Abstract
Protein misfolding and aggregation is a major hallmark of neurodegenerative disorders such as Alzheimer's disease (AD), Parkinson's disease (PD) and Huntington's disease (HD). Until recently, the consensus was that each aggregation-prone protein was characteristic of each disorder [α-synuclein (α-syn)/PD, mutant huntingtin (Htt)/HD, Tau and amyloid beta peptide/AD]. However, growing evidence indicates that aggregation-prone proteins can actually co-aggregate and modify each other's behavior and toxicity, suggesting that this process may also contribute to the overlap in clinical symptoms across different diseases. Here, we show that α-syn and mutant Htt co-aggregate in vivo when co-expressed in Drosophila and produce a synergistic age-dependent increase in neurotoxicity associated to a decline in motor function and life span. Altogether, our results suggest that the co-existence of α-syn and Htt in the same neuronal cells worsens aggregation-related neuropathologies and accelerates disease progression.
Introduction
The presence of protein aggregates in the brain is a common hallmark for several neurodegenerative diseases, such as Alzheimer's disease (AD), Parkinson's disease (PD) and Huntington's disease (HD). The specific subcellular location and composition of protein aggregates are characteristics for each disorder. PD hallmarks are the formation of Lewy bodies, intracytoplasmic inclusions of misfolded proteins containing mainly α-synuclein (α-syn) and the demise of dopaminergic neurons in the substantia nigra. In AD, deposits of amyloid-beta and tau proteins in the brain lead to hippocampal degeneration, cognitive impairment and dementia. In HD, mutant huntingtin (Htt) with polyglutamine (polyQ) repeat expansion accumulates in cytoplasmic and intranuclear aggregates leading to neurodegeneration in the striatum (1).
Although each neurodegenerative disorder has its characteristic pathophysiology, current evidence indicates that there is also significant overlap between apparently different disorders. For example, α-syn is the protein that characteristically aggregates in PD, but it was originally discovered as a constituent of amyloid plaques in AD (2) and, later on, was found in protein aggregates in diverse pathologies of the central nervous system, such as HD, trisomy of chromosome 21, progressive supranuclear palsy and frontotemporal dementia (3–7). Tau, an AD-associated protein, was detected in protein aggregates in patients with PD, sporadic dementia with Lewy bodies and multiple system atrophy, as well as in some animal models for synucleinopathies (8–14).
This apparent convergence of the molecular and cellular phenomena is accompanied by an overlap in the symptoms. For instance, patients suffering from diseases that affect movement control and coordination, as is the case of PD and HD, may also exhibit dementia in more advanced stages of disease (15). Conversely, patients afflicted by dementia can also show PD- or HD-like motor symptoms (16).
There is growing evidence that co-occurrence of aggregate-prone proteins may decisively influence the pathophysiology and severity of neural disorders. Tau and α-syn interact and co-aggregate, and this is associated with an increase in neurotoxicity in cellular and Drosophila models (17,18). Htt has been recently shown to co-aggregate with proteins associated with synucleinopathies and tauopathies (19–22). Mutant Htt induces Tau hyperphosphorylation and aggregation, preventing its association to the microtubular network and producing large ring-like aggregates close to the microtubular network (19,20). DJ-1, which is associated with familial PD, interacts and co-aggregates with α-syn and Htt, modulating their toxicity in models of PD and HD (20,22).
We have previously shown that α-syn modifies the dynamics and aggregation pattern of mutant Htt in cells in culture (23). Here, we expand those studies and report that co-existence of α-syn and mutant Htt in vivo strongly enhances PD- and HD-related neuropathology in Drosophila melanogaster, suggesting that the interplay between the two proteins deserves further investigation in the context of HD and PD.
Results
Co-expression of mutant Htt and α-syn alters Htt aggregation pattern
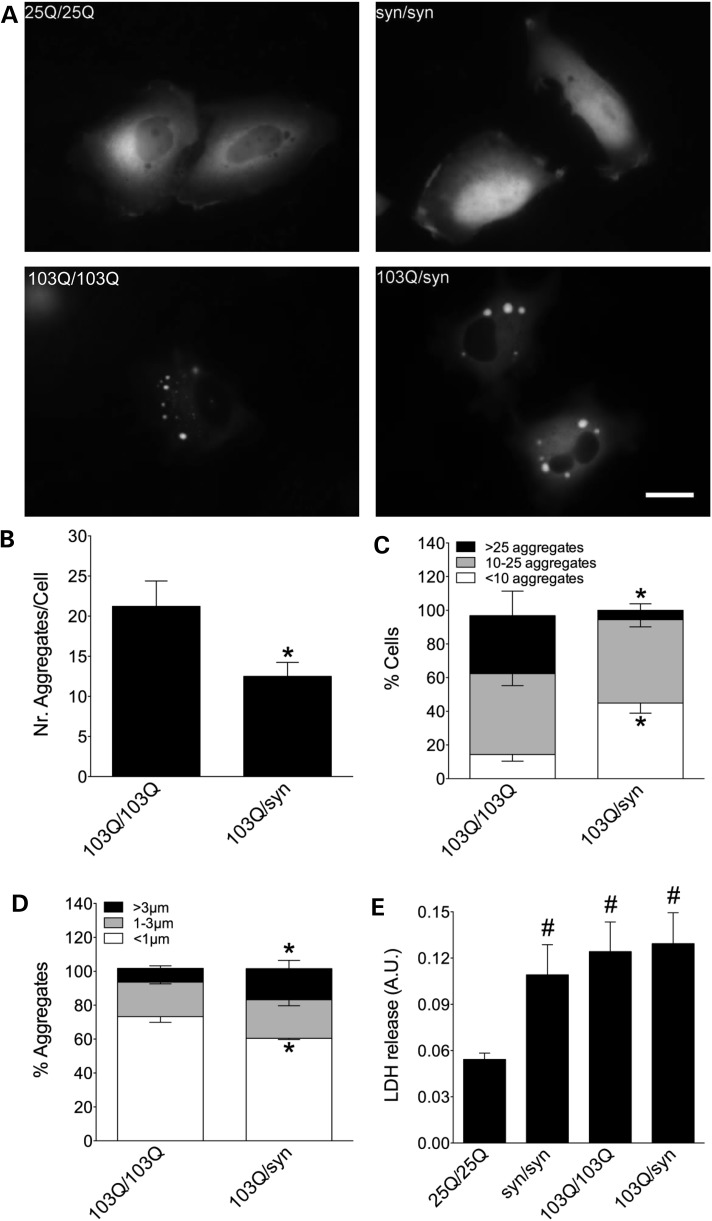
Expression of normal (25Q) Htt or α-syn bimolecular fluorescence complementation (BiFC) pairs in human (H4) cells produced mostly homogeneous fluorescence, Htt being more often restricted to the cytosol and α-syn spreading both through nucleus and cytosol (Fig. 1A). On the other hand, mutant (103Q) Htt BiFC pairs produced protein aggregates. The combination of mutant Htt/α-syn also produced aggregates, but they seemed fewer and larger than pure 103Q aggregates. This was confirmed quantitatively (Fig. 1B–D), as the number of aggregates per cell was reduced 2-fold in mutant Htt/α-syn combinations (Fig. 1B), and the number of cells with <10 aggregates grew at the expense of cells with >25 aggregates (Fig. 1C). Finally, the percentage of aggregates larger than 3 µm increased in mutant Htt/α-syn combinations at the expense of aggregates smaller than 1 µm (Fig. 1D). The proportion of cells with 10–25 aggregates and of aggregates between 1 and 3 µm remained unchanged.
Figure 1.
Co-expression of mutant Htt and α-syn alters Htt aggregation pattern. (A) H4 cells transfected with different combinations of α-syn- and Htt-Venus BiFC constructs. Cells transfected with a Htt25Q-Venus BiFC pair of plasmids show homogeneous fluorescence indicative of oligomeric species, while a Htt103Q-Venus BiFC pair produces both oligomeric species and large intracellular fluorescent aggregates with variable size and morphology. Htt location is primarily cytosolic. α-Syn-Venus BiFC pair produces homogeneous fluorescence distributed throughout all cellular compartments, including the nucleus. When α-syn and Htt103Q BiFC constructs were combined, there is a change in the aggregation pattern of both proteins, quantified in B–D. Co-transfection of Htt103Q with α-syn BiFC constructs decreases the average number of aggregates per cell (B and C) and increases the average size of aggregates (D). (E) The α-syn pair or the Htt103Q pair are more toxic than the wild-type Htt pair and combining α-syn with Htt103Q does not enhance toxicity in this model. *Significant versus 103Q/103Q, P < 0.01; #Significant versus 25Q/25Q, P < 0.001. Scale bar, 20 µm. AU, arbitrary units.
Both mutant Htt and α-syn increased cell death 24 h after transfection of H4 cells with the corresponding combinations of BiFC constructs (Fig. 1E). However, and in spite of the clear changes induced by α-syn on mutant Htt aggregation pattern, we did not observe changes at the viability level, even 72 h after transfection (data not shown). Longer experiments are extremely difficult to carry out in this experimental setup.
Htt103Q-mCherry and α-syn-EGFP co-localize and co-aggregate in dopaminergic neurons and in photoreceptors
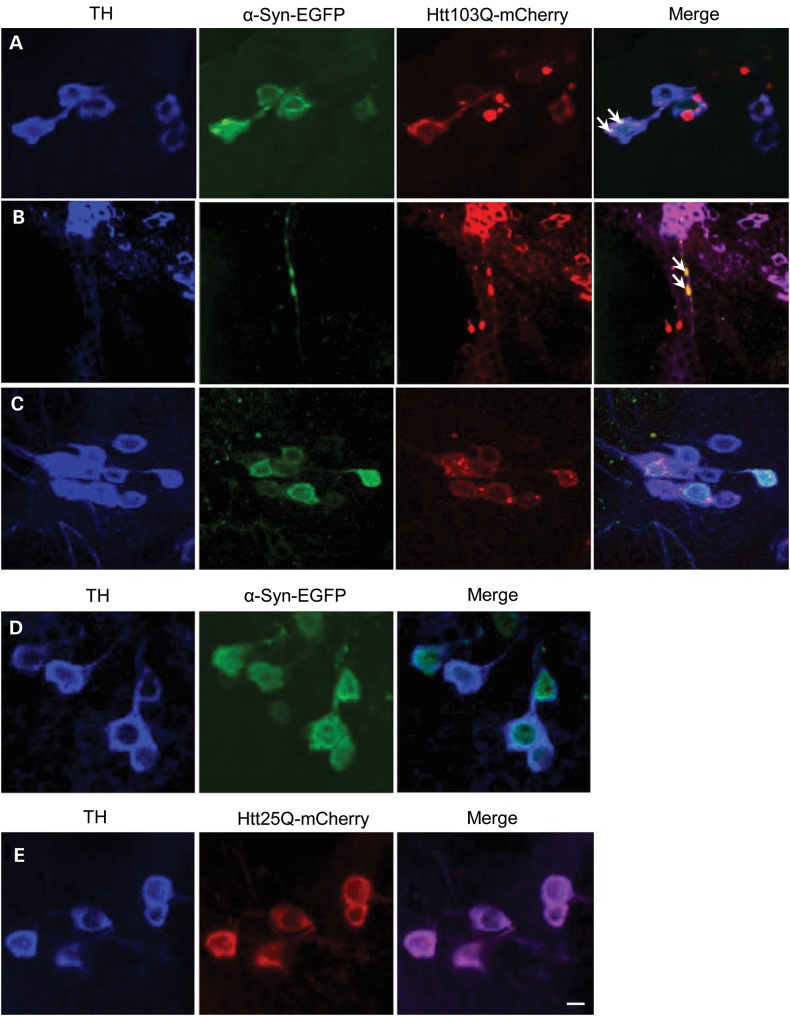
In order to confirm our results with human cells in culture and to be able to measure neurotoxic effects of Htt/α-syn combinations during longer periods of time, we performed assays using transgenic overexpression in Drosophila. Co-expression of a mutant version of Htt containing 103 glutamines (Htt103Q-mCherry) and wild-type α-syn-EGFP in dopaminergic neurons (TH-GAL4) showed co-localization and co-aggregation of these two proteins (Fig. 2), confirming our results in cultured human cells. In terms of subcellular location, we found that these α-syn-EGFP/Htt103Q-mCherry aggregates accumulate both in cell bodies (Fig. 2A) and neurites (Fig. 2B) of dopaminergic neurons. In contrast, flies co-expressing α-syn-EGFP with a wild-type version of Htt (Htt25Q-mCherry) do not show aggregates (Fig. 2C), suggesting that only mutant Htt stimulates the deposition of α-syn in these aggregates. Flies expressing α-syn-EGFP or wild-type Htt25Q-mCherry alone did not show aggregates (Fig. 2D and E).
Figure 2.
Htt103Q-mCherry and α-syn-EGFP co-localize and co-aggregate when expressed in dopaminergic neurons. (A) Co-localization of Htt103Q-mCherry and α-syn-EGFP aggregates in cell bodies of dopaminergic neurons (arrows). (B) Co-localization of Htt103Q-mCherry and α-syn-EGFP aggregates in neurites of dopaminergic neurons (arrows). (C) Wild-type Htt (Htt25Q-mCherry) does not form aggregates with α-syn-EGFP. (D and E) The expression of α-syn-EGFP alone or Htt25Q-mCherry does not induce the formation of aggregates. The images in A–E show dopaminergic neurons in the paired posterior lateral 1 (PPL1) cluster marked by an anti-TH antibody (against tyrosine hydroxylase). Genotypes: (A, B) TH-GAL4, UAS-α-syn-EGFP/ UAS-Htt103Q-mCherry, (C) TH-GAL4, UAS-α-syn-EGFP/UAS-Htt25Q-mCherry, (D) TH-GAL4, UAS-α-syn-EGFP and (E) TH-GAL4, UAS-Htt25Q-mCherry. Scale bars represent 10 µm.
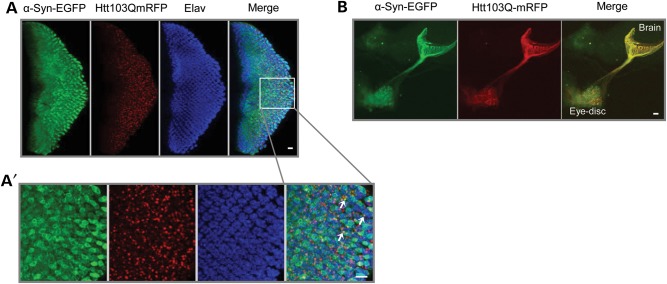
Using sGMR-GAL4 we induced the co-expression of Htt103Q-mCherry and α-syn-EGFP in eye imaginal discs of third instar larvae (Fig. 3). We observed co-localization and co-aggregation of α-syn and mutant Htt in the cytoplasm of the photoreceptors (Fig. 3A and A′), and also in the axonal projections of the photoreceptors in the larval brain (Fig. 3B).
Figure 3.
Htt103Q-mCherry and α-syn-EGFP co-localize and co-aggregate in the eye discs of third instar larvae. (A) Third instar larval eye disc from double transgenics stained with the pan-neuronal marker anti-elav. α-Syn-EGFP co-localized and co-aggregated with Htt103Q-mCherry aggregates in the cytoplasm of larval photoreceptors. (A′) Magnified area delimited by the box in (A). Arrows indicate foci of co-aggregated proteins. (B) Htt103Q-mCherry and α-syn-EGFP co-localized in the photoreceptors' axonal projections in the larval brain. Genotype: sGMR-GAL4, UAS-α-syn-EGFP/UAS-Htt103Q-mCherry. Scale bars represent 10 µm.
Htt103Q-mCherry and α-syn-EGFP are physically interacting
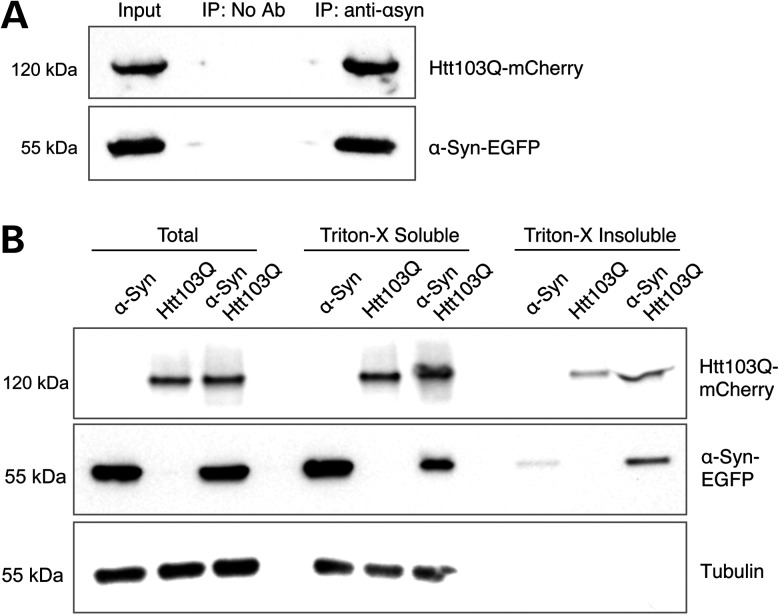
Mutant Htt103Q-mCherry was co-immunoprecipitated with α-syn-EGFP, using an antibody against the green fluorescent protein (GFP) tag, further demonstrating that mutant Htt103Q-mCherry and α-syn-EGFP interact and co-aggregate in Drosophila cells (Fig. 4A). In order to analyze the solubility status of these proteins when they are co-expressed, we performed a Triton-X solubility experiment using protein extracts from 8 days old adult heads. We found that the levels of α-syn-EGFP and Htt103Q-mCherry in the Triton insoluble fraction increased when these proteins are co-expressed, in comparison when they are expressed alone (Fig. 4B). This result indicates that co-expression of α-syn-EGFP and Htt103Q-mCherry causes an increase in the formation of insoluble aggregates containing these two proteins.
Figure 4.
Htt103Q-mCherry and α-syn-EGFP are physically interacting. (A) Immunoprecipitation of α-syn-EGFP with an antibody against the EGFP tag pulled down Htt103Q-mCherry from dopaminergic neurons, showed by immunoblotting analysis. (B) Immunoblotting of Total, Triton-X soluble and Triton-X insoluble fractions of α-syn-EGFP and Htt103Q-mCherry from adult heads of flies co-expressing these two proteins. The levels of α-syn-EGFP and Htt103Q-mCherry in the insoluble fraction are higher when these proteins are co-expressed. Genotypes: (A) TH-GAL4, UAS-α-syn-EGFP/UAS-Htt103Q-mCherry. (B) α-Syn: sGMR-GAL4, UAS-α-syn-EGFP. Htt103Q: sGMR-GAL4, UAS-Htt103Q-mCherry. α-Syn/Htt103Q-mCherry: sGMR-GAL4, UAS-α-syn-EGFP/UAS-Htt103Q-mCherry.
Co-expression of Htt103Q-mCherry and α-syn-EGFP produces premature and severe degeneration in the photoreceptors
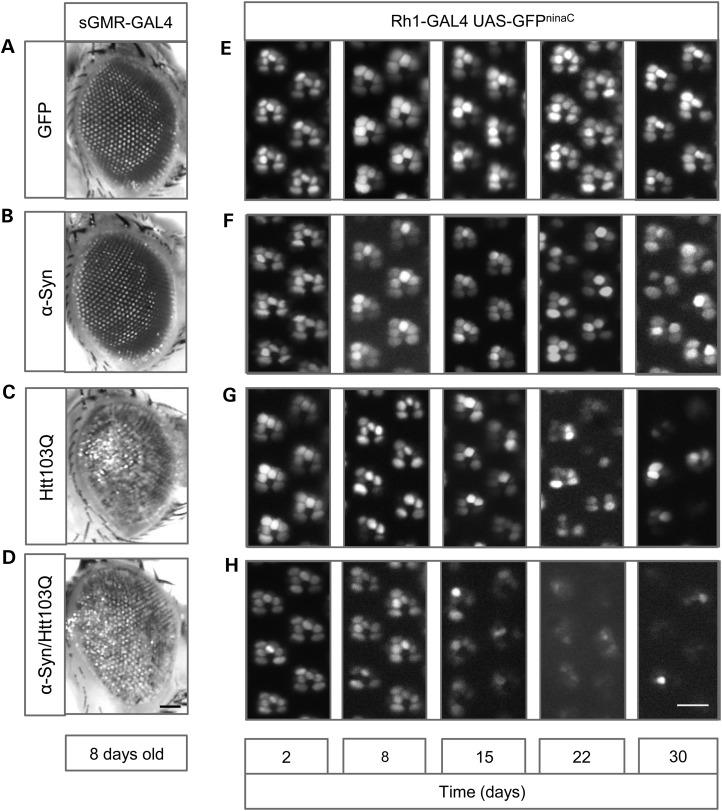
Next, we expressed Htt103Q-mCherry and α-syn-EGFP in the eye using sGMR-GAL4 and analyzed the external morphology of the adult eye in 8 days old flies (Fig. 5A–D). Expression of the GFP tag alone had no effect on eye development or viability as the eyes looked normal (Fig. 5A). Flies expressing α-syn-EGFP alone also developed a normal eye, identical to control flies (Fig. 5B). On the other hand, flies expressing mutant Htt103Q-mCherry, alone or in combination with α-syn-EGFP, showed striking retinal degeneration, producing a strong rough eye phenotype (Fig. 5C and D). Co-expression of mutant Htt103Q-mCherry and α-syn-EGFP did not worsen the rough eye phenotype in comparison with Htt103Q-mCherry alone. However, co-expression of Htt103Q-mCherry and α-syn-EGFP specifically in the photoreceptors R1–R6, using Rh1-GAL4, showed a stronger and premature degeneration phenotype (Fig. 5G) in comparison with Htt103Q-mCherry alone (Fig. 5H). In this case, control GFP or α-syn-EGFP alone did not show any degeneration phenotype (Fig. 5E and F). These results indicate that co-expression of α-syn-EGFP and mutant Htt103Q-mCherry enhances neurodegeneration of the Drosophila photoreceptors in vivo.
Figure 5.
Co-expression of Htt103Q-mCherry and α-syn-EGFP produces premature and severe degeneration in the photoreceptors. (A–D) External morphology of the adult eye. (A and B) Expression of GFP or α-syn-EGFP does not affect external eye morphology. (C and D) Co-expression of Htt103Q-mCherry and α-syn-EGFP produces a rough eye phenotype identical to the one induced by the single expression of Htt103Q-mCherry. (E–H) Photoreceptors neurodegeneration in the adult eye, analyzed by water immersion microscopy assay of the retinas. (E) Expression of GFP did not induce degeneration of the photoreceptors, with the six photoreceptors being present in the ommatidial clusters; (F, G) flies expressing α-syn-EGFP or Htt103Q-mCherry showed progressive degeneration of the photoreceptors. (H) Co-expression of Htt103Q-mCherry and α-syn-EGFP induced severe and early degeneration of the photoreceptors. Genotypes are indicated in the boxes on top and lateral of the figures: (A) sGMR-GAL4, UAS-GFP, (B) sGMR-GAL4, UAS-α-syn-EGFP, (C) sGMR-GAL4, UAS-Htt103Q-mCherry, (D) sGMR-GAL4, UAS-α-syn-EGFP/UAS-Htt103Q-mCherry, (E) Rh1-GAL4, UAS-GFP, (F) Rh1-GAL4, UAS-α-syn-EGFP, (G) Rh1-GAL4, UAS-Htt103Q-mCherry and (H) Rh1-GAL4, UAS-α-syn-EGFP/UAS-Htt103Q-mCherry. Scale bars represent 50 µm (A–D) and 10 µm (E–H).
Co-expression of Htt103Q-mCherry and α-syn-EGFP in the nervous system causes severe motor dysfunction and a decrease in life span
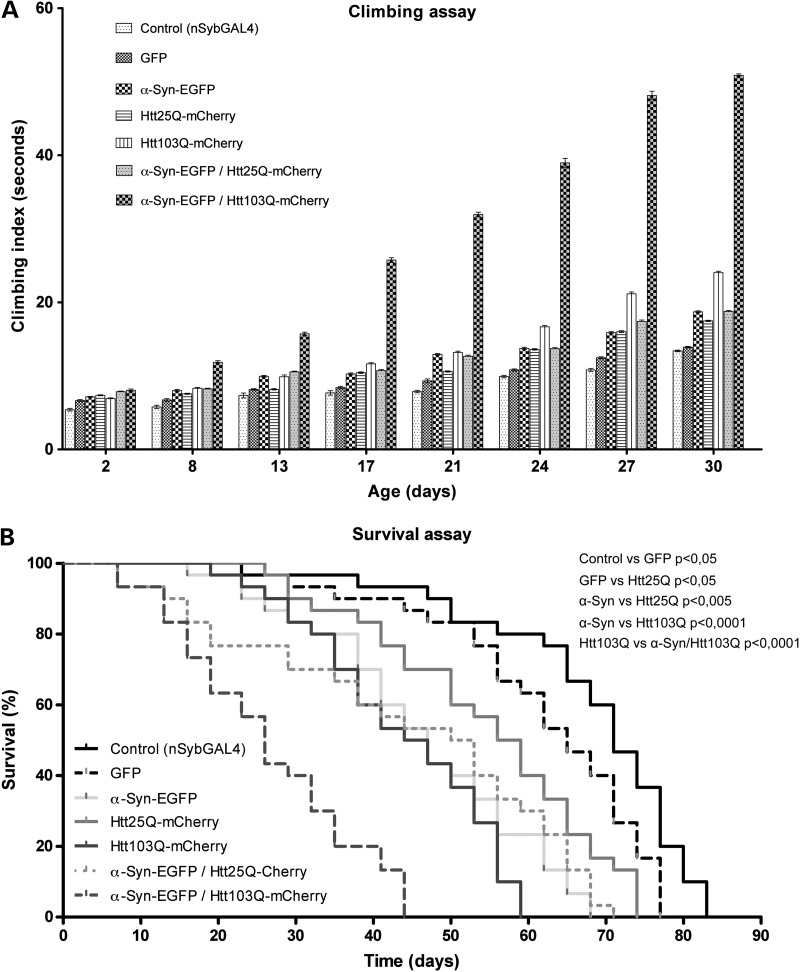
We evaluated the motor function and life span of flies co-expressing mutant Htt103Q-mCherry and α-syn-EGFP in the central nervous system, using nSyb-GAL4 (Fig. 6). To test motor function we used ‘climbing assays’, where flies of different genotypes were tapped to the bottom of the vial and allowed to climb up the walls (Fig. 6A). We recorded the climbing time when five flies crossed a 15-cm finish line. Eight days old flies co-expressing Htt103Q-mCherry and α-syn-EGFP took, on average, four more seconds to reach the finish line than the other genotypes tested. The motor ability of flies co-expressing Htt103Q-mCherry/α-syn-EGFP deteriorated more and faster than the other genotypes, with the difference in climbing times reaching 32 s by day 30.
Figure 6.
Co-expression of Htt103Q-mCherry and α-syn-EGFP in the nervous system causes severe motor dysfunction and a decrease in life span. (A) Flies co-expressing Htt103Q-mCherry and α-syn-EGFP under the control of nSyb-GAL4 show a strong impairment of the motor abilities compared with the rest of the genotypes tested. The Y-axis represents the time (in seconds), it took for five males to climb 15 cm (mean ± SEM). Statistically significant values, comparing each of genotypes, were calculated by doing two-way ANOVA with Bonferroni post-test. With very few exceptions, all the differences detected between the genotypes tested in this assay were statistically significant with a P < 0.001 (except the difference between the genotypes Htt25Q-mCherry and α-Syn-EGFP/Htt25Q-mCherry which is significant for a P < 0.01). The differences not statistically significant (P > 0.05) were GFP versus Htt25Q-mCherry at day 13; α-Syn-EGFP versus Htt103Q-mCherry at day 13; α-Syn-EGFP versus α-Syn-EGFP/Htt25Q-mCherry at days 24 and 30; Htt103Q-mCherry versus α-Syn-EGFP/Htt25Q-mCherry at day 8. (B) Flies co-expressing Htt103Q-mCherry and α-syn-EGFP under the control of nSyb-GAL4 have a life span significantly shorter in comparison with the rest of the genotypes tested. Values on the Y-axis represent the percentage of flies alive at each time point analyzed. The maximum survival (in days) is indicated for each genotype (the mean values indicate the number of days it took for half of the flies to die): control (nSyb-GAL4) 83 (mean = 70), GFP (nSyb-GAL4, UAS-GFP) 77 (mean = 64), α-syn-EGFP (nSyb-GAL4, UAS-α-syn-EGFP) 68 (mean = 45), Htt25Q-mCherry (nSyb-GAL4, UAS- Htt25Q-mCherry) 74 (mean = 56), Htt103Q-mCherry (nSyb-GAL4, UAS-Htt103Q-mCherry) 59 (mean = 44), α-syn-EGFP/Htt25Q-mCherry (nSyb-GAL4, UAS-α-syn-EGFP/UAS-Htt25Q-mCherry) 71 (mean = 50) and α-syn-EGFP/Htt103Q-mCherry (nSyb-GAL4, UAS-α-syn-EGFP/UAS-Htt103Q-mCherry) 44 (mean = 25). Statistical significance is indicated in the graph (log-rank, Mantel–Cox test).
Flies co-expressing Htt103Q-mCherry and α-syn-EGFP also showed a dramatic decrease in life span, with 44 days of maximum survival (Fig. 6B). Flies co-expressing Htt25Q-mCherry and α-syn-EGFP had a maximum survival of 71 days. Maximum survival of flies expressing GFP, α-syn-EGFP, Htt25Q-mCherry or Htt103Q-mCherry alone ranged from 59 to 83 days. Interestingly, neurotoxicity, motor dysfunction and fly death started around days 8–15 and their increase occurred in parallel, indicating an association between these events. These results suggest that co-expression of α-syn and mutant Htt synergistically enhances each other's toxicity, eventually accelerating the progression of the disorder.
Discussion
Here, we show that α-syn and mutant Htt, two proteins associated with PD and HD, respectively, can interact and co-aggregate in vivo. Furthermore, their co-expression produced a synergistic deterioration of neural tissue, motor function and life span in Drosophila. The effects of α-syn/Htt co-expression under the different promoters are summarized in Table 1. While co-expression of other proteins associated with human neurodegenerative diseases has also shown deleterious effects in different models (17,18,23,24), this is the first demonstration that co-expression of α-syn and mutant Htt can enhance neurodegeneration in Drosophila.
Table 1.
GAL4 lines used in this study and respective phenotypes
| GAL4 line | Tissue | Phenotype (co-expression of α-syn and mutant Htt) |
|---|---|---|
| TH-GAL4 | Dopaminergic neurons | Co-localization, interaction and co-aggregation of α-syn and Htt |
| nSyb-GAL4 | Nervous system | Impairment of motor abilities and reduced life span |
| sGMR-GAL4 | Eye | Co-localization, co-aggregation and retinal degeneration |
| Rh1-GAL4 | Photoreceptors R1-R6 | Premature degeneration of the photoreceptors |
Our results support the idea that the co-occurrence of these two different aggregation-prone proteins in neural cells can produce striking changes in the pathology, symptoms and disease progression. Moreover, our study may give some clues why some patients suffering with a specific neuropathology may show, at the symptomatic level, a significant overlap between different human neurodegenerative diseases. It is possible that a HD patient containing a mutant version of the Htt gene (expansion of the trinucleotide CAG) and thereby suffering from the consequences of mutant Htt aggregation and toxicity may be also in risk of developing aggregation of α-syn and therefore being affected by some of the symptoms associated with PD. This could occur as a consequence of the propensity of mutant Htt and α-syn to interact, co-aggregate and interfere with their normal biological roles in cells. In addition, the two most affected brain regions in patients with PD and HD, the substantia nigra and striatum, respectively, are anatomically and functionally interconnected, reinforcing the interest of studying the crosstalk between these two neuropathologies. Finally, the models established in this work may constitute useful tools to screen and discover new candidate drugs against PD and HD.
Material and Methods
Cell culture and BiFC plasmids
Maintenance of H4 human glioma cells and Htt and α-syn BiFC constructs were previously described in detail (23,25). Briefly, in our BiFC systems, Htt and α-syn were fused to two non-fluorescent halves of the Venus protein (Venus 1, amino acids 1–157; and Venus 2, amino acids 158–239). When the Htt and α-syn dimerize, the Venus halves get together and reconstitute a functional fluorophore. Fluorescence is therefore proportional to the amount of Htt/α-syn dimers and oligomers. H4 cells were transfected with the corresponding BiFC pairs and 24 h later pictures were taken on a Zeiss Axiovert 200M. A total of 150 cells from three independent experiments were analyzed. Graphs of Figure 1 show mean ± SD of three independent experiments.
Drosophila stocks
To generate UAS-α-Syn-EGFP, we fused α-syn with EGFP and cloned into pUAST using the BglII and Acc65I restriction sites. The transgenic flies were generated by BestGene, USA. We generated two different UAS-Htt-mCherry lines, one encoding a wild-type version of Htt (exon 1) with a 25 polyQ tail and the other encoding a mutant version of Htt with a 103 polyQ tail. These two constructs were cloned into pWalium10-roe and transgenic lines were generated using phiC31 integrase-mediated DNA integration (BestGene Strain #9723, attP acceptor site in 28E7).
Four different drivers were obtained from the Bloomington Stock Center (Indiana University, Bloomington, IN, USA): nSyb-GAL4 (active in the entire nervous system, under the control of the Synaptobrevin promoter), TH-GAL4 (active in dopaminergic neurons, under the control of the tyrosine hydroxylase promoter), GMR-GAL4 (active in the eye, under the control of the glass multiple reporter) and Rh1-GAL4 (active in the photoreceptors R1–R6, under the control of the rhodopsin1 promoter). Drosophila stocks were maintained at 25°C on standard cornmeal media in an incubator with a 12 h light/dark cycle.
Immunohistochemistry and microscopy
Brain preparations for confocal microscopy imaging were done as previously described (26). Briefly, adult flies were anesthetized with CO2 and the brains were isolated from the head cuticles and fixed in phosphate buffered saline (PBS) containing 4% paraformaldehyde.
Dopaminergic neurons were stained by incubation for 48 h at 4°C with mouse anti-TH antibody (Immunostar, Hudson, WI, USA) diluted 1:50 in PBST (1× PBS + 0.3% Triton X-100) containing 5% (v/v) normal goat serum. Three 10-min washes with PBST were done before incubation with a secondary anti-mouse Cy5 (Jackson ImmunoResearch), also diluted in PBST-containing 5% (v/v) normal goat serum.
Brain samples were analyzed and images were collected using a LSM 710 Meta Zeiss confocal microscope. Images were acquired with a resolution of 1024 × 1024, with a slice thickness of 1 µm and a line-average of 4. Z-projections were generated using ImageJ and the images were processed using Adobe Photoshop.
The degenerative effects caused by co-expression of α-syn and mutant Htt were assessed in the photoreceptors of adult eyes. To analyze external eye morphology, we used a Leica Z16 APO macroscope and a Leica DFC 420C camera. To analyze photoreceptor degeneration, we performed water immersion microscopy as previously described (27). Images were obtained using a Leica DM5500 B microscope and an Andor LucaR camera.
Immunoprecipitation, Triton-X solubility and immunoblotting analysis
Flies were transferred to 50-ml tubes, frozen in liquid nitrogen and immediately decapitated by vigorous vortexing. Isolated heads were collected to 1.5-ml tubes and maintained in dry ice. Proteins were extracted in lysis buffer supplemented with Complete Protease Inhibitor Cocktail tablets from Roche (Basel, Switzerland). Total protein was quantified using the DC Protein Assay, from Bio-Rad (CA, USA). In the immunoprecipitation experiments, α-syn-EGFP was pulled down from 1 mg of total protein extract, using GFP-Trap_A beads or blocked agarose beads (No Ab, no antibody negative control), following manufacturer's instructions (Chromotek, Munich, Germany). α-syn-EGFP pulldown and Htt103Q-mCherry co-immunoprecipitation were analyzed by immunoblotting using anti-GFP (3H9) and anti-RFP (5F8) antibodies from Chromotek, both diluted 1:1000 in PBS. Input lane corresponds to 30 µg of total protein extract and co-IP lane corresponds to one-fifth of the immunoprecipitated material.
The Triton-X solubility experiment was performed as previously described (28). Briefly, 200 µg of total protein extract was incubated with 1% Triton X-100 on ice during 30 min. Triton soluble and insoluble proteins fractions were separated by a 60-min centrifugation step at 15 000g at 4°C. The supernatant, containing the soluble proteins fraction (Triton-X soluble), was carefully collected and the pellet, containing the insoluble proteins fraction (Triton-X insoluble), was resuspended in 40 µl of 2% sodium dodecyl sulfate Tris–HCl buffer pH 7.4 by pipetting and sonication for 10 s. For the immunoblotting analysis, equal volumes of each fraction were loaded and the presence of α-syn-EGFP and Htt103Q-mCherry in the total, soluble and insoluble fractions was detected using anti-GFP (3H9) and anti-RFP (5F8). Additionally, anti-α-tubulin (AA4.3) from Developmental Studies Hybridoma Bank (IA, USA), diluted 1:500 in PBS, was used as loading control.
Climbing assays and survival assays
Motor function was analyzed by startle-induced locomotion and negative geotaxis response assays, commonly called climbing assays, as previously described (29). Briefly, groups of 10 males of the same age of each genotype of interest were placed into 18-cm-long vials, at room temperature for environmental acclimatization, and 30 min later they were gently tapped to the bottom of the vial (a minimum number of 30 males per genotype was tested). We recorded the climbing time when five flies crossed the 15-cm finish line. For each genotype we tested three independent groups of males and performed five trials for each time point. Results are the average climbing time ± SEM of these independent experiments.
For survival assays, flies were maintained in a humid incubator at 25°C under a 12 h light/dark cycle. Thirty adult females of the same age were placed in three vials (10 flies per vial) containing fresh food. Each 3 days the flies were transferred into vials with fresh food and the number of living flies was registered.
Statistical analysis
Statistical analyses were done using GraphPad Prism software version 5 (San Diego, CA, USA). For BiFC we performed a one-way ANOVA followed by a post-hoc Tukey test for average comparison. For climbing assays, we performed a two-way ANOVA followed by a Bonferroni post-test. For the survival assays, we performed a Log-rank (Mantel–Cox) test.
Funding
This work was supported by grant FCT-ANR/NEU-NMC/0006/2013 from Fundação para a Ciência e a Tecnologia, Portugal. G.M.P. was supported by a doctoral fellowship from the Fundação para a Ciência e a Tecnologia (SFRH/BD/61477/2009). J.B.S. and F.H. were supported by fellowships from the Fundação para a Ciência e a Tecnologia (SFRH/BD/85275/2012 and SFRH/BPD/63530/2009, respectively). F.H. and T.F.O. were also supported by seed funds from the European Huntington's Disease Network (EHDN). T.F.O. is supported by the DFG Center for Nanoscale Microscopy and Molecular Physiology of the Brain.
Acknowledgements
We thank Maria Luísa Vasconcelos, Rui Martinho and the Bloomington Stock Centre for Drosophila stocks. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing plasmid vectors used in this study. We thank Developmental Studies Hybridoma Bank for antibodies.
Conflict of Interest statement. None declared.
References
- 1.Soto C., Estrada L.D. Protein misfolding and neurodegeneration. Arch. Neurol. 2008;65:184–189. doi: 10.1001/archneurol.2007.56. [DOI] [PubMed] [Google Scholar]
- 2.Yoshimoto M., Iwai A., Kang D., Otero D.A., Xia Y., Saitoh T. NACP, the precursor protein of the non-amyloid beta/A4 protein (A beta) component of Alzheimer disease amyloid, binds A beta and stimulates A beta aggregation. Proc. Natl. Acad. Sci. USA. 1995;92:9141–9145. doi: 10.1073/pnas.92.20.9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilhelmsen K.C., Forman M.S., Rosen H.J., Alving L.I., Goldman J., Feiger J., Lee J.V., Segall S.K., Kramer J.H., Lomen-Hoerth C., et al. 17q-linked frontotemporal dementia-amyotrophic lateral sclerosis without tau mutations with tau and alpha-synuclein inclusions. Arch. Neurol. 2004;61:398–406. doi: 10.1001/archneur.61.3.398. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton R.L. Lewy bodies in Alzheimer's disease: a neuropathological review of 145 cases using alpha-synuclein immunohistochemistry. Brain Pathol. 2000;10:378–384. doi: 10.1111/j.1750-3639.2000.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raghavan R., Khin-Nu C., Brown A., Irving D., Ince P.G., Day K., Tyrer S.P., Perry R.H. Detection of Lewy bodies in Trisomy 21 (Down's syndrome) Can. J. Neurol. Sci. 1993;20:48–51. doi: 10.1017/s0317167100047405. [DOI] [PubMed] [Google Scholar]
- 6.Judkins A.R., Forman M.S., Uryu K., Hinkle D.A., Asbury A.K., Lee V.M., Trojanowski J.Q. Co-occurrence of Parkinson's disease with progressive supranuclear palsy. Acta Neuropathol. 2002;103:526–530. doi: 10.1007/s00401-001-0483-7. [DOI] [PubMed] [Google Scholar]
- 7.Charles V., Mezey E., Reddy P.H., Dehejia A., Young T.A., Polymeropoulos M.H., Brownstein M.J., Tagle D.A. Alpha-synuclein immunoreactivity of huntingtin polyglutamine aggregates in striatum and cortex of Huntington's disease patients and transgenic mouse models. Neurosci. Lett. 2000;289:29–32. doi: 10.1016/s0304-3940(00)01247-7. [DOI] [PubMed] [Google Scholar]
- 8.Galpern W.R., Lang A.E. Interface between tauopathies and synucleinopathies: a tale of two proteins. Ann. Neurol. 2006;59:449–458. doi: 10.1002/ana.20819. [DOI] [PubMed] [Google Scholar]
- 9.Clarimon J., Molina-Porcel L., Gomez-Isla T., Blesa R., Guardia-Laguarta C., Gonzalez-Neira A., Estorch M., Ma Grau J., Barraquer L., Roig C., et al. Early-onset familial lewy body dementia with extensive tauopathy: a clinical, genetic, and neuropathological study. J Neuropathol. Exp. Neurol. 2009;68:73–82. doi: 10.1097/NEN.0b013e3181927577. [DOI] [PubMed] [Google Scholar]
- 10.Galloway P.G., Grundke-Iqbal I., Iqbal K., Perry G. Lewy bodies contain epitopes both shared and distinct from Alzheimer neurofibrillary tangles. J. Neuropathol. Exp. Neurol. 1988;47:654–663. doi: 10.1097/00005072-198811000-00008. [DOI] [PubMed] [Google Scholar]
- 11.Giasson B.I., Forman M.S., Higuchi M., Golbe L.I., Graves C.L., Kotzbauer P.T., Trojanowski J.Q., Lee V.M. Initiation and synergistic fibrillization of tau and alpha-synuclein. Science. 2003;300:636–640. doi: 10.1126/science.1082324. [DOI] [PubMed] [Google Scholar]
- 12.Ishizawa T., Mattila P., Davies P., Wang D., Dickson D.W. Colocalization of tau and alpha-synuclein epitopes in Lewy bodies. J. Neuropathol. Exp. Neurol. 2003;62:389–397. doi: 10.1093/jnen/62.4.389. [DOI] [PubMed] [Google Scholar]
- 13.Wills J., Jones J., Haggerty T., Duka V., Joyce J.N., Sidhu A. Elevated tauopathy and alpha-synuclein pathology in postmortem Parkinson's disease brains with and without dementia. Exp. Neurol. 2010;225:210–218. doi: 10.1016/j.expneurol.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haggerty T., Credle J., Rodriguez O., Wills J., Oaks A.W., Masliah E., Sidhu A. Hyperphosphorylated Tau in an alpha-synuclein-overexpressing transgenic model of Parkinson's disease. Eur. J. Neurosci. 2011;33:1598–1610. doi: 10.1111/j.1460-9568.2011.07660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perry R.J., Hodges J.R. Spectrum of memory dysfunction in degenerative disease. Curr. Opin. Neurol. 1996;9:281–285. doi: 10.1097/00019052-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Tsolaki M., Kokarida K., Iakovidou V., Stilopoulos E., Meimaris J., Kazis A. Extrapyramidal symptoms and signs in Alzheimer's disease: prevalence and correlation with the first symptom. Am. J. Alzheimers Dis. Other Demen. 2001;16:268–278. doi: 10.1177/153331750101600512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy B., Jackson G.R. Interactions between Tau and alpha-synuclein augment neurotoxicity in a Drosophila model of Parkinson's disease. Hum. Mol. Genet. 2014;23:3008–3023. doi: 10.1093/hmg/ddu011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badiola N., de Oliveira R.M., Herrera F., Guardia-Laguarta C., Goncalves S.A., Pera M., Suarez-Calvet M., Clarimon J., Outeiro T.F., Lleo A. Tau enhances alpha-synuclein aggregation and toxicity in cellular models of synucleinopathy. PLoS One. 2011;6:e26609. doi: 10.1371/journal.pone.0026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blum D., Herrera F., Francelle L., Mendes T., Basquin M., Obriot H., Demeyer D., Sergeant N., Gerhardt E., Brouillet E., et al. Mutant huntingtin alters Tau phosphorylation and subcellular distribution. Hum. Mol. Genet. 2015;24:76–85. doi: 10.1093/hmg/ddu421. [DOI] [PubMed] [Google Scholar]
- 20.Sajjad M.U., Green E.W., Miller-Fleming L., Hands S., Herrera F., Campesan S., Khoshnan A., Outeiro T.F., Giorgini F., Wyttenbach A. DJ-1 modulates aggregation and pathogenesis in models of Huntington's disease. Hum. Mol. Genet. 2014;23:755–766. doi: 10.1093/hmg/ddt466. [DOI] [PubMed] [Google Scholar]
- 21.Gratuze M., Noel A., Julien C., Cisbani G., Milot-Rousseau P., Morin F., Dickler M., Goupil C., Bezeau F., Poitras I., et al. Tau hyperphosphorylation and deregulation of calcineurin in mouse models of Huntington's disease. Hum. Mol. Genet. 2015;24:86–89. doi: 10.1093/hmg/ddu456. [DOI] [PubMed] [Google Scholar]
- 22.Zondler L., Miller-Fleming L., Repici M., Goncalves S., Tenreiro S., Rosado-Ramos R., Betzer C., Straatman K.R., Jensen P.H., Giorgini F., et al. DJ-1 interactions with alpha-synuclein attenuate aggregation and cellular toxicity in models of Parkinson's disease. Cell Death Dis. 2014;5:e1350. doi: 10.1038/cddis.2014.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrera F., Outeiro T.F. Alpha-synuclein modifies huntingtin aggregation in living cells. FEBS Lett. 2012;586:7–12. doi: 10.1016/j.febslet.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 24.Corrochano S., Renna M., Carter S., Chrobot N., Kent R., Stewart M., Cooper J., Brown S.D., Rubinsztein D.C., Acevedo-Arozena A. Alpha-synuclein levels modulate Huntington's disease in mice. Hum. Mol. Genet. 2012;21:485–494. doi: 10.1093/hmg/ddr477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrera F., Tenreiro S., Miller-Fleming L., Outeiro T.F. Visualization of cell-to-cell transmission of mutant huntingtin oligomers. PLoS Curr. 2011;3:RRN1210. doi: 10.1371/currents.RRN1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J.S., Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat. Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- 27.Pichaud F., Desplan C. A new visualization approach for identifying mutations that affect differentiation and organization of the Drosophila ommatidia. Development. 2001;128:815–826. doi: 10.1242/dev.128.6.815. [DOI] [PubMed] [Google Scholar]
- 28.Tenreiro S., Reimao-Pinto M.M., Antas P., Rino J., Wawrzycka D., Macedo D., Rosado-Ramos R., Amen T., Waiss M., Magalhaes F., et al. Phosphorylation modulates clearance of alpha-synuclein inclusions in a yeast model of Parkinson's disease. PLoS Genet. 2014;10:e1004302. doi: 10.1371/journal.pgen.1004302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.M., et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]