic analysis of TSC-associated PRCC from two TSC patients
| Patient | Germline variant in TSC2 | Tumor number | Second hit mutation in TSC2 (with mutant allele frequency) | Mutations in other genes (with mutant allele frequency) |
|---|---|---|---|---|
| Female | c.2714G>A; p.R905Q; chr16:2126143 | 1 (left kidney) | c.4051G>T; p.E1351* (23%); chr16:2134274 |
PROS1 c.1294C>T; p.R432W (20%)a; chr3:93605209 TACC3 c.2188G>T; p.E730* (17%); chr4:1742678 NPFFR2 c.925C>T; p.R309W (22%)a; chr4:73012885 XYLT1 c.1962_1963insC (29%); chr16:17228394-17228395 |
| 2 (right kidney) | c.4178_4179delCT (20%); chr16:2134401-2134402 |
GALNTL6 c.268C>T; p.P90S (30%); chr4:173232785 NUP214 c.3395C>G; p.P1132R (19%); chr9:134053773 TLL2 c.1771C>A; p.R591S (24%)a; chr10:98146791 GLUD2 c.655G>T; p.V219F (19%); chrX:120182193 |
||
| 3 (left kidney) | c.3094C>T; p.R1032* (34%); chr16:2129160 |
USP34 c.7749+1A>C (34%); chr2:61450193 MS4A7 c.644C>G; p.P215R (37%); chr11:60160255 NDE1 c.329T>A; p.L110* (11%); chr16:15771749 |
||
| 4 (left kidney) | c.5138G>A; p.R1713H (23%); chr16:2138118 |
ZNF507 c.1506_1513delAATGCCTA (26%); chr19:32845242-32845249 |
||
| 5 (left kidney) | LOH: p.R905Q (80%) |
ZRANB1 c.276G>T; p.M92I (16%); chr10:126631338 PANX1 c.212C>T; p.S71F (27%); chr11:93886687 PHF20 c.1226T>A; p.M409K (29%); chr20:34459695 RASA1 c.926T>G; p.V309G (33%)b; chr5:86633817 PNISR c.1795A>G; p.R599G (18%); chr6:99849039 |
||
| Male | c.2714G>A; p.R905Q; chr16:2126143 | 1 (right kidney) | c.2355+1G>T (11%); chr16:2122985 | NA |
| 2 (left kidney) | Analysis failed due to not sufficient amount of DNA |
aMutations in these genes at these amino acids have been reported previously (COSMIC http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/, TumorPortal http://cancergenome.broadinstitute.org).
bRASA1 has been identified as a cancer gene both in the ‘PanCan’ dataset and in head and neck cancer (http://cancergenome.broadinstitute.org) (15).
Whole-exome sequencing performed on the five fresh frozen samples confirmed the presence of each second TSC2 mutation and LOH in the respective samples (Table 1, read frequency for the c.2714A nt was 80% in the first sample), and revealed relatively few somatic mutations in each tumor. A median of four (range 1–5) nonsynonymous somatic mutations were identified at allele fractions >10%. These affected 17 genes, none of which was mutated in more than one sample (Table 1). Only one of these 17 genes, RASA1, has been reported as significantly mutated in cancer, (http://cancergenome.broadinstitute.org) (15), both in the ‘PanCan’ dataset and in head and neck cancer, though at very low frequency, 2 and 4%, respectively. RASA1 encodes p120RasGAP, which acts as a suppressor of RAS function, and mutations in RASA1 are a cause of capillary malformation-arteriovenous malformation (16,17). Three mutations in other genes (PROS1, NPFFR2 and TLL2; Table 1) have been seen previously in various human cancers, though none are recognized as cancer genes (COSMIC http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/, TumorPortal http://cancergenome.broadinstitute.org).
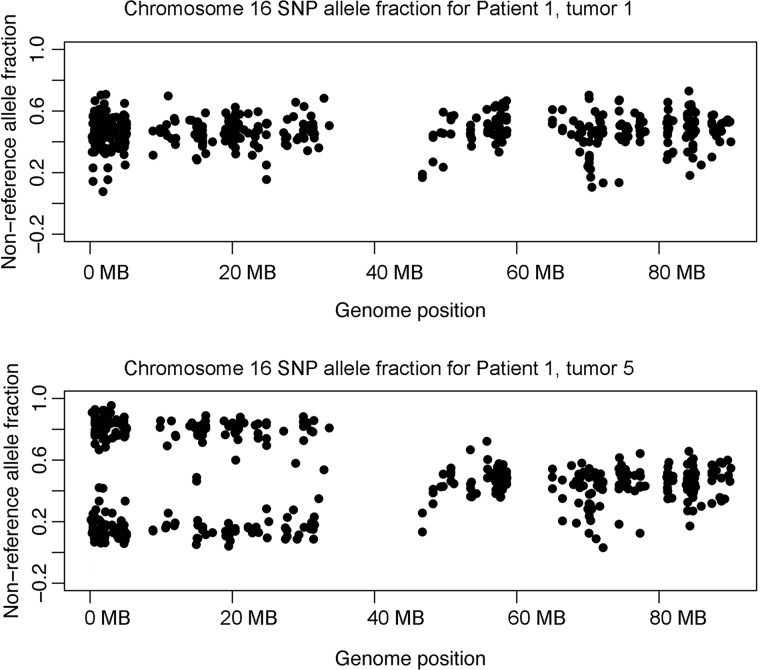
Somatic copy number changes were analyzed comparing the read depth of these whole-exome sequence data to a set of controls using CapSeg (github.com/aaronmck/CapSeg). No copy number changes were seen. Global LOH analysis considering SNP allele frequencies for all heterozygous SNPs in this individual indicated there was LOH for SNPs across the length of chromosome 16p in the same tumor sample (Table 1, # 5) that showed LOH for the TSC2 c.2714G>A, p.(R905Q) mutation (Fig. 2). No evidence for LOH by SNP allele distortion was seen elsewhere in the genome in that sample or for any genomic region in any of the other samples. This implies that the single tumor with LOH on 16p had copy neutral LOH, with replacement of chromosome 16p bearing the wild-type TSC2 allele with the mutant-bearing chromosome 16p.
Figure 2.
Chromosome 16 SNP allele fraction plot for two renal tumors from patient 1. A plot of the non-reference allele fraction for heterozygous SNPs on chromosome 16 is shown for patient 1, tumor 1 (top) and patient 1, tumor 5 (bottom). Note that nearly all SNP allele fractions for tumor 5 from 16p (0–35 MB) were in the range of 10–30 or 70–90%, consistent with loss of one allele or the other. This is not seen for 16q (45–90 MB).
Second TSC patient with multifocal RCC
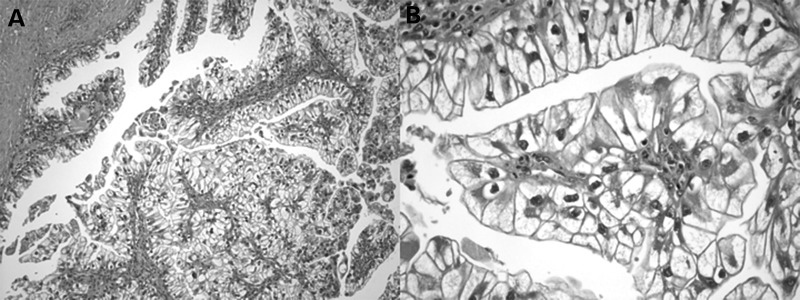
Patient 2 was diagnosed with TSC at age 5 after he developed partial seizures and was found to have numerous hypomelanotic ‘confetti’ macules. Family history was notable for many individuals with similar hypomelanotic macules and seizures, inherited in an autosomal dominant pattern, but no family history of RCC. Beginning at age 10, he developed fleshy tumors on his forearms and around his waist that were never biopsied and are of unknown histology. He has never had facial angiofibroma or periungual fibroma, dilated retinal examinations have all been normal, and his most recent brain MRI in 2014 showed no cortical tubers. In 2009, he was found to have bilateral, multifocal renal masses and had a partial right nephrectomy followed 2 months later by a partial left nephrectomy. All lesions resected from each kidney showed the same pathology of TSC-associated PRCC (9) (Fig. 3). He has had repeated cryoablation procedures in the interim for additional renal tumors until April 2014 when he was started on everolimus. His most recent abdominal MRI showed no interval growth of existing lesions and no new lesions. Germline mutational testing revealed that he had the same c.2714G>A, p.(R905Q) mutation seen in the Patient 1.
Figure 3.
Microscopic pathology of a papillary tumor seen in the second TSC patient. (A) 100×; (B) 1000×.
FFPE DNA from two tumors was subjected to NGS for analysis of mutations in TSC1 and TSC2. One tumor was found to have a splice mutation at 11% allele frequency in TSC2, while the other tumor sample had insufficient DNA for this analysis (Table 1). Three additional tumors available as FFPE DNA as well as the sample that failed were analyzed by Sanger sequencing. None showed evidence of LOH at the c.2714G>A, p.(R905Q) site and none had the TSC2 splice mutation.
Discussion
Renal cell carcinoma has been recognized in TSC for several decades (7,10,11,18). In contrast to other genetic syndromes in which kidney cancer risk is a prominent feature, the increased risk for development of RCC in TSC appears to be confined to those of age <50, while the lifetime incidence of RCC in TSC is similar to that of the general population, 2–4% (19). We recently had the opportunity to review clinical features and histopathologic subtypes in a collection of 19 TSC patients with 46 distinct renal epithelial tumors (9). A marked skewing towards younger age of onset was also seen in this collection, with 17 of 19 (90%) patients having age <50. The most common type of RCC we identified (24 of 46, 52%) had a unique morphology, including prominent papillary architecture and lack of expression of SDH subunit B, and was called TSC-associated PRCC, a subtype with morphologic features distinct from the more common PRCC (9). Both of the individuals studied here had multifocal occurrence of TSC-associated PRCC, with multiple tumors in each kidney. Multifocality for this type of RCC was also common in our larger series with three of seven other patients having multifocal lesions, including two others in whom bilateral tumors were identified (9). Although our series likely does not reflect the true population frequency of multifocal disease due to ascertainment effects, multifocality of RCC in TSC is clearly relatively common.
These observations raised the question of the molecular mechanism of epithelial tumor formation in TSC. Since the vast majority (>90%) of TSC subjects do not develop renal epithelial neoplasms, following the Poisson distribution, one would expect that multifocal renal epithelial tumors in RCC would be rare, seen in <<1% of patients. Furthermore, if the expected number of RCC occurring in a TSC patient is 1 (an overestimate), then the probability of seeing 15 such independent tumors in one patient, as reported here, is 2.8 × 10−13 (Poisson distribution with λ = 1). Thus, these observations suggest that in certain TSC patients there is a predisposition to multifocal RCC, whereas in the majority these tumors are not seen at all.
One mechanism to explain the high frequency of RCC multifocality in TSC would be if the tumors seen were not due to independent tumorigenic events, but rather reflected an unusual growth pattern, such that the multiple lesions seen in a single kidney, or even bilaterally were part of the same clonal tumor expansion. The tendency toward uniform tumor histology, as seen in the two cases presented here, would of course fit this model.
In contrast, we show here that five separate tumors of papillary morphology from a single patient had five distinct second hit genetic events leading to biallelic inactivation of TSC2. Furthermore, the possibility that an early genetic event elsewhere in the genome was enabling for multifocal tumor development was considerably reduced by whole-exome sequencing analysis that identified a very small number of additional somatic genetic events in each tumor, and failed to identify any genetic alterations seen in more than one tumor. We note that whole-exome sequencing does not cover all exons perfectly, though 81–88% of target nt had a read depth of ≥20× in the samples analyzed here, so there is a possibility that a mutation occurring in a region covered poorly by the exome sequencing might be present in all tumors and be enabling for tumor development.
Another mechanism to explain RCC multifocality in TSC is that some alternative enabling event for renal tumorigenesis is present in a small fraction of TSC individuals, either on a systemic or germline basis or a mosaic basis in the some fraction of cells in the developing kidney. Such an event might be epigenetic silencing of a gene required for cellular senescence in response to two-hit inactivation of TSC2, such as CDKN2A or TP53. Indeed, one can predict that second hit events are quite likely to occur in TSC2 in many cells in every TSC patient with a TSC2 mutation, and hence ask the question: why does not every patient with TSC develop multifocal RCC? Epigenetic silencing events that enable growth of renal cells with two-hit inactivation of TSC2 would not have been detected by the analyses performed here. Furthermore, many other events might influence tumor development in the setting of two-hit inactivation of TSC2 leading to the marked variability in apparent predisposition to renal tumor formation in TSC.
Our data clearly support the model that the multiple tumors developing in these two patients were due to a relatively large number (‘a shower’) of second hit genetic events in TSC2, that occurred in individual progenitor cells bilaterally, enabling tumor development. The nature of additional predisposing events is unknown.
It is notable that very few additional genetic events beyond second hit mutations of TSC2 were observed in these tumors. The only known ‘cancer gene’ identified with a mutation in this set of cancers was RASA1. The RASA1 p.V309G mutation was likely to disrupt function. However, the mutant allele frequency, 33%, is relatively low, in contrast to the allele frequency seen at the TSC2 point mutation, 80%, suggesting that the other allele was intact, so that only single allele loss of function was present. A nonsense mutation in TACC3 was also identified in one sample (Table 1). TACC3 has previously been reported to interact with TSC2 at the nuclear membrane (20), though it is hard to imagine how mutations in both genes would facilitate tumor development. Although a limited dataset, these genetic findings emphasize the distinction between this type of renal cancer, and the classic other forms, as none of the genes commonly mutated in clear cell carcinoma (VHL, PBRM1, KDM5C, PTEN, SETD2, BAP1, MTOR, TP53 and PIK3CA) (21), PRCC (MET) (22) or chromophobe cancer (TP53, PTEN) (23) were mutated in these tumors.
As noted previously (9), all of the TSC-associated PRCC identified in our earlier larger study did not express the SDHB protein by immunohistochemistry. SDH/mitochondrial complex II is a critical respiratory enzyme that resides in the mitochondria and catalyzes the oxidation of succinate to fumarate (24). Inherited or somatic bi-allelic mutation in any of the five SDH genes (SDHA, SDHB, SDHC, SDHD and SDHAF2) will cause loss of expression of SDHB, and each of the five genes is considered a tumor suppressor gene, although relatively few patients have been identified with germline mutations (25,26). Succinate accumulates in the absence of SDH activity, and inhibits HIF-α prolyl hydroxylases, leading to stabilization and activation of HIF-1α, with diverse downstream effects (27). It is possible that this effect in these tumors contributes to tumorigenesis and provides a functional mechanism similar to mutation and loss of expression of VHL. Careful scrutiny of the whole-exome sequence data for the SDH genes was performed to investigate the potential genetic basis of the lack of expression of SDHB seen in these tumors. There was good read depth coverage for all exons for the five SDH genes in these five samples, and no mutations of any kind were observed. Thus, the mechanism of lack of expression of this critical mitochondrial complex protein in these tumors is not explained by standard genetic events, and may instead reflect the cell of origin or epigenetic/chromatin effects. Such an epigenetic event is another candidate predisposing mechanism for multifocal renal tumor development in some TSC patients.
Finally, it is notable that both of the patients studied here had the same germline mutation in TSC2 c.2714G>A, p.(R905Q). This is a relatively common TSC2 mutation occurring through the classic C>T deamination reaction at a methylated CpG site. Forty individuals carrying this mutation have been reported previously, including 25 from a large family in which some members did not meet diagnostic criteria for TSC despite full evaluation (14). Eleven members of the large family were >10 years of age, and had renal imaging performed which did not show evidence of RCC (14). The additional 15 individuals with this mutation, not in the large family, also appeared overall to be mildly affected, similar to the two patients described here. However, renal imaging data was not available for those 15 individuals (14), and hence presence of RCC is unknown. Although relatively common, this mutation has been seen in <1% of TSC patients overall, and hence it remains an interesting coincidence that both of the patients available to us with multifocal RCC had this same mutation. Germline mutation data were available for three other patients in our pathologic series (9) with TSC-associated PRCC. None of those patients had the TSC2 R905Q mutation, but all three had a non-truncating mutation (two missense mutations and one in-frame deletion mutation). This suggests the possibility that non-truncating mutations in which there is residual TSC2 protein expression may be a predisposing factor for development of these TSC renal tumors. This possibility might be explained by previous observations that the TSC protein complex has effects on mTORC2 function, in the absence of GAP activity for mTORC1 (28). However, additional study is required to explore the hypothesis that there is an association between non-truncating TSC1/TSC2 mutations and TSC-associated PRCC.
Materials and Methods
Renal cell carcinoma
Tumor tissue samples and normal kidney were obtained at the time of surgical resection from two TSC patients. Tumor samples were stored as either fresh frozen or fixed with 10% formalin and embedded in paraffin. All samples were studied by a single specialist GU pathologist (C Wu). Frozen tissues were embedded in optimum cutting temperature compound for preparation of sections and standard pathologic evaluation (9). Paraffin-embedded materials were subject to histologic evaluation by similar means. All tumor samples used for DNA preparation were assessed to have at least 50% tumor cell content by histologic review. DNA was prepared from frozen tissues and paraffin blocks of tumor using the Puregene Genomic DNA Isolation kit (Qiagen) and the BiOstic FFPE Tissue DNA Isolation Kit (MO BIO Laboratories), respectively.
Microsatellite markers kg8 and STR7 were evaluated by PCR using fluorescently labeled primers followed by capillary electrophoresis, as previously described (29).
NGS and validation
Targeted NGS of the coding exons and intronic regions of TSC1 and TSC2 was performed as described previously on DNA extracted from fresh frozen tumor samples (30,31). Briefly, long-range PCR (3–8 kb amplicons) was performed on DNA prepared from the frozen tumor samples to amplify all of the coding exons and most of the intronic sequence of each of TSC1 and TSC2. Amplicons were purified and used to prepare a small fragment library for Illumina sequencing (31). Libraries from different samples were generated with different indices and then mixed at an equimolar ratio and sequenced on an Illumina GAIIx or HiSeq2000 sequencer (Illumina, San Diego, CA, USA) for 75 nt paired-end reads.
Primary sequence data were deconvoluted using the index sequences to individual sample files and converted to FASTQ format, aligned to the human genome using bwa-0.5.8c (Burrows-Wheeler Alignment) (32), filtered to eliminate reads of low quality and to reduce redundancy to a uniform 50 reads starting at each nucleotide position of interest in each direction. The data were then analyzed for sequence variants using tools from the Genome Analysis Toolkit (GATK) (33), including IndelGenotyperV2 and UnifiedGenotyper, to identify both indels and single-nucleotide variants. A second approach was used in parallel to analyze the sequence data, with capture of read calls at all positions using Samtools Pileup (SAMtools) (34), followed by custom processing in Python and Matlab to determine base call frequency at each position in each read orientation. The output from these analyses was manually reviewed in comparison with those from other samples, including controls, to exclude artifacts derived from the sequencing process. All variants seen at a frequency of ≥1% more than that seen in other samples were directly reviewed using the Integrative Genomics Viewer (35) to help confirm bona fide variant calls and to exclude sequencing artifacts. The median read depth for each coding exon of TSC2 was 7213 for the four samples analyzed, and the minimum for a single exon-sample pair was 3459. Novel or biologically significant single-nucleotide variants and indels with minor allele fractions ≥10% were confirmed using Sanger bidirectional sequencing.
NGS on two DNA samples extracted from FFPE samples was performed at the Center for Cancer Genome Discovery at Dana Farber Cancer Institute. Methods are similar to those described above (36), but included use of an Agilent SureSelect hybrid capture probe set for the entire (coding, intronic and 10 kb up and downstream regions) TSC1 and TSC2 genes. Bioinformatic analysis was similar to the above. The median read depth for each coding exon of TSC1 and TSC2 was 535 for one sample, with a minimum of 275. The second sample failed in this analysis due to insufficient amount of DNA.
Whole-exome sequencing
Exome sequencing was performed by the Broad Institute Genomics Platform and analyzed using a standard analytic pipeline. Briefly, reads were aligned using bwa, followed by indel realignment and quality score recalibration using the Genome Analysis Toolkit. Somatic mutations were identified using muTect (37) and Indelocator, which compared sequence variants called in the tumors with those called in normal kidney, and were then annotated using Oncotator (http://www.broadinstitute.org/oncotator). Exome capture targeted 32,950,014 nt, and mean read depth ranged from 58 to 94 reads per target nt. 81–88% of target nt had a read depth of ≥20×.
Copy number profiles were derived from fractional coverage values of each exon compared with a panel of normals using CapSeg (github.com/aaronmck/CapSeg), with the assistance of Lee Lichtenstein, Broad Institute, Cambridge, MA. Somatic LOH was detected using custom R scripts to visualize tumor allele frequencies of all common SNPs identified in the germline analysis of the matched normal sample.
Funding
This work is supported by NIH grants 1P01CA120964 and 2R37NS031535.
Acknowledgements
The authors thank the patients involved in this study for their participation; Ms. Kate Loranger at the Cancer Risk Program of the UCSF Helen Diller Family Comprehensive Cancer Center, Paul Van Hummelen at the Center for Cancer Genome Discovery at DFCI, Yvonne Li at Dana Farber Cancer Institute and Elizabeth Thiele and Alexandra Geffrey at Massachusetts General Hospital.
Conflict of Interest statement: None declared.
References
- 1.Kwiatkowski D.J., Thiele E.A., Whittemore V.H. Tuberous Sclerosis Complex. Weinheim, Germany: Wiley-VCH; 2010. [Google Scholar]
- 2.Cook J.A., Oliver K., Mueller R.F., Sampson J. A cross sectional study of renal involvement in tuberous sclerosis. J. Med. Genet. 1996;33:480–484. doi: 10.1136/jmg.33.6.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bissler J., Henske E.P. Renal manifestations of tuberous sclerosis complex. In: Kwiatkowski D.J., Whittemore V.H., Thiele E.A., editors. Tuberous Sclerosis Complex. Weinheim, Germany: Wiley-VCH; 2010. pp. 311–326. [Google Scholar]
- 4.Ewalt D.H., Sheffield E., Sparagana S.P., Delgado M.R., Roach E.S. Renal lesion growth in children with tuberous sclerosis complex. J. Urol. 1998;160:141–145. [PubMed] [Google Scholar]
- 5.Dixon B.P., Hulbert J.C., Bissler J.J. Tuberous sclerosis complex renal disease. Nephron Exp. Nephrol. 2011;118:e15–e20. doi: 10.1159/000320891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampson J.R., Maheshwar M.M., Aspinwall R., Thompson P., Cheadle J.P., Ravine D., Roy S., Haan E., Bernstein J., Harris P.C. Renal cystic disease in tuberous sclerosis: role of the polycystic kidney disease 1 gene. Am. J. Hum. Genet. 1997;61:843–851. doi: 10.1086/514888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Washecka R., Hanna M. Malignant renal tumors in tuberous sclerosis. Urology. 1991;37:340–343. doi: 10.1016/0090-4295(91)80261-5. [DOI] [PubMed] [Google Scholar]
- 8.Pea M., Bonetti F., Martignoni G., Henske E.P., Manfrin E., Colato C., Bernstein J. Apparent renal cell carcinomas in tuberous sclerosis are heterogeneous: the identification of malignant epithelioid angiomyolipoma. Am. J. Surg. Pathol. 1998;22:180–187. doi: 10.1097/00000478-199802000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Yang P., Cornejo K.M., Sadow P.M., Cheng L., Wang M., Xiao Y., Jiang Z., Oliva E., Jozwiak S., Nussbaum R.L., et al. Renal cell carcinoma in tuberous sclerosis complex. Am. J. Surg. Pathol. 2014;38:895–909. doi: 10.1097/PAS.0000000000000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinblatt M.E., Kahn E., Kochen J. Renal cell carcinoma in patients with tuberous sclerosis. Pediatrics. 1987;80:898–903. [PubMed] [Google Scholar]
- 11.Sampson J.R., Patel A., Mee A.D. Multifocal renal cell carcinoma in sibs from a chromosome 9 linked (TSC1) tuberous sclerosis family. J. Med. Genet. 1995;32:848–850. doi: 10.1136/jmg.32.11.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hidai H., Chiba T., Takagi Y., Taki A., Nagashima Y., Kuroko K. Bilateral chromophobe cell renal carcinoma in tuberous sclerosis complex. Int. J. Urol. 1997;4:86–89. doi: 10.1111/j.1442-2042.1997.tb00147.x. [DOI] [PubMed] [Google Scholar]
- 13.Al-Saleem T., Wessner L.L., Scheithauer B.W., Patterson K., Roach E.S., Dreyer S.J., Fujikawa K., Bjornsson J., Bernstein J., Henske E.P. Malignant tumors of the kidney, brain, and soft tissues in children and young adults with the tuberous sclerosis complex. Cancer. 1998;83:2208–2216. [PubMed] [Google Scholar]
- 14.Jansen A.C., Sancak O., D'Agostino M.D., Badhwar A., Roberts P., Gobbi G., Wilkinson R., Melanson D., Tampieri D., Koenekoop R., et al. Unusually mild tuberous sclerosis phenotype is associated with TSC2 R905Q mutation. Ann. Neurol. 2006;60:528–539. doi: 10.1002/ana.21037. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eerola I., Boon L.M., Mulliken J.B., Burrows P.E., Dompmartin A., Watanabe S., Vanwijck R., Vikkula M. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am. J. Hum. Genet. 2003;73:1240–1249. doi: 10.1086/379793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Revencu N., Boon L.M., Mendola A., Cordisco M.R., Dubois J., Clapuyt P., Hammer F., Amor D.J., Irvine A.D., Baselga E., et al. RASA1 mutations and associated phenotypes in 68 families with capillary malformation-arteriovenous malformation. Hum. Mutat. 2013;34:1632–1641. doi: 10.1002/humu.22431. [DOI] [PubMed] [Google Scholar]
- 18.Taylor R.S., Joseph D.B., Kohaut E.C., Wilson E.R., Bueschen A.J. Renal angiomyolipoma associated with lymph node involvement and renal cell carcinoma in patients with tuberous sclerosis. J. Urol. 1989;141:930–932. doi: 10.1016/s0022-5347(17)41057-3. [DOI] [PubMed] [Google Scholar]
- 19.Bjornsson J., Short M.P., Kwiatkowski D.J., Henske E.P. Tuberous sclerosis-associated renal cell carcinoma. Am. J. Path. 1996;149:1201–1208. [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez-Baldo L., Schmidt S., Maxwell C.A., Bonifaci N., Gabaldon T., Vidalain P.O., Senapedis W., Kletke A., Rosing M., Barnekow A., et al. TACC3-TSC2 maintains nuclear envelope structure and controls cell division. Cell Cycle. 2010;9:1143–1155. doi: 10.4161/cc.9.6.11018. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albiges L., Guegan J., Le Formal A., Verkarre V., Rioux-Leclercq N., Sibony M., Bernhard J.C., Camparo P., Merabet Z., Molinie V., et al. MET is a potential target across all papillary renal cell carcinomas: result from a large molecular study of pRCC with CGH array and matching gene expression array. Clin. Cancer Res. 2014;20:3411–3421. doi: 10.1158/1078-0432.CCR-13-2173. [DOI] [PubMed] [Google Scholar]
- 23.Davis C.F., Ricketts C.J., Wang M., Yang L., Cherniack A.D., Shen H., Buhay C., Kang H., Kim S.C., Fahey C.C., et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rustin P., Munnich A., Rotig A. Succinate dehydrogenase and human diseases: new insights into a well-known enzyme. Eur. J. Hum. Genet. 2002;10:289–291. doi: 10.1038/sj.ejhg.5200793. [DOI] [PubMed] [Google Scholar]
- 25.Ricketts C.J., Forman J.R., Rattenberry E., Bradshaw N., Lalloo F., Izatt L., Cole T.R., Armstrong R., Kumar V.K., Morrison P.J., et al. Tumor risks and genotype-phenotype-proteotype analysis in 358 patients with germline mutations in SDHB and SDHD. Hum. Mutat. 2010;31:41–51. doi: 10.1002/humu.21136. [DOI] [PubMed] [Google Scholar]
- 26.Ricketts C., Woodward E.R., Killick P., Morris M.R., Astuti D., Latif F., Maher E.R. Germline SDHB mutations and familial renal cell carcinoma. J. Natl. Cancer Inst. 2008;100:1260–1262. doi: 10.1093/jnci/djn254. [DOI] [PubMed] [Google Scholar]
- 27.Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H., et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J., Dibble C.C., Matsuzaki M., Manning B.D. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol. Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malinowska I., Kwiatkowski D.J., Weiss S., Martignoni G., Netto G., Argani P. Perivascular epithelioid cell tumors (PEComas) harboring TFE3 gene rearrangements lack the TSC2 alterations characteristic of conventional PEComas: further evidence for a biological distinction. Am. J. Surg. Pathol. 2012;36:783–784. doi: 10.1097/PAS.0b013e31824a8a37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badri K.R., Gao L., Hyjek E., Schuger N., Schuger L., Qin W., Chekaluk Y., Kwiatkowski D.J., Zhe X. Exonic mutations of TSC2/TSC1 are common but not seen in all sporadic pulmonary lymphangioleiomyomatosis. Am. J. Respir. Crit. Care Med. 2013;187:663–665. doi: 10.1164/ajrccm.187.6.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyburczy M.E., Wang J.A., Li S., Thangapazham R., Chekaluk Y., Moss J., Kwiatkowski D.J., Darling T.N. Sun exposure causes somatic second-hit mutations and angiofibroma development in tuberous sclerosis complex. Hum. Mol. Genet. 2014;23:2023–2029. doi: 10.1093/hmg/ddt597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hettmer S., Teot L.A., van Hummelen P., MacConaill L., Bronson R.T., Dall'Osso C., Mao J., McMahon A.P., Gruber P.J., Grier H.E., et al. Mutations in Hedgehog pathway genes in fetal rhabdomyomas. J. Pathol. 2013;231:44–52. doi: 10.1002/path.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]