Abstract
The essential micronutrient manganese is enriched in brain, especially in the basal ganglia. We sought to identify neuronal signaling pathways responsive to neurologically relevant manganese levels, as previous data suggested that alterations in striatal manganese handling occur in Huntington's disease (HD) models. We found that p53 phosphorylation at serine 15 is the most responsive cell signaling event to manganese exposure (of 18 tested) in human neuroprogenitors and a mouse striatal cell line. Manganese-dependent activation of p53 was severely diminished in HD cells. Inhibitors of ataxia telangiectasia mutated (ATM) kinase decreased manganese-dependent phosphorylation of p53. Likewise, analysis of ATM autophosphorylation and additional ATM kinase targets, H2AX and CHK2, support a role for ATM in the activation of p53 by manganese and that a defect in this process occurs in HD. Furthermore, the deficit in Mn-dependent activation of ATM kinase in HD neuroprogenitors was highly selective, as DNA damage and oxidative injury, canonical activators of ATM, did not show similar deficits. We assessed cellular manganese handling to test for correlations with the ATM-p53 pathway, and we observed reduced Mn accumulation in HD human neuroprogenitors and HD mouse striatal cells at manganese exposures associated with altered p53 activation. To determine if this phenotype contributes to the deficit in manganese-dependent ATM activation, we used pharmacological manipulation to equalize manganese levels between HD and control mouse striatal cells and rescued the ATM-p53 signaling deficit. Collectively, our data demonstrate selective alterations in manganese biology in cellular models of HD manifest in ATM-p53 signaling.
Introduction
Huntington's disease (HD) is a devastating neurological disorder characterized by motor, psychological and cognitive impairments and premature death (1). Symptoms stem primarily from central nervous system (CNS) neurodegeneration—most notably death of medium spiny neurons (MSNs) in the caudate and putamen. HD is caused by an expansion of a CAG triplet-repeat region in exon 1 of the Huntingtin gene. Although HD is a monogenic, autosomal-dominant disease, environmental factors play a major role in modifying age of disease onset. CAG repeat length contributes to just over half of the variability in age of onset, and the majority of the remaining age of onset variability was attributed to unknown environmental factors in a landmark genetic study of a large Venezuelan kindred (2). The minority contribution from genetic modifiers has been shown by studies of candidate gene polymorphisms, which have demonstrated >12 genes that may alter Huntington's disease age of onset including GRIK2, APOE, PPARGC1A, HAP1 and—notably for this study—TP53 (3). To further support the large impact of the environment, monozygotic twins with HD have shown differences in both age of onset (differences up to 7 years) and symptomatic manifestation, in spite of identical repeat lengths (4–6). Despite the strong evidence for environmental modification in HD pathobiology, few specific environmental modifiers have been discovered. Aside from environmental enrichment in HD mouse models, metals (copper, iron, cadmium and manganese) are important environmental modifiers of HD (7–11). We have previously shown differential toxicological sensitivity to manganese (Mn2+) and cadmium (Cd2+), but not other metal ions tested (Fe3+, Cu2+, Pb2+, Co2+, Zn2+, Ni2+) in an immortalized mouse striatal model of HD (STHdhQ7/Q7 and STHdhQ111/Q111) (11,12). Expression of mutant Huntingtin conferred a survival advantage under cytotoxic manganese exposure conditions due to a substantial decrease in net manganese uptake, whereas mutant Huntingtin increased sensitivity to cadmium cytotoxicity (11).
Manganese, an essential element, is required for several enzymatic processes (13), and several of these processes are altered in HD (e.g. oxidative stress, glutamate cycling and the urea cycle) (14). On the other hand, excess manganese causes cytotoxicity via increased oxidative stress, in part, from direct inhibition of mitochondrial respiration (15). Thus, appropriate homeostatic regulation of manganese levels is needed to ensure biological function but avoid toxicity (16). The globus pallidus, caudate nucleus and other areas of the basal ganglia contain the highest level of manganese in the brain and are the areas most susceptible to HD degeneration (17,18). Pre-symptomatic HD mice (Tg-YAC128Q) have elevated sensitivity to manganese exposure, with increased dendritic spine loss in MSNs (19). HD patients also have reduced manganese levels in cortical regions, and manganese-exposed HD mouse models have reduced manganese accumulation in the striatum, the most vulnerable region in HD (11,20). Collectively, these data suggested a potential alteration in brain manganese-handling in HD. In this study, we assessed whether biological responses to manganese are altered by pathogenic CAG repeat expansions in Huntingtin using both human induced pluripotent stem cell (iPSC)-derived early striatal-like (ventralized) forebrain lineage neuroprogenitors and mouse STHdh immortalized striatal cells (21–24). Another recent study has also taken advantage of parallel use of hiPSC-derived and mouse STHdh model systems, demonstrating excessive mitochondrial fragmentation in both the STHdhQ111/Q111 cell line and iPSC-derived medium spiny neurons (25). Previous HD iPSC based studies have demonstrated increased caspase activity after growth factor withdrawal, increased lysosomal activity and reduced ATP levels in neuroprogenitors and accumulation of vacuoles in astrocytes (26–29). Many of these observations further validate disease differences shown in previous models systems (30–36).
We postulated that manganese exposure and mutant Huntingtin may impinge upon common intracellular signaling pathways. Manganese exposure increases AKT and ERK phosphorylation in the rat striatum, and mouse striatal and microglial cultures (11,37,38). Manganese exposure in non-human primates elicited alterations in p53-dependent transcripts and increased p53 immunoreactivity in the frontal cortex (39). Additionally, in PC12 cells, manganese can increase p21 mRNA expression, an established transcriptional target of p53 (40). Expression of mutant Huntingtin has also been shown to alter AKT (11,37,41,42), p53 (43,44), ERK (45,46), mTOR (47), AMPK (48) and GSK3β (49) signaling. However, most of the manganese studies were performed at acutely cytotoxic levels of manganese. To test the hypothesis that expression of mutant Huntingtin would alter intracellular signaling in response to neurologically relevant manganese levels, we assessed the response of several signaling pathways to sub-cytotoxic levels of manganese in human and mouse striatal-like neuroprogenitor models of HD.
Results
Generation and validation of human HD patient and control iPSC lines
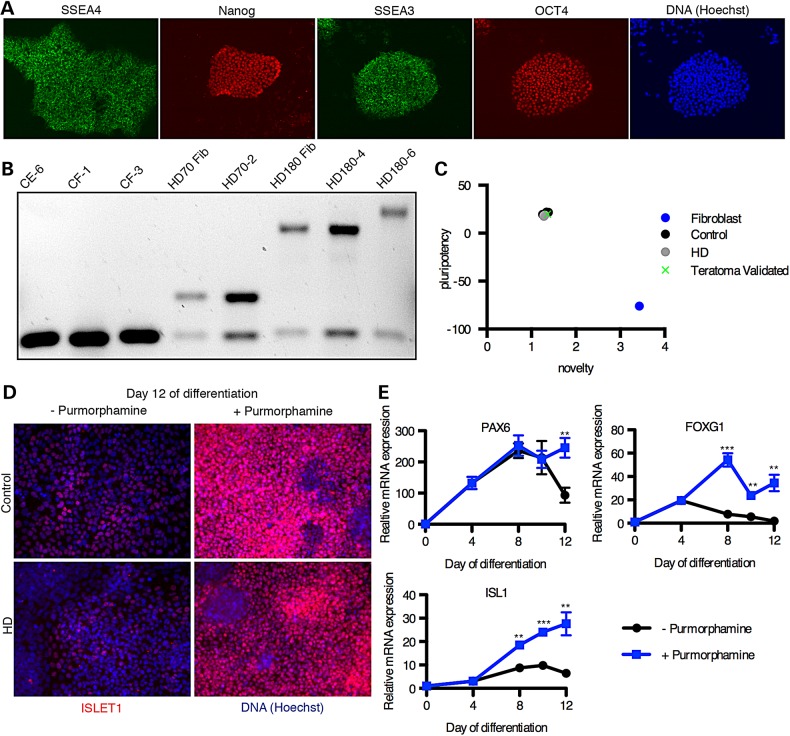
We generated iPSC lines from dermal fibroblasts (Coriell Cell Repositories [GM21756 and GM09197]) of two juvenile-onset HD patients with large Huntingtin CAG repeat lengths (70 and 180). We also derived iPSC lines from fibroblasts of two control subjects with no history of neurological conditions (CE and CF) (see Materials and Methods). Integration-free iPSC lines were generated by electroporating fibroblasts with episomal plasmid vectors (21). The iPSC lines were validated for expression of protein markers of pluripotency such as OCT4 and Nanog (Fig. 1A). HD genetic status was confirmed by assessing the CAG repeat length in exon 1 of Huntingtin in both the iPSC lines and the originating fibroblast lines (Fig. 1B). We identified one clonal line, HD180 clone 6 (referred to as HD180-6), that demonstrated an expansion of the CAG repeat from 180 to >210 in the pathogenic allele. To our knowledge, this is the first reported expansion of the Huntingtin gene CAG repeat region in an iPSC line; however, iPSC lines from patients with Friedrich's ataxia often demonstrate expansion of the GAA triplet repeat due to high expression of MSH2, which is known to mediate somatic triplet expansion in the Huntingtin gene (50,51). To validate the quality of our iPSC lines, we performed PCR on DNA isolated from each clonal iPSC line to ensure the absence of plasmid integration. The episomal vectors are typically lost over successive cell cycles; however, they can integrate into the genome at a low frequency (21). Primers were targeted to the WHP Posttranscriptional Regulatory Element (WPRE) region of the plasmid that would constitute a novel sequence in the human genome. Lines maintaining the WPRE region were excluded from our studies. A normal euploid karyotype was confirmed for all validated lines. Lastly, we performed an mRNA microarray with the PluriTest bioinformatics assay to test for conformity to pluripotency (Fig. 1C). All iPSC lines used in this study were confirmed by this test with very little variance between lines and high similarity to a previously published teratoma-validated control line (52).
Figure 1.
Differentiation of iPSCs into early striatal-like neuroprogenitors. (A) Immunofluorescent staining of HD iPSC line HD70-2 for markers of pluripotency. (B) PCR for the Huntingtin gene including the CAG repeat region. The lower bands are the non-pathogenic alleles, and the larger bands are pathogenic. All iPSC lines show similar sizes to the fibroblast lines from which the iPSCs were generated except HD180-6, which shows an expanded CAG repeat size. (C) The PluriTest analysis was used to validate the mRNA microarray profile from our iPSC lines, and all lines (CE-6, CF-1, CF-3, HD70-2, HD180-4, and HD180-6 are plotted here) had similar pluripotency profiles, including a teratoma-validated iPSC line. (CA-6) (D) Control (CA-24) and HD (HD180-2) iPSCs were differentiated to early forebrain neuroprogenitors by the dual-SMAD small molecule technique with and without purmorphamine (SHH agonist) and were immunostained for ISL1 (red), a marker of striatal progenitors, with nuclei labeled with Hoechst dye (blue). (E) Expression of markers of neurodevelopment was measured by qRT-PCR from the CF-3 control cells differentiated with and without purmorphamine. **P < 0.01 and ***P < 0.001 by t-test. N = 4. Mean ± SEM.
Differentiation to neuroprogenitors expressing early striatal markers
We used the established ‘dual-SMAD inhibition’ method of neuralization via SB431542 and DMH1 to generate telencephalic neural progenitors from our iPSC cultures (22,23,52). These early neuroprogenitors expressed the neuroectodermal marker, PAX6 and the telencephalic marker, FOXG1 (Fig. 1E) (53). Addition of sonic hedgehog (SHH) or a SHH agonist, purmorphamine, has been shown to ventralize these neuroprogenitors to express markers of the lateral ganglionic eminence (LGE) from which striatal MSNs develop (23). Addition of purmorphamine to the neuroprogenitor cultures from Day 4 of differentiation onward resulted in a significant increase in expression of Islet1 (ISL1) protein and mRNA over time (Fig. 1D and E). In the developing telencephalon, ISL1 is expressed specifically in the earliest post-mitotic cells of the striatum (54). A recently published method for generating fully validated MSNs uses an analogous technique in the first stage (24). Our ISL1+ neuroprogenitors provided a nearly homogenous cell population (∼90% ISL1+ cells by immunofluorescence microscopy) regardless of the genotype (HD or control) to begin assessing the effects of manganese. Furthermore, quantitative reverse-transcriptase PCR (qPCR) for lSL1 demonstrated statistically identical expression levels between control and HD neuroprogenitors (Supplementary Material, Fig. S1). These human striatal-like neuroprogenitors are at a homologous developmental time point as the STHdhQ7/Q7 and STHdhQ111/Q111 cells, which were isolated from the mouse striatum on Embryonic day 14 (E14), a developmental time point wherein ISL1 expression defines the developing striatum (12,55), thus, allowing for more valid comparisons between these two model systems.
Manganese exposure evokes strong p53 response in human neuroprogenitors and mouse striatal cells
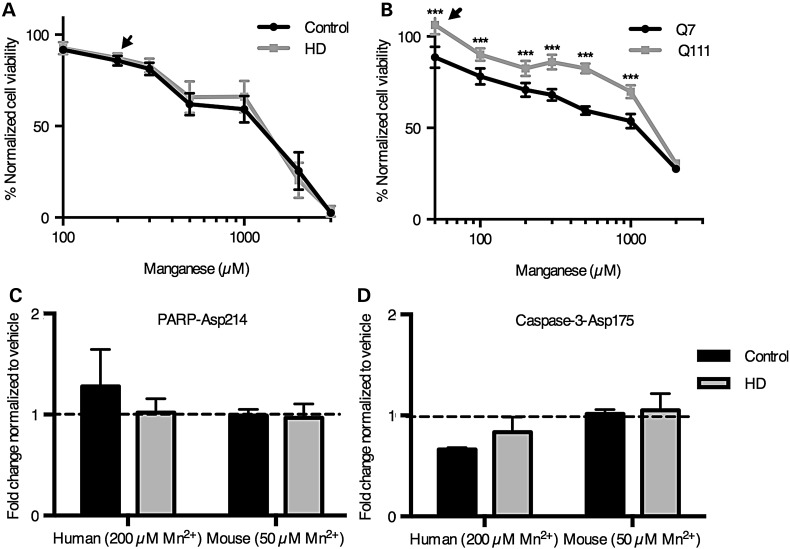
To elucidate the effect of sub-toxic manganese exposure on intracellular signaling pathways in both the human and mouse cell models, we performed the PathScan ELISA-based signaling pathway array (PathScan, Cell Signaling Technology, Danvers, MA). The sub-toxic manganese exposure (24 h) concentrations in the human and mouse neuroprogenitors were determined via the CellTiter-Blue cell viability assay (Promega, Madison, WI) (Fig. 2A and B). We selected 200 µm manganese exposure for the human ISL1+ neuroprogenitors, and 50 µm manganese for the mouse STHdh cells because these concentrations caused similar, but minimal (<15%), loss of viability across the two model systems (Fig. 2A). These concentrations are at, or just above, the threshold associated with brain toxicity in vivo (56). Furthermore, these manganese levels are within the range we previously found to elicit a strong HD/manganese interaction in the mouse STHdh model (11,57). As expected from our cytotoxicity assays, proteolytic activation of Caspase-3 and PARP-1, enzymes activated during apoptotic cell death, showed no change between vehicle and manganese exposures in the human neuroprogenitors, indicating minimal apoptotic cell death in our PathScan array experiments (Fig. 2B and C). Additionally, western blot analysis of manganese-exposed STHdh cells for caspase-3 cleavage did not show a significant increase above baseline until 200 µM exposure (P = 0.0077 and 0.0138; Q7 and Q111 respectively), well above the 50 µM exposure paradigm (Supplementary Material, Fig. S2). Likewise, detection of caspase-3 cleavage in hiPSC-derived neuroprogenitors was not observed until 2 mm Mn exposure (Supplementary Material, Fig. S3). We suspect the slight decrease in detected viability by the CellTiter-Blue assay at these manganese exposure levels (Fig. 2A and B arrowheads) is due to decreased cell proliferation over the 24 h exposure period, rather than cell death given the absence of necrotic and apoptotic markers.
Figure 2.
Manganese-induced loss of viability in human neuroprogenitors is unaffected by mutant Huntingtin and not via apoptotic signaling. (A and B) The cell viability assay, CellTiterBlue, was performed on human neuroprogenitors (A) and mouse STHdh striatal cells (B). The signals were normalized by defining the vehicle-treated cells as 100% viability. Arrowheads denote concentrations chosen for the Pathscan array. (N = 6 for control and 5 for HD in human neuroprogenitors and N = 6 wells in mouse striatal cells). (B) Cleavage of PARP was quantified by the Pathscan Intracellular Signaling array from samples treated with manganese (N = 2 for human and 3 for mouse). (C) Cleavage of Caspase-3 was quantified by the Pathscan Intracellular Signaling array from samples treated with manganese (N = 2 for human and 3 for mouse). Values are relative to vehicle-treated samples. For genotype comparisons ***P < 0.001 by t-test. Bars, mean + SEM (A, C and D) and SD (B).
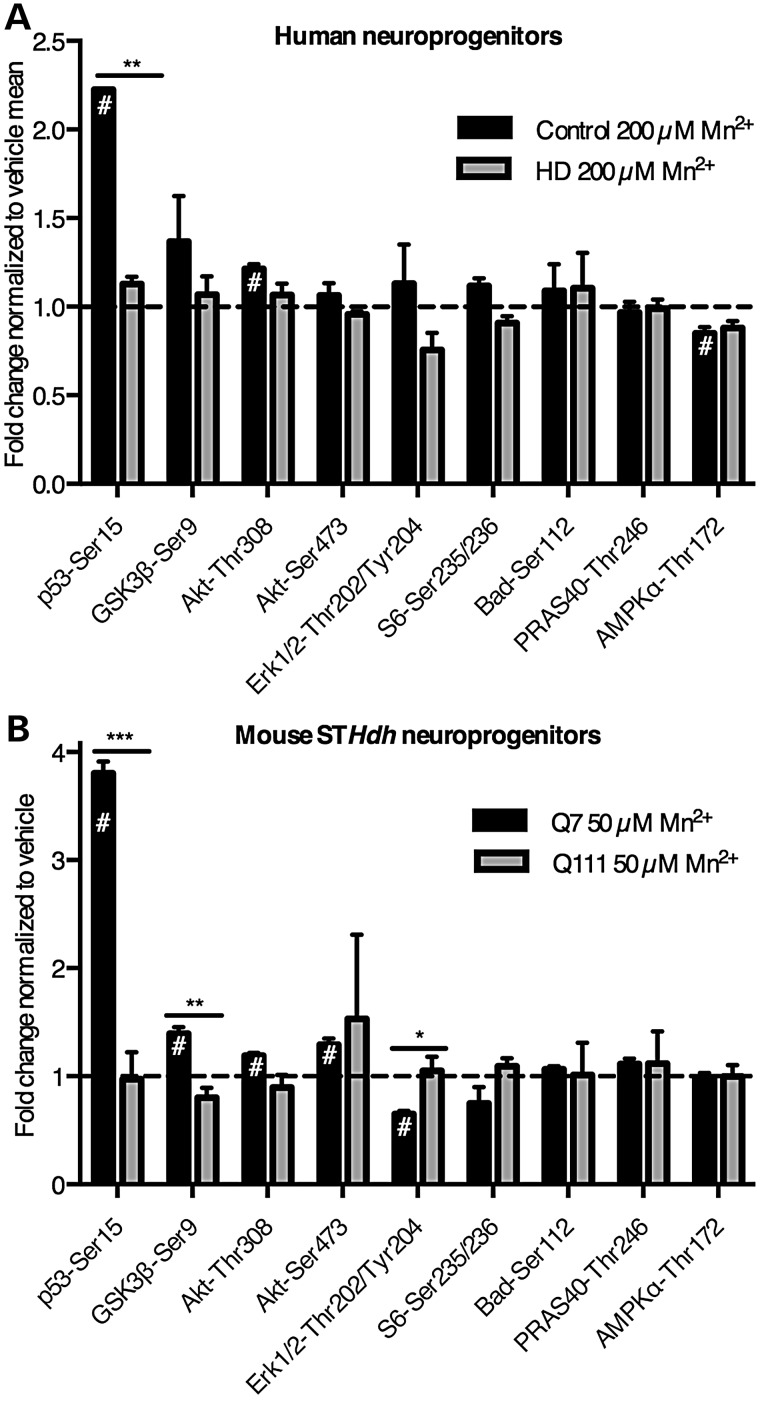
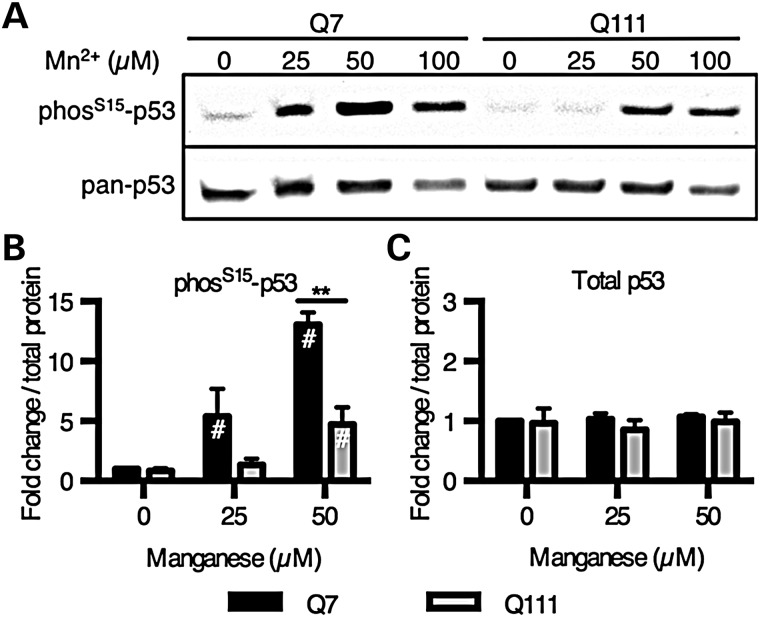
The PathScan array was used to assess manganese-induction of 18 unique signaling events (phosphorylation or proteolytic cleavage) in control and HD human neuroprogenitors and mouse STHdh cells (Fig. 3). Phosphorylation of several signaling proteins displayed a manganese-dependent increase in control cells (Human: p53 and AKT; mouse: p53, GSK3β and AKT). Phosphorylation of p53 at serine-15 [p53(S15)] and phosphorylation of AKT at threonine-308 were significantly changed in both model systems, with phosphorylation of p53(S15) showing the most robust change in both human and mouse models (Fig. 3A and B). Interestingly, manganese-induced phosphorylation of p53(S15) was severely diminished in HD neuroprogenitors in both human (223% in control versus 113% in HD; P< 0.01) and mouse striatal-like neuroprogenitors (380% in control versus 97% in HD; P < 0.001).
Figure 3.
PathScan ELISA array shows manganese-induced phospho-p53(S15) at sub-cytotoxic manganese concentrations. (A) Human cells were exposed for 24 h to 200 µm Mn2+. N = 2 for control (CE-6 and CF-1, once each) and HD (HD70-2 and HD180-6, once each). (B) Mouse STHdh striatal cells (Q7 and Q111) cells were exposed for 24 h to 50 µm Mn2+. (A and B) Phosphorylation and cleavage events were measured by the Fluorescent PathScan Intracellular Signaling Array sandwich ELISA. Background from a no protein sample was subtracted, and signals were divided by the mean of the untreated samples by genotype. The array was imaged and quantified using an Odyssey IR imager. Antibodies whose signal was <4 times the background were not plotted. For both human and STHdh cells this included phosphorylation at Stat1-Tyr701, HSP27-Ser78, p38-Thr180/Tyr182, PARP-1 cleavage and caspase-3 cleavage. For human cells (A) phosphorylation at Stat3-Tyr70 was below detection, and phosphorylation of mTOR-Ser2448, p70 S6 Kinase-Thr389 and SAPK/JNK-Thr183/Tyr185 were below detection for the mouse striatal cells (B). #P < 0.05 compared with vehicle-treated by t-test. For genotype comparisons *P < 0.05 and **P < 0.01 by t-test. Bars, mean + SEM.
Phosphorylation of p53 by manganese is dose-dependent and deficient in HD mutant cells
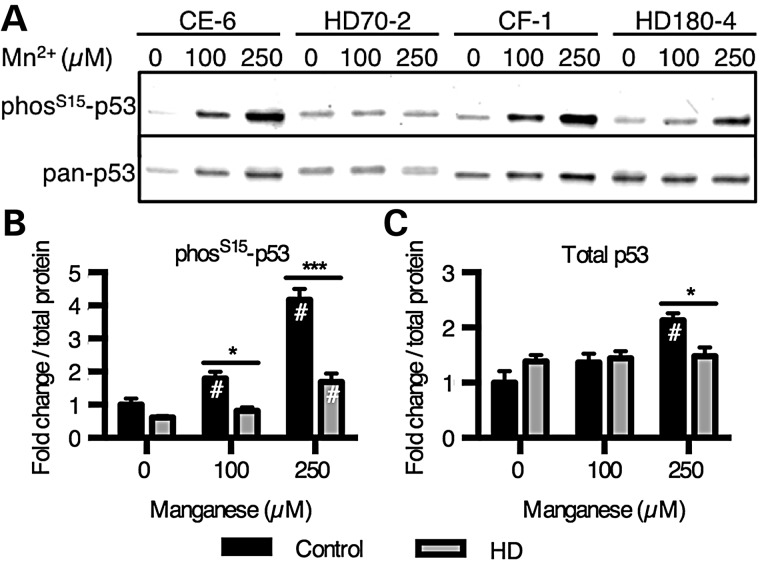
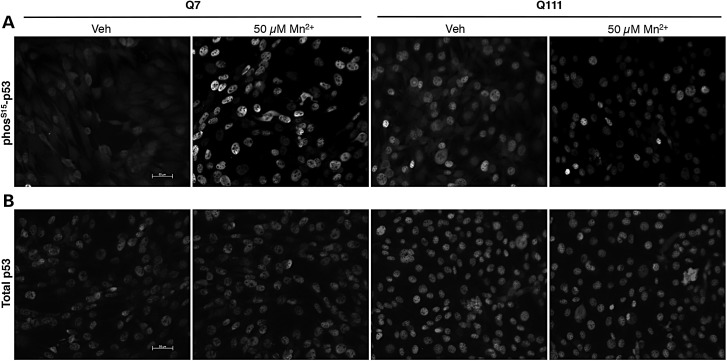
Western blot analysis was used to further assess the manganese and HD/manganese interaction effects on p53(S15) phosphorylation. Manganese-induced phosphorylation of p53(S15) was concentration-dependent (Fig. 4A and B). The control lines (CE-6 and CF-1) had substantially greater phosphorylation with manganese treatment (at 100 and 250 µm) than the HD mutant lines (HD70–2 and HD180-6) (e.g. 418% in control versus 169% in HD at the 250 µm manganese exposure, P < 0.01) (Fig. 4A and B). Human HD neuroprogenitors showed a small, but significant, manganese-induced p53(S15) phosphorylation, that was significantly blunted compared with control cells. Levels of total p53 were also significantly increased by manganese exposure in the control cells (P < 0.01), but not enough to account for the differences in phosphorylation (2.1-fold increase in total p53 versus 4.2-fold increase in phosphorylated p53[S15]) (Fig. 4C). The mouse striatal cell model showed a similar dose-dependent phosphorylation of p53(S15) with a strong deficit in HD cells (Fig. 5A and B). Additionally, the same response with nuclear localization of p53 can be seen by immunostaining for phos-p53(S15) (Fig. 6A) and total p53 (Fig. 6B). Control mouse neuroprogenitors also showed a manganese-dependent increase in the p53 transcriptional target p21 (CDKN1A) mRNA expression (6.87-fold increase above vehicle, P < 0.001), while the HD mutant cells did not (with only a non-significant 1.82-fold increase above vehicle) (Supplementary Material, Fig. S4). Elevated expression of this canonical p53 transcriptional target is highly consistent with manganese exposure increasing p53 transcriptional activity.
Figure 4.
HD striatal progenitors show reduced manganese-dependent p53(S15) phosphorylation. (A) Human cells exposed to manganese were probed by western blot for phospho-p53(S15) and total p53. (B and C) Western blot data were quantified and demonstrate significant differences in p53 phosphorylation between genotypes. N = 4 for control (CE-6 and CF-1, twice each) and HD (HD70-2 and HD180-4, twice each). #P < 0.05 compared with vehicle-treated by t-test. *P < 0.05 and ***P < 0.001 by t-test. Bars, mean + SEM.
Figure 5.
HD mouse striatal cells show reduced manganese-dependent p53(S15) phosphorylation. (A) Mouse STHdh striatal cells exposed to manganese for 24 h were probed by western blot for phospho-p53(S15) and total p53. (B and C) Western blot data were quantified and demonstrate significant differences in p53 phosphorylation between genotypes (N = 3). #P < 0.05 compared with vehicle-treated by t-test. The p53 phosphorylation data for 100 µm Mn was not quantified because of increased cell viability loss seen in Figure 2B. **P < 0.01 by t-test. Bars, mean + SEM.
Figure 6.
HD mouse striatal cells show reduced manganese-dependent p53(S15) phosphorylation by immunostaining with nuclear p53 localization. Mouse STHdh striatal cells were exposed to vehicle or 50 µm manganese for 24 h followed by immunofluorescence for either phos-p53(S15) (A) or total p53 (B).
ATM activity is associated with manganese-dependent p53 activation
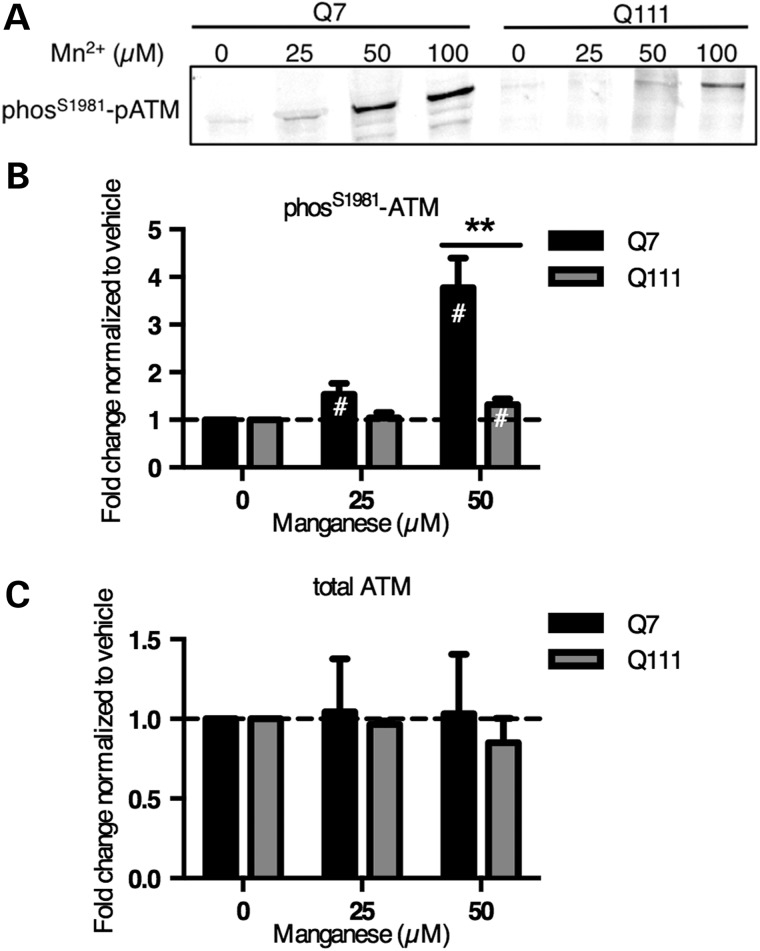
ATM, AMPK, ERK1/2 and p38 are known to phosphorylate p53(S15) (58–61). We did not see a manganese-dependent increase in phosphorylation of AMPK-α (threonine-172), ERK1/2 (threonine-202/tyrosine-204) or p38 (threonine-180/tyrosine-182) by the PathScan array, but ATM kinase was not on the array. High concentrations of manganese (5 mm) have been reported to activate ATM (ataxia telangiectasia mutated) to induce phosphorylation of p53(S15) in cell-free kinase assays (58,59,62,63). First, we measured the level of autophosphorylation of ATM at serine 1981, which is thought to be indicative of kinase activity, in manganese-treated mouse STHdh striatal cell lysates (64). The same manganese-dependent increase in phosphorylation seen with p53(S15) was also observed with ATM(S1981) (Fig. 7A and B). Manganese does not significantly alter levels of total ATM (Fig. 7C and Supplementary Material, Fig. S5). The HD cells (STHdhQ111/Q111) had a similar deficiency in response that was seen with p53(S15).
Figure 7.
HD mouse striatal cells show reduced manganese-dependent ATM(S1981) phosphorylation. (A) Mouse STHdh striatal cells exposed to manganese for 24 h were probed by western blot for phospho-ATM(S1981). (B) Western blot data were quantified and demonstrate significant differences in ATM phosphorylation between genotypes (N = 6). (C) Western blot quantification of total ATM showed no statistical effect of manganese exposure (N = 3). #P < 0.05 compared with vehicle-treated by t-test. The p53 phosphorylation data for 100 µm Mn was not quantified because of increased cell viability loss seen in Figure 2B. **P < 0.01 by t-test. Bars, mean + SEM.
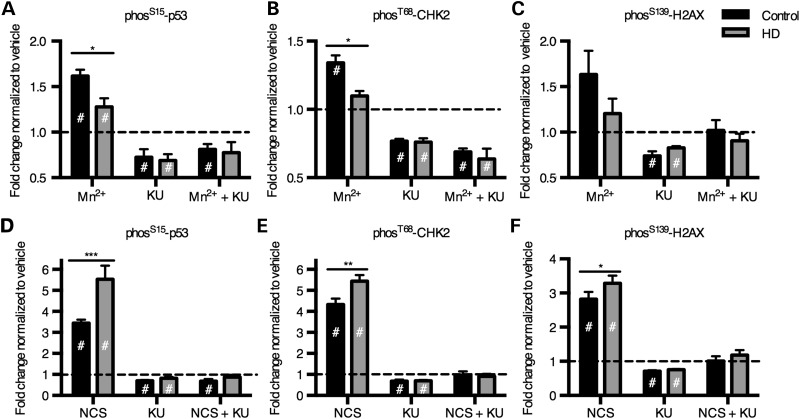
To further implicate ATM as an upstream kinase for manganese-induced p53 phosphorylation, we tested the hypothesis that blocking ATM kinase activity during manganese exposure would inhibit p53(S15) phosphorylation. We observed by both an In-Cell Western assay (Fig. 8A) and ELISA Array (Supplementary Material, Fig. S6) that the selective ATM inhibitor, KU-55933, completely blocked the manganese-dependent phosphorylation of p53 in the human neuroprogenitors (65). The effect of the inhibitor was quite specific to p53 phosphorylation, as no significant effects were observed for other signaling proteins on the PathScan array (Supplementary Material, Fig. S5). KU-55933 also inhibited manganese-dependent p53(S15) phosphorylation in the mouse STHdh striatal cells (Supplementary Material, Fig. S7).
Figure 8.
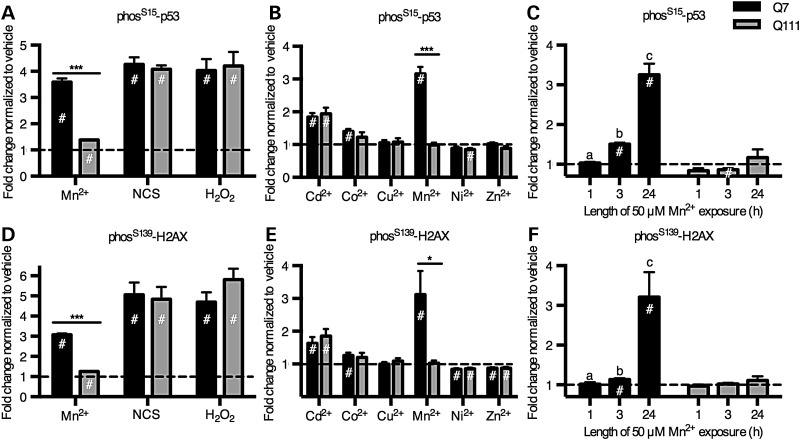
Manganese induces phosphorylation of 3 ATM targets, which is blocked by ATM kinase inhibitor, KU-55933. ‘In-Cell Western’ was performed on human neuroprogenitors for phosS15-p53 (A and D), phosT68-CHK2 (B and E) and phosS139-H2AX (i.e. γ-H2AX) (C and F) with exposure to Mn at 200 µm (A–C) or DNA-damaging agent, neocarzinostatin at 100 ng/ml (NCS) (D–F). The cells were also exposed to ATM inhibitor, KU-55933 at 1 µm. N = 4 for control (CE-6, CF-3; two experiments each) and N = 4 for HD (HD70-2, HD180-4; 2 experiments each). #P < 0.05 compared with vehicle-treated by t-test. *P < 0.05, **P < 0.01 and ***P < 0.001 for t-test between genotypes.
DNA damage is a canonical activator of the ATM-p53 signaling pathway. To test whether the ATM-p53 pathway is broadly deficient in its ability to respond to any pathway-relevant stressor in HD neuroprogenitors, we exposed the cells to neocarzinostatin, a double-stranded DNA break (DSB)-inducing agent known to activate p53 via ATM (58). In contrast to the HD deficit in manganese-induced p53 activation, the human HD neuroprogenitors were hypersensitive to p53(S15) activation by neocarzinostatin (Fig. 8D), and this phosphorylation was completely blocked by the ATM inhibitor, KU-55933, confirming activation occurred via ATM kinase. If ATM kinase is activated by manganese, then other ATM targets may be activated by manganese as well. Therefore, we measured phosphorylation of two other established targets of ATM kinase, CHK2 at threonine-68 (T68) and H2AX at serine-139 (S139, i.e. γ-H2AX). Using the In-Cell Western assay, control neuroprogenitors had a significant increase in phosphorylation of CHK2(T68) with manganese exposure, but CHK2(T68) was non-responsive in HD neuroprogenitors (Fig. 8B). H2AX(S139) showed the same trend but with greater variance at the same sample size (N = 4) (Fig. 8C). In both cases, the ATM inhibitor KU-55933 completely blocked manganese-dependent phosphorylation. Both ATM kinase targets were activated by the DNA-damaging agent, neocarzinostatin, with elevated phosphorylation in HD cells compared with control (Fig. 8E and F). These data demonstrate a deficit in ATM activation at neurologically relevant levels of manganese in HD neuroprogenitors; yet, a fully responsive (or even more responsive in the case of human HD neuroprogenitors) ATM kinase activity from a canonical activator of this pathway, DSB DNA damage.
The HD-dependent deficit in ATM activation is specific to manganese
Oxidative stress was recently discovered to activate ATM via a mechanism independent from the DNA damage-dependent activation of ATM (63). To test whether ATM kinase is responding to manganese via an oxidative stress-dependent mechanism, we exposed the STHdhQ7/Q7 and STHdhQ111/Q111 cells to 250 µm hydrogen peroxide for 1 h and measured the phosphorylation of p53(S15) and H2AX(S139) by In-Cell Western assay. We observed an equivalent phosphorylation of ATM targets by hydrogen peroxide in HD and control cells (Fig. 9A and D), with levels of phosphorylation of p53(S15) and H2AX(S139) equivalent to those observed after neocarzinostatin exposure (100 ng/ml for 1 h). We corroborated these findings from the In-Cell Western assay by performing western blots and found a similar pattern of response with a genotype difference with manganese exposure but an equivalent response to both neocarzinostatin and hydrogen peroxide (Supplementary Material, Fig. S8B and D). These observations suggest that the mechanism of HD-dependent deficiency in p53 response is not the same as the mechanisms by which the oxidative stressor hydrogen peroxide or the DNA-damaging agent neocarzinostatin activate ATM. In mouse STHdh cells, the ATM inhibitor, KU-55933, significantly blocked phosphorylation of p53 due to manganese, neocarzinostatin, and hydrogen peroxide exposure (Supplementary Material, Fig. S7). We have also shown that both model systems (human and mouse) can have normal or elevated levels of ATM-p53 activity via stressors other than manganese.
Figure 9.
Mutant Huntingtin only inhibits manganese-dependent ATM target phosphorylation. (A and D) Mouse STHdh striatal cells (Q7 and Q111) were exposed to manganese (50 µm) for 24 h, neocarzinostatin (NCS, 100 ng/ml) for 1 h or hydrogen peroxide (H2O2, 250 µm) for 1 h. (B and E) Mouse striatal cells were exposed for 24 h to several divalent metal cations all at 50 µm, except 100 µm Cu2+ and 20 µm Cd2+. The means are normalized by the vehicle mean for that line to allow for easier genotype comparisons. (C and F) Mouse striatal cells were also exposed to manganese (50 µm) for increasing amounts of time. Significant differences are designated as: a is significantly different from b and b is significantly different from c. If no letters are listed, then none of the means are significantly different from one another. # indicates significantly different from vehicle treated (P < 0.05 by Tukey's post hoc). N = 3 for means from 3 to 5 wells for independent replicate experiments (A–C). N = 4 for means from 3 to 5 wells for independent replicate experiments (D–F). *P < 0.05 and ***P < 0.001 for t-test between genotypes. Bars, mean ± SEM.
We tested the hypothesis that the HD/manganese interaction effect on the ATM-p53 pathway is related to a generic metal response pathway by exposing the mouse striatal neuroprogenitors to other divalent metal ions (Cd2+, Co2+, Cu2+, Ni2+, and Zn2+) and assessing ATM phosphorylation targets (Fig. 9B and E). The same concentration (50 µM) was used for all the metals except copper (100 µm) and cadmium (20 µm), as higher concentrations of cadmium resulted in cytotoxicity (11). We found that other metals either significantly increased (Cd2+, Co2+), decreased (Ni2+, Zn2+) or did not affect (Cu2+) p53(S15) and H2AX(S139) phosphorylation; however, except for cadmium (Cd2+), the changes in p53 phosphorylation were quite small compared with that observed with manganese. None of these other metals, however, showed an HD-dependent deficit in p53(S15) phosphorylation.
We tested the hypothesis that the HD-dependent differences in ATM activity at 24 h may be due to altered response kinetics to manganese (e.g. peak response of p53 activation occurring at different times post-exposure). We performed a time-course experiment on the STHdh cells with 50 µm manganese (see Fig. 9C and F [for 1, 3 and 24 h exposures] and Supplementary Material, Figure S9 [for 1, 3, 6 and 24 h exposures]) and saw a continuous accumulation of p53(S15) and H2AX(S139) phosphorylation over the 24 h period in the control line (STHdhQ7/Q7). The HD line (STHdhQ111/Q111) did not show manganese-induced phosphorylation of p53 or H2AX at any of the time points, indicating that the difference in ATM activity observed between control and HD lines was not the result of a difference in temporal response patterns. Together, these observations demonstrate a manganese-specific deficit in ATM activation in HD not observed with other known activators of ATM kinase.
Cellular manganese accumulation is diminished in HD neuroprogenitors and HD striatal cells
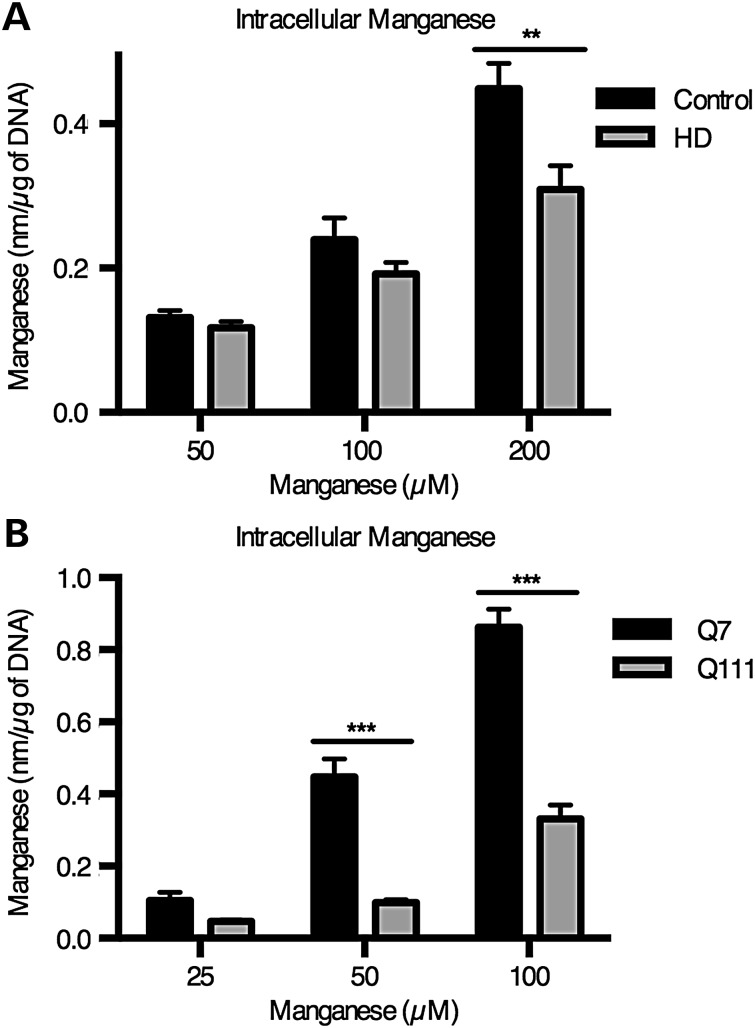
Because the HD-dependent deficit in manganese-induced ATM activation was remarkably selective to manganese and because manganese is sufficient to induce ATM activity in cell-free kinase assays, we hypothesized that mutant Huntingtin alters intracellular manganese homeostasis (62,63). We have previously reported decreased manganese accumulation in the mouse STHdhQ111/Q111 cells compared with the control, STHdhQ7/Q7 (11,57). However, this phenotype occurred with a significant decrease in manganese-induced cytotoxicity in the mutant cells (STHdhQ111/Q111), which we did not see in the human neuroprogenitors used in this study (Fig. 2A). To measure intracellular manganese content, we used a fluorescence-based assay (CFMEA) that we have previously described (66,67). We found that human HD neuroprogenitors accumulated significantly less manganese (31% reduction compared with control cells, P < 0.01) after a 24 h manganese exposure at 200 µm (Fig. 10A). A similar, though larger magnitude difference in manganese accumulation was observed in mouse neuroprogenitors (78% reduction of manganese in STHdhQ111/Q111 cells versus control cells; Fig. 10B). Thus, both human and mouse neuroprogenitors with pathogenic alleles of Huntingtin display deficits in net manganese accumulation at concentrations associated with deficits in manganese-dependent ATM-p53 signaling.
Figure 10.
Reduced accumulation of manganese in neuroprogenitors expressing mutant Huntingtin. (A) Accumulation of intracellular manganese was quantified by cellular fura-2 manganese extraction assay (CFMEA) in human neuroprogenitors that were exposed for 24 h to manganese. For human N = 4 for control (CE-6 and CF-1, twice each) and HD (HD70-2 and HD180-4, twice each). (B) The same procedure was performed on mouse STHdh cells (Q7 and Q111) with N = 3. For genotype comparisons **P < 0.01 and ***P < 0.001 by t-test. Bars, mean + SEM.
Pharmacological equalization of cellular manganese content rescues the ATM-p53 signaling deficit
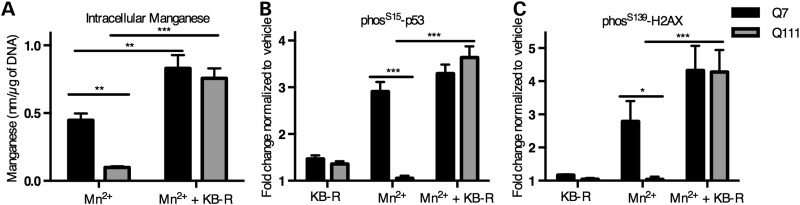
To determine if the differential manganese accumulation in HD is the basis of the differential manganese-induced ATM activation, we pharmacologically manipulated intracellular manganese content. To this end, we used the sodium/calcium exchanger (NCX) inhibitor KB-R7943 that blocks manganese efflux in mouse tissues (68) to alter cellular manganese content in the control and HD STHdh cells, given the magnitude of the manganese accumulation deficit. Co-incubation of the mouse STHdh cells with 10 µm KB-R7943 and 50 µm manganese resulted in equivalent manganese accumulation between control STHdhQ7/Q7 and HD STHdhQ111/Q111 lines (Fig. 11A, P = 0.585). Using these conditions to equalize cellular manganese levels, we found that KB-R7943 also equalized the phosphorylation levels of p53(S15) and H2AX(S139) between HD and control neuroprogenitors (Fig. 11B and C; P = 0.31 for p53[S15] and P = 0.95 for H2AX[S139]). These data demonstrate that normalizing cellular manganese levels mitigates the HD-dependent deficit in manganese-induced ATM target phosphorylation.
Figure 11.
Pharmacological equalization of manganese content results in similar p53(S15) and pH2AX(S139) phosphorylation. (A) Mouse STHdh cells (Q7 and Q111) were treated for 24 h with manganese (50 µm) with or without the NCX inhibitor KB-R7943. Intracellular manganese accumulation was measured by the CFMEA technique. N = 3 independent experiments. (B and C) The same treatment paradigm was used for In-Cell Western analysis to quantify the phosphorylation of p53 and H2AX. N = 4 independent experiments for (B) and N = 3 independent experiments for (C). For genotype comparisons *P < 0.05, **P < 0.01 and ***P < 0.001 by t-test. Bars, mean + SEM.
Discussion
Our observations demonstrate that a manganese-handling deficit occurs in HD striatal-like neuroprogenitors selectively impairing ATM-p53 signaling under low-level (sub-cytotoxic) manganese exposure (>85% viability; Fig. 2A) with no detectable caspase-3 or PARP-1 cleavage (Fig. 2B and C). We used an ELISA-based array for 18 cellular signaling events and found that phosphorylation of p53(S15) showed the largest response (2.2-fold increase in human; 3.8-fold increase in mouse; Fig. 3A and B), and this response was significantly reduced in HD neuroprogenitors (∼78% decrease in human response [P < 0.001]; 69% decrease in mouse response [P < 0.01]) (Fig. 4B and C). AKT(T308) phosphorylation was also induced by manganese but to a lesser extent (1.2-fold for human and mouse, P < 0.05) (Fig. 3A and B). Manganese-dependent phosphorylation of CHK2(T68) and H2AX(S139), two canonical targets of ATM kinase, were also found to be deficient in HD cells (Fig. 8A–C). DNA damage and oxidative stress induced by hydrogen peroxide, two known mechanisms to activate ATM activity, did not show HD-dependent differences in the mouse striatal cells. DNA-damage resulted in hyper-activation of ATM targets in human HD neuroprogenitors (Fig. 8E and F). Mirroring our findings in the mouse striatal cell line, the human neuroprogenitors had a small but significant defect in cellular manganese accumulation relative to control (Fig. 10A). Combined, our results demonstrate altered manganese biology in HD patient-derived neuroprogenitors and identify a novel manganese-responsive cell signaling event, ATM activation, that is impaired in HD. Our data support the hypothesis that a neuronal manganese homeostatic defect may occur in HD.
Our data establish ATM-p53 as a major manganese response pathway. Alterations in p53-dependent gene expression following manganese exposure have been observed in a non-human primate model (39,40). Here, we provide evidence for manganese-induced p53(S15) phosphorylation by western blot, ELISA and In-Cell Western assays in both human and mouse neuronal cells. Further, this signaling pathway was activated more robustly than all other intracellular signaling events assayed. The identification of ATM as the upstream manganese-induced kinase of p53 is based on ATM autophosphorylation, pharmacological kinase inhibition and other established ATM phosphorylation targets (CHK2[T68] and H2AX[S139]). DNA damage and oxidative stress have both been shown to activate ATM, but unlike manganese-dependent ATM activation, DNA damage and hydrogen peroxide had the same or greater response in HD cells compared with controls, consistent with previous reports on p53 in HD model systems (58,59,63). Among the metal ions tested, only cadmium induced ATM activation to a similar magnitude as manganese. However, unlike manganese, cadmium is a known DNA-damaging agent at micromolar concentrations providing a clearly established route to ATM activation (69). Manganese, on the other hand, is known to activate ATM phosphorylation of p53(S15) in purified kinase activity assays in the absence of DNA (62,63). Thus, our data strongly suggest that manganese activates ATM via a mechanism independent of DNA damage. Additionally, our results do not support a mechanism via direct redox cycling (Fenton reaction) because many of the other metals, which had minimal effects on ATM activity, are known to be more redox active than manganese (e.g. copper). As neither a DNA damage nor oxidative stress mechanisms clearly explain the manganese-dependent ATM activation, we propose that manganese activates ATM via a novel mechanism that is specific to manganese compared with all other metal ions tested.
As mentioned in the Introduction, manganese is known to be neurotoxic at high exposure levels. Our data demonstrate that manganese mediates the activation of ATM kinase activity at neurologically relevant concentrations (50–200 µm) in living cells (56), much lower than the 5–10 mm used in previously published cell-free purified kinase assays (62,63). Although our concentrations in the human model border the highest levels seen in toxic human exposures, we do not see overt manganese-dependent toxicity in our Islet1 neuroprogenitors until much higher concentrations. One potential explanation is reduced manganese uptake and accumulation in neuroprogenitors compared with mature neurons. We have recently published data demonstrating just such a difference between Day 11 floor plate neuroprogenitors and Day 25 post-mitotic dopaminergic neurons derived from control human pluripotent stem cells (hiPSCs) (70). Therefore, the concentrations needed to elicit an ATM response in mature neurons may well be below what was necessary in our neuroprogenitors.
The HD-dependent deficit in manganese-induced ATM-p53 activation is one of many examples implicating this pathway with HD pathophysiology. Previous work has shown increased p53 accumulation and phosphorylation in HD patient and mouse brains as well as HD cell lines under basal conditions, while knockdown of p53 can ameliorate both mitochondrial and behavioral HD phenotypes (25,43). Possible direct interactions between mutant Huntingtin and both p53 and ATM have also been reported ((43,44,71). Both p53 and ATM phosphorylation have been reported to be elevated in an HD iPSC model (72). This pathway was also hyper-activated by DNA damage in our human HD neuroprogenitors, mirroring the published findings of camptothecin (a DNA-damaging agent) treatment of HD muscle cells (73). Genetic variation in the TP53 gene (which encodes p53 protein) accounts for ∼13% of age-of-onset variability in HD not accounted for by the CAG repeat length (74). The pro-apoptotic function of activated p53 is its best characterized biological role; however, at low levels of activation, p53 transcriptional activity plays a largely pro-survival role by up-regulating genes involved in energetic balance, oxidative defense and metabolic state (75). The HD-dependent alterations in p53 responsiveness to a non-lethal stressor could provide a mechanism for the age-dependent degeneration seen in HD. Namely, the accumulation of cellular damage over time due to insufficient neuroprotection by the manganese-dependent ATM-p53 pathway.
Our data support a model in which mutant Huntingtin impinges on the ATM-p53 response to manganese exposure by altering manganese transport/homeostasis. We found lower cellular manganese content in the HD human ISL1+ neuroprogenitors than control cells following low-level exposures (Fig. 10A). The cause of this difference is not yet explained; however, iron transporters thought to be important for manganese transport do not seem to play a substantial role in the differential manganese accumulation in the STHdh model system (57). Equalizing cellular manganese content in HD and control cells by pharmacologically blocking manganese efflux (68) eliminated the differential response in p53 phosphorylation (Fig. 11B), providing strong evidence that the deficit in manganese-induced ATM activation in HD cells was dependent on cellular manganese-handling. Remarkably, the magnitude of the deficit in the ATM-p53 signaling pathways in mouse and human HD models is similar despite the substantially stronger manganese accumulation deficit in the mouse model system. One potential explanation could be differences in compartmentalization of intracellular manganese between the mouse and human models. Manganese has been indicated to accumulate primarily in the nucleus of neurons where a majority of ATM protein is localized (76,77).
Epidemiological studies indicate a significant role for environmental factors in HD progression (2). The defect we observe in manganese-handling/homeostasis in HD patient-derived neuroprogenitors substantiates previous reports of reduced accumulation of manganese in both mouse striatal cell models of HD and the striata of manganese-exposed Tg-YAC128Q HD model mice, as well as altered manganese and iron localization in HD patient brains (11,20,57). Decreased cellular manganese in HD could explain at least some HD pathogenic mechanisms. For example, decreased activity of manganese-dependent enzymes, including superoxide dismutase (MnSOD), arginase, glutamine synthetase and pyruvate carboxylase could contribute to the oxidative stress, altered neuronal energetics, and excitotoxicity pathological phenotypes reported in HD (14). Further, differential manganese nutritional status of patients could be an important modifier of disease progression. Manganese deficiency is known to alter glutamate cycling, the urea cycle, insulin signaling and cellular energetics; all of these biochemical pathways are dependent on manganese-dependent enzymes and have been shown to be dysfunctional in Huntington's disease models (34,78–80). In conclusion, these findings suggest the possibility that alterations in brain manganese homeostasis may contribute to HD pathophysiology, which may open new therapeutic opportunities.
Materials and Methods
Reagents and plasmids
The immortalized murine striatal cell line models of HD, both wild-type (STHdhQ7/Q7) and mutant (STHdhQ111/Q111), were obtained from Coriell Cell Repository (Cambden, NJ). The ATM kinase inhibitor, KU55933, was from Tocris (Bristol, UK), hydrogen peroxide was from Sigma-Aldrich (St. Louis, MO), neocarzinostatin was from Sigma-Aldrich. Heavy metal cations used in this study were Cd (Cd2+ as CdCl2 hydrate), Cu (Cu2+ as CuSO4 pentahydrate), Mn (Mn2+ as MnCl2 tetrahydrate), Ni (Ni2+ as NiCl2 hexahydrate), Co (Co2+ as CoCl2 hexahydrate) and Zn (Zn2+ as ZnCl2). Plasmids used for reprogramming (#s 27077, 27078 and 27080) were obtained from Addgene (Cambridge, MA).
Antibodies
Antibodies used for immunofluorescence, immunoblotting and the In-cell western assay are listed in Table 1.
Table 1.
Antibodies
| Antigen | Species | Company | Catalog # | Model system | WB | ICW | IF |
|---|---|---|---|---|---|---|---|
| phos-ATM(Ser1981) | Mo | Cell Signaling | 4526 | Hu, Mo | 1:1000 | ||
| ATM | Rb | Cell Signaling | 2873 | Hu, Mo | 1:1000 | ||
| Caspase-3 | Rb | Cell Signaling | 9665 | Mo | 1:1000 | ||
| Huntingtin | Mo | Millipore | MAB2166 | Mo | 1:200 | ||
| Islet 1 & 2 | Mo | DSHB | 39.4D5 | Hu | 1:100 | ||
| Nanog | Gt | R&D Systems | AF1997 | Hu | 1:20 | ||
| Oct3/4 | Rb | Cell Signaling | 2750 | Hu | 1:200 | ||
| PAX6 | Rb | Covance (Biolegend) | PRB-278P | Hu | 1:100 | ||
| phospho-p53(Ser15) | Mo | Cell Signaling | 9286 | Hu, Mo | 1:1000 | 1:400 | |
| phospho-p53(Ser15) | Rb | Cell Signaling | 9284 | Hu, Mo | 1:1000 | 1:400 | |
| p53 | Mo | Santa Cruz | sc-126 | Hu | 1:1000 | ||
| p53 | Rb | Santa Cruz | sc-6243 | Mo | 1:1000 | ||
| phospho-H2A.X | Mo | EMD Millipore | 05-636 | Hu, Mo | 1:1000 | 1:400 | |
| phospho-CHK2(Thr68) | Rb | Cell Signaling | 2661 | Hu | 1:400 | ||
| SSEA3 | Rt | Millipore | MAB4303 | Hu | 1:500 | ||
| SSEA4 | Mo | DSHB | MC-813-70 | 1:500 | |||
| Anti Mouse IRDYE 800CW | Dk | LI-COR | 926–32 212 | 1:10 000 | 1:800 | ||
| Anti Rabbit IRDYE 800CW | Dk | LI-COR | 926–32 213 | 1:800 | |||
| Anti Rabbit IRDye 680LT | Dk | LI-COR | 926–68 023 | 1:10 000 | |||
| Anti (mo, rb, gt or rt) Dylight 488 or 549 | Dk | Jackson | 1:800 |
Antibodies used throughout this study are listed in the table with information about the species, vendor and model system used in as well as the dilution factor used for each assay type.
Generation of human iPSC lines
Human dermal fibroblasts were obtained from either Dr Kevin Ess (CE and CF) or from Coriell Cell Repository (GM21756 and GM09197 which are HD70 and HD180, respectively). Primary dermal fibroblasts were obtained by skin biopsy from healthy adult subjects (CE and CF) with no known family history of neurodegenerative disease after appropriate patient consent/assent under the guidelines of an approved IRB protocol (Vanderbilt #080369). Cells were grown to ∼80% confluency, trypsinized and counted 6 × 105 cells were then electroporated with the CXLE plasmid vectors using the Neon Transfection System (Life Technologies, Carlsbad, CA) according to the conditions described in Okita et al. 2011. Cells were replated in normal 10% FBS medium (formulation described in Cell Culture) and grown with daily media changes for 7 days. The cells were then trypsinized and replated at 1 × 105 cells per 100 mm dish on top of SNL feeder cells. The next day, the medium was changed to KOSR ES medium (described in 52) and replaced daily for ∼30 days before colonies were picked and propagated.
Karyotype analysis
Human iPSC lines were submitted for standard g-band karyotype analysis by Genetics Associates, Inc., (Nashville, TN). For each line, 20 metaphase spreads were analyzed and normal lines used in this study has euploid in 20 of 20 cells.
Pluritest
All iPSC lines used in this study were validated using the bioinformatics assay, ‘Pluritest’, developed by Muller et al. (81). Human iPSC mRNA was isolated using the Qiagen RNeasy kit according to the manufacturer's recommendations (Qiagen, Valencia, CA). Microarrays were performed by Expression Analysis, Inc., (Durham, NC) on Illumina Human HT-12 v4.0 expression BeadChips (Illumina, Inc., San Diego, CA). Pluripotency of the iPSC lines was determined by uploading binary microarray scanner output files (.idat file) into the freely available Pluritest Internet application (http://www.pluritest.org).
Cell culture
STHdhQ7/Q7 and STHdhQ111/Q111 immortalized murine striatal cells and human dermal fibroblasts were cultured in Dulbecco's Modified Eagle Medium (D6546, Sigma-Aldrich) supplemented with 10% FBS (Atlanta Biologicals, Flowery Branch, GA), 2 mm GlutaMAX (Life Technologies), Penicillin–Streptomycin, 0.5 mg/ml G418 Sulfate (Life Technologies), MEM non-essential amino acids solution (Life Technologies), 14 mm HEPES (Life Technologies). They were incubated at 33°C and 5% CO2. Cells were passaged before reaching >90% confluency. The cells were split by trypsinization using 0.05% Trypsin–EDTA solution (Life Technologies) incubated for 5 min. One day prior to exposure, cells were plated in the appropriate cell culture plate type at 8 × 105 cells/ml for Q7 and 1 × 106 cells/ml for Q111.
Human iPSC lines were maintained in mTeSR1 medium (StemCell technologies, Vancouver, BC) on Matrigel (BD Biosciences, San Jose, CA) coated 6-well plates. Media was replaced daily. Cells were passaged at ∼50% confluency. Before passage, spontaneously differentiated cells were manually removed using a glass scraper (52). Then colonies were washed once in DMEM/F12 (Life Technologies) and incubated for 5–10 min with dispase (StemCell Technologies). The colonies were then washed twice with DMEM/F12. The colonies were then scraped into mTeSR1, triturated several times to achieve smaller cell clump size and replated at a dilution of 1:3–1:5 onto Matrigel.
Neural progenitor differentiation
Human iPSC colonies grown on Matrigel in mTeSR1 were dissociated by incubating for 15 min in Accutase (Innovative Cell Technologies, San Diego, CA). Cells were diluted, centrifuged and resuspended in mTeSR1 medium containing 10 µm ROCK inhibitor, Y-27632 (Tocris). Cells were then replated onto Matrigel-coated 12-well plates at a concentration of 1 × 105 cells/ml. The medium was changed 24 h later to mTeSr1 without ROCK inhibitor and the cells were fed each subsequent day until cells reach >90% confluency (typically 4 days later). Medium was then replaced with Knockout DMEM/F12 (Life Technologies) supplemented with 20% Knockout Serum Replacement (Life Tech), 2 mm Glutamax (Life Tech), MEM non-essential amino acids (Life Tech) and 55 µm 2-mercaptoethanol) containing 0.5 µm DMH1 (gift of Dr Charles C. Hong) and 10 µm SB431542 (Tocris). After 72 h, the medium was supplemented with 25% N2 medium (DMEM/F12 [with l-glutamine and HEPES] (Life Tech), 1× N2 supplement (Life Tech) and 4.5 g/l d-glucose) and 0.65 µm purmorphamine (Stemgent, Cambridge, MA) and SB431542 was no longer added. After an additional 48 h, the N2 medium content was increased to 50%, and DMH1 was no longer added. After another 48 h, an additional 25% N2 medium was added (25% neuralization medium, 75% N2 medium). Cultures were dissociated 10 days after the initial addition of neuralization medium via a 20 min incubation with Accutase. For all experiments using human iPSC-derived neural progenitors, cells were replated onto Matrigel-coated dishes at 7 × 105cells/ml in 100% N2 medium containing 0.65 µm purmorphamine and 10 µm ROCK inhibitor, Y-27632. The next day the experimental exposure was added in 100% N2 medium containing purmorphamine without ROCK inhibitor. Assays were performed 24 h later.
CellTiter-Blue cytotoxicity assay
Human neural progenitors or mouse striatal cells (STHdhQ7/Q7, STHdhQ111/Q111) were grown on 96-well plates. The day after replating, they were exposed to toxicants in the cell type appropriate neuralization medium. At 2 h prior to the completion of the 24 h exposure period or after toxicant removal and washes following 1 h exposures, 20 µl of CellTiter-Blue reagent (Promega) was added to each well. Prior to this addition, cell lysis buffer was added to several wells to provide an accurate fluorescence background for 0% viable cells. The plates were then incubated for 2 h at 37°C. Fluorescence was measured using excitation of 570 nm and emission of 600 nm on a POLARstar Omega microplate reader (BMG Labtech, Ortenberg, Germany).
Cellular Fura-2 manganese extraction assay (CFMEA)
CFMEA was performed exactly as described previously (67). Matrigel-coated wells show a manganese concentration-dependent background that is not observed in uncoated wells. A manganese concentration-specific background was, therefore, subtracted from the experimental signals. Manganese content was normalized to DNA content of each culture.
PCR
PCR was performed for both validation of the loss of the episomal plasmid vector and for validating the HD mutation in the iPSC lines. DNA was isolated using the DNeasy kit (Qiagen) and quantified using a Nanodrop 1000 spectrophotometer (Nanodrop Instruments). Primers and template DNA were mixed with GoTaq Green 2× Master Mix (Promega). The PCR amplification was performed on a MyCycler Thermal Cycler (Bio-Rad, Hercules, CA). For amplification of the mutant Huntingtin gene CAG repeat region, 1× Q buffer (Qiagen) and 5% DMSO were added. DNA products were run on 1 or 2% agarose gels in TBE buffer and stained with ethidium bromide.
Quantitative RT-PCR
Cells were lysed using RLT buffer with 1% BME and homogenized on QIAshredder columns (Qiagen). Total RNA was isolated using the RNeasy kit with on column RNase-free DNase treatment (Qiagen). Total RNA was quantified using a Nanodrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA). First strand cDNA synthesis reactions were performed on 1 µg of total RNA using the SuperScript III kit with random hexamers (Life Technologies). Quantitative PCR was performed using the Power SYBR Green Master Mix (Life Technologies) as 10 µl reactions in 384-well plates with 10 ng of cDNA on a 7900HT fast real-time PCR system (Applied Biosystems, Carlsbad, CA). Primers are listed in Table 2. Actin was used as a normalizing control gene.
Table 2.
Primer sequences
| Primer name | Gene | Sequence | Tm | Product size |
|---|---|---|---|---|
| AT83 | FOXG1 F | CAACGGCATCTACGAGTTCA | 59.9 | 57 |
| AT84 | FOXG1 R | TGCTTGTTCTCGCGGTAGTA | 59.7 | |
| AT95 | PAX6 F | CAGCTTCACCATGGCAAATA | 59.7 | 77 |
| AT96 | PAX6 R | GCAGCATGCAGGAGTATGAG | 59.6 | |
| AT103 | Actin F | CTGTGGCATCCACGAAACTA | 59.7 | 84 |
| AT104 | Actin R | AGCACTGTGTTGGCGTACAG | 60.0 | |
| AT277 | Islet-1 F | ACGGTGGCTTACAGGCTAAC | 59.3 | 56 |
| AT278 | Islet-1 R | TTTCCAAGGTGGCTGGTAAC | 60.0 | |
| 1594 | HTT CAG Repeat F | CCGCTCAGGTTCTGCTTTTA | 60.5 | 278 |
| 1595 | HTT CAG Repeat R | GGCTGAGGAAGCTGAGGAG | 60.2 | (19 CAG repeats) |
| Insertion1 | WPRE F | CAGGCAACGTGGCGTGGTGT | 70.0 | 265 |
| Insertion2 | WPRE R | GGACGTCCCGCGCAGAATCC | 71.5 | |
| AT335 | Mouse Actin F | CAGCCTTCCTTCTTGGGTAT | 58.3 | 83 |
| AT336 | Mouse Actin R | CGGATGTCAACGTCACACTT | 59.6 | |
| AT327 | Mouse p21 F | GAGGCCCAGTACTTCCTCTG | 58.9 | 78 |
| AT328 | Mouse p21 R | AGAGTGCAAGACAGCGACAA | 59.8 |
Primers used for PCR and qRT-PCR are listed with both the ‘in-house’ primer name, gene name, sequence, melting temperature and the product size.
Pathscan ELISA array
The assay was performed following the manufacturer’s instructions. In brief, cells were washed once with ice-cold PBS and lysed with ice-cold sandwich ELISA array lysis buffer (Cell Signaling, Danvers, MA) containing 1 mm PMSF for 5 min on ice. The lysates were then pipetted into pre-chilled tubes. Lysate protein concentration was calculated using the DC protein assay (Promega). After diluting to equivalent concentrations, lysates were added to the prepared PathScan Intracellular signaling array (Cell Signaling, Danvers, MA). The fluorescent signal was assessed using the Odyssey Infrared Imaging system (LI-COR, Lincoln, NE). Two wells were incubated in dilution buffer only and provided the background signal for each pathway. Only signal intensities ≥4-fold the background signal are reported, except for cleavage of caspase-3 and PARP-1. Since human data was generated on a single slide, the data was divided by the mean of the untreated sample intensities to achieve a fold change value. For the mouse PathScan, each of the three data points for each signaling pathway were generated on a separate slide on separate days. Therefore, the values for each day were divided by the untreated sample intensity for that day. Therefore, the untreated means have no variance since all values are 1. 95% confidence intervals were used to calculate statistical significance. Human cell analysis was performed by t-test.
Immunoblot analysis
Protein samples were prepared by scraping cells into ice-cold PBS, centrifuging and adding RIPA buffer containing protease (Sigma-Aldrich) and phosphatase inhibitor cocktails 2 and 3 (Sigma, Sigma-Aldrich) to the pellet. After gentle homogenization, cells were centrifuged at 4°C for 10 min at 20 000g. The resulting DNA containing pellet was removed from the lysate, and the protein concentration was quantified using the DC assay (Bio-Rad, Hercules, CA) with a BSA standard curve. Samples were mixed with 5× SDS loading buffer containing 1% 2-mercaptoethanol and boiled for 5 min. Fifteen micrograms of protein was loaded for each sample onto a 4–15% pre-cast gel SDS-PAGE gel (BioRad) and run at 90 V for 120 min. The protein bands were then transferred onto nitrocellulose membranes using iBlot Gel Transfer Device (Life Technologies), and the protein was transferred onto a nitrocellulose membrane. The remaining gel was stained with IRDye Blue protein stain (LI-COR, Lincoln, NE). Since the gels retained ∼1/3 of the original protein after transferring with the iBlot, we imaged the stained gel on the Li-Cor Odyssey Imaging System and quantified the intensity of the entire lane from ∼150–20 kDa. This value was used to normalize the values of immunostained bands. The membrane was blocked in Odyssey Blocking Buffer for hour prior to the addition of the primary antibodies. The primary antibodies were diluted in Odyssey Blocking Buffer containing 0.1% TWEEN and incubated overnight. After washing 5 times for 5 min in TBST, membranes were incubated with secondary antibodies at 1:10 000 (LiCor, Lincoln, NE) for 1 h. Membranes were imaged using the Li-Cor Odyssey Imaging System, and quantification was performed using Image Studio Lite (LiCOR, Lincoln, NE).
In-Cell western assay
Cells were plated in 96-well µClear black-walled plates (Greiner Bio-One, Frickenhausen, Germany) at the appropriate density for the particular line. After exposing cells to toxicants, they were washed once in room temperature PBS (without calcium and magnesium). The cells were then fixed with 4% paraformaldehyde in PBS for 30 min at room temperature, washed 5 times for in PBS with 0.1% Triton-X 100, and blocked for 1.5 h in 150 µl of Odyssey blocking buffer. The cells were then incubated with primary antibody at 1:400 in Odyssey blocking buffer (LiCor, Lincoln, NE) with 50 µl in each well for 2.5 h. After washing four times in PBS with 0.1% Tween-20 for 5 min, cells were incubated for 1 h in the appropriate LiCor IRdye800 secondary antibody at 1:800 dilution in Odyssey blocking buffer along with 1:500 of CellTag normalization dye (LI-COR, Lincoln, NE). An additional round of 5 washes for 5 min in PBS with Tween-20 was performed after which all buffer was removed. Plates were imaged with the Li-Cor Odyssey Imaging System and intensities were calculated for each well with Image Studio software. Cultures that were not incubated with primary antibodies served as background. Antibody signals were normalized using the CellTag signal (a measure of total cells). We have included exemplary images from both human (Supplementary Material, Fig. S9) and mouse (Supplementary Material, Fig. S10) In-Cell western assays.
Immunofluorescence microscopy
Cells were fixed in paraformaldehyde for 30 min at room temperature. After permeabilization with 0.2% Triton-X 100 for 20 min at room temperature, cells were incubated in PBS containing 5% donkey serum and 0.05% Triton-X100 for 2 h at room temperature or overnight. Cell were incubated in primary antibody overnight (see table to antibody dilutions), washed in PBS with 0.05% Triton-X100 and incubated for 3 h with secondary antibody. Images were obtained with a Zeiss ObserverZ1 microscope and AxioVs40 software (version 4.7.2).
Supplementary Material
Funding
This work was supported by the National Institutes of Health (ES016931 to A.B., ES010563 to A.B./M.A., NS077632 to A.T. and GM07347 to K.K.).
Supplementary Material
Acknowledgements
We thank Bryan Cawthon and Bingying Han for extensive technical assistance in generation and propagation of the iPSC lines. We also thank Gary Li, Mihir Odak, Megan Culbreth, Molly Overmyer and Kyle Horning for additional technical assistance; Terry Jo Bichell and Dr Christopher Wright for helpful discussions; Dr Charles Hong for providing helpful reagents; and Drs Gunnar Kwakye and Blairanne Williams for their previously published work that established the basis for this study.
Conflict of Interest statement. The authors of the manuscript declare no competing financial interests related to this paper.
References
- 1.Zuccato C., Valenza M., Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 2.Wexler N.S. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gusella J.F., MacDonald M.E. Huntington’s disease: the case for genetic modifiers. Genome Med. 2009;1:80. doi: 10.1186/gm80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgiou N., Bradshaw J.L., Chiu E., Tudor A., O'Gorman L., Phillips J.G. Differential clinical and motor control function in a pair of monozygotic twins with Huntington’s disease. Mov. Disord. 1999;14:320–325. doi: 10.1002/1531-8257(199903)14:2<320::aid-mds1018>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 5.Anca M., Gazit E., Loewenthal R., Ostrovsky O., Frydman M., Giladi N. Different phenotypic expression in monozygotic twins with Huntington disease. Am. J. Med. Genet. A. 2004;124:89–91. doi: 10.1002/ajmg.a.20328. [DOI] [PubMed] [Google Scholar]
- 6.Friedman J.H., Trieschmann M.E., Myers R.H., Fernandez H.H. Monozygotic twins discordant for Huntington disease after 7 years. Arch. Neurol. 2005;62:995–997. doi: 10.1001/archneur.62.6.995. [DOI] [PubMed] [Google Scholar]
- 7.Glass M., Van Dellen A., Blakemore C., Hannan A., Faull R. Delayed onset of huntington’s disease in mice in an enriched environment correlates with delayed loss of cannabinoid CB1 receptors. Neuroscience. 2004;123:207–212. doi: 10.1016/s0306-4522(03)00595-5. [DOI] [PubMed] [Google Scholar]
- 8.Fox J.H., Kama J.A., Lieberman G., Chopra R., Dorsey K., Chopra V., Volitakis I., Cherny R.A., Bush A.I., Hersch S. Mechanisms of copper ion mediated Huntington's disease progression. PLoS One. 2007;2:e334. doi: 10.1371/journal.pone.0000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao G., Fan Q., Wang X., Zhou B. Huntington disease arises from a combinatory toxicity of polyglutamine and copper binding. Proc. Natl. Acad. Sci. USA. 2013;110:14995–15000. doi: 10.1073/pnas.1308535110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wild E.J., Tabrizi S.J. Huntington's disease phenocopy syndromes. Curr. Opin. Neurol. 2007;20:681–687. doi: 10.1097/WCO.0b013e3282f12074. [DOI] [PubMed] [Google Scholar]
- 11.Williams B.B., Li D., Wegrzynowicz M., Vadodaria B.K., Anderson J.G., Kwakye G.F., Aschner M., Erikson K.M., Bowman A.B. Disease-toxicant screen reveals a neuroprotective interaction between Huntington's disease and manganese exposure. J. Neurochem. 2010;112:227–237. doi: 10.1111/j.1471-4159.2009.06445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V.C., Sharp A.H., Persichetti F., Cattaneo E., MacDonald M.E. Dominant phenotypes produced by the HD mutation in STHdhQ111 striatal cells. Hum. Mol. Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 13.Aschner J.L., Aschner M. Nutritional aspects of manganese homeostasis. Mol. Aspects Med. 2005;26:353–362. doi: 10.1016/j.mam.2005.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowman A.B., Kwakye G.F., Herrero Hernández E., Aschner M. Role of manganese in neurodegenerative diseases. J. Trace Elem. Med. Biol. 2011;25:191–203. doi: 10.1016/j.jtemb.2011.08.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S., Fu J., Zhou Z. In vitro effect of manganese chloride exposure on reactive oxygen species generation and respiratory chain complexes activities of mitochondria isolated from rat brain. Toxicol. In Vitro. 2004;18:71–77. doi: 10.1016/j.tiv.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Santamaria A. Manganese exposure, essentiality & toxicity. Indian J. Med. Res. 2008;128:484. [PubMed] [Google Scholar]
- 17.Larsen N.A., Pakkenberg H., Damsgaard E., Heydorn K. Topographical distribution of arsenic, manganese, and selenium in the normal human brain. J. Neurol. Sci. 1979;42:407–416. doi: 10.1016/0022-510x(79)90173-4. [DOI] [PubMed] [Google Scholar]
- 18.Prohaska J.R. Functions of trace elements in brain metabolism. Physiol. Rev. 1987;67:858–901. doi: 10.1152/physrev.1987.67.3.858. [DOI] [PubMed] [Google Scholar]
- 19.Madison J.L., Wegrzynowicz M., Aschner M., Bowman A.B. Disease-toxicant interactions in manganese exposed Huntington disease mice: early changes in striatal neuron morphology and dopamine metabolism. PLoS One. 2012;7:e31024. doi: 10.1371/journal.pone.0031024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosas H.D., Chen Y.I., Doros G., Salat D.H., Chen N.-K., Kwong K.K., Bush A., Fox J., Hersch S.M. Alterations in brain transition metals in Huntington disease: an evolving and intricate story. Arch. Neurol. 2012;69:887–893. doi: 10.1001/archneurol.2011.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K.-I. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 22.Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma L., Hu B., Liu Y., Vermilyea S.C., Liu H., Gao L., Sun Y., Zhang X., Zhang S.-C. Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell. 2012;10:455–464. doi: 10.1016/j.stem.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carri A.D., Onorati M., Lelos M.J., Castiglioni V., Faedo A., Menon R., Camnasio S., Vuono R., Spaiardi P., Talpo F. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 2013;140:301–312. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 25.Guo X., Disatnik M.-H., Monbureau M., Shamloo M., Mochly-Rosen D., Qi X. Inhibition of mitochondrial fragmentation diminishes Huntington's disease–associated neurodegeneration. J. Clin. Invest. 2013;123:5371–5388. doi: 10.1172/JCI70911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang N., An M.C., Montoro D., Ellerby L.M. Characterization of human Huntington's disease cell model from induced pluripotent stem cells. PLoS Curr. 2010;2 doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camnasio S., Carri A.D., Lombardo A., Grad I., Mariotti C., Castucci A., Rozell B., Riso P.L., Castiglioni V., Zuccato C. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington's disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol. Dis. 2012;46:41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 28.Consortium H.I. Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juopperi T.A., Kim W.R., Chiang C.-H., Yu H., Margolis R.L., Ross C.A., Ming G., Song H. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington's disease patient cells. Mol. Brain. 2012;5:17. doi: 10.1186/1756-6606-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castiglioni V., Onorati M., Rochon C., Cattaneo E. Induced pluripotent stem cell lines from Huntington's disease mice undergo neuronal differentiation while showing alterations in the lysosomal pathway. Neurobiol. Dis. 2012;46:30–40. doi: 10.1016/j.nbd.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 31.Gines S., Seong I.S., Fossale E., Ivanova E., Trettel F., Gusella J.F., Wheeler V.C., Persichetti F., MacDonald M.E. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington's disease knock-in mice. Hum. Mol. Genet. 2003;12:497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 32.Weydt P., Pineda V.V., Torrence A.E., Libby R.T., Satterfield T.F., Lazarowski E.R., Gilbert M.L., Morton G.J., Bammler T.K., Strand A.D. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1α in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Mochel F., Durant B., Meng X., O'Callaghan J., Yu H., Brouillet E., Wheeler V.C., Humbert S., Schiffmann R., Durr A. Early alterations of brain cellular energy homeostasis in Huntington disease models. J. Biol. Chem. 2012;287:1361–1370. doi: 10.1074/jbc.M111.309849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seong I.S., Ivanova E., Lee J.-M., Choo Y.S., Fossale E., Anderson M., Gusella J.F., Laramie J.M., Myers R.H., Lesort M. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 35.Nagata E., Sawa A., Ross C.A., Snyder S.H. Autophagosome-like vacuole formation in Huntington’s disease lymphoblasts. Neuroreport. 2004;15:1325–1328. doi: 10.1097/01.wnr.0000127073.66692.8f. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., de Vries R., Arias E., Harris S., Sulzer D. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordova F.M., Aguiar A.S., Jr, Peres T.V., Lopes M.W., Gonçalves F.M., Remor A.P., Lopes S.C., Pilati C., Latini A.S., Prediger R.D. In vivo manganese exposure modulates Erk, Akt and Darpp-32 in the striatum of developing rats, and impairs their motor function. PLoS One. 2012;7:e33057. doi: 10.1371/journal.pone.0033057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bae J.-H., Jang B.-C., Suh S.-I., Ha E., Baik H.H., Kim S.-S., Lee M.-Y., Shin D.-H. Manganese induces inducible nitric oxide synthase (iNOS) expression via activation of both MAP kinase and PI3 K/Akt pathways in BV2 microglial cells. Neurosci. Lett. 2006;398:151–154. doi: 10.1016/j.neulet.2005.12.067. [DOI] [PubMed] [Google Scholar]
- 39.Guilarte T.R., Burton N.C., Verina T., Prabhu V.V., Becker K.G., Syversen T., Schneider J.S. Increased APLP1 expression and neurodegeneration in the frontal cortex of manganese-exposed non-human primates. J. Neurochem. 2008;105:1948–1959. doi: 10.1111/j.1471-4159.2008.05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao F., Zhang J.-B., Cai T.-J., Liu X.-Q., Liu M.-C., Ke T., Chen J.-Y., Luo W.-J. Manganese induces p21 expression in PC12 cells at the transcriptional level. Neuroscience. 2012;215:184–195. doi: 10.1016/j.neuroscience.2012.04.027. [DOI] [PubMed] [Google Scholar]
- 41.Colin E., Régulier E., Perrin V., Dürr A., Brice A., Aebischer P., Déglon N., Humbert S., Saudou F. Akt is altered in an animal model of Huntington's disease and in patients. Eur. J. Neurosci. 2005;21:1478–1488. doi: 10.1111/j.1460-9568.2005.03985.x. [DOI] [PubMed] [Google Scholar]
- 42.Humbert S., Bryson E.A., Cordelières F.P., Connors N.C., Datta S.R., Finkbeiner S., Greenberg M.E., Saudou F. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev. Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- 43.Bae B.-I., Xu H., Igarashi S., Fujimuro M., Agrawal N., Taya Y., Hayward S.D., Moran T.H., Montell C., Ross C.A. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Steffan J.S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y.-Z., Gohler H., Wanker E.E., Bates G.P., Housman D.E., Thompson L.M. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Apostol B.L., Illes K., Pallos J., Bodai L., Wu J., Strand A., Schweitzer E.S., Olson J.M., Kazantsev A., Marsh J.L. Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum. Mol. Genet. 2006;15:273–285. doi: 10.1093/hmg/ddi443. [DOI] [PubMed] [Google Scholar]
- 46.Fan J., Gladding C.M., Wang L., Zhang L.Y., Kaufman A.M., Milnerwood A.J., Raymond L.A. P38 MAPK is involved in enhanced NMDA receptor-dependent excitotoxicity in YAC transgenic mouse model of Huntington disease. Neurobiol. Dis. 2012;45:999–1009. doi: 10.1016/j.nbd.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 47.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 48.Ju T.-C., Chen H.-M., Lin J.-T., Chang C.-P., Chang W.-C., Kang J.-J., Sun C.-P., Tao M.-H., Tu P.-H., Chang C. Nuclear translocation of AMPK-α1 potentiates striatal neurodegeneration in Huntington's disease. J. Cell Biol. 2011;194:209–227. doi: 10.1083/jcb.201105010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carmichael J., Sugars K.L., Bao Y.P., Rubinsztein D.C. Glycogen synthase kinase-3β inhibitors prevent cellular polyglutamine toxicity caused by the Huntington,s disease mutation. J. Biol. Chem. 2002;277:33791–33798. doi: 10.1074/jbc.M204861200. [DOI] [PubMed] [Google Scholar]
- 50.Ku S., Soragni E., Campau E., Thomas E.A., Altun G., Laurent L.C., Loring J.F., Napierala M., Gottesfeld J.M. Friedreich’s ataxia induced pluripotent stem cells model intergenerational GAA⋅ TTC triplet repeat instability. Cell Stem Cell. 2010;7:631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manley K., Shirley T.L., Flaherty L., Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 52.Neely M.D., Litt M.J., Tidball A.M., Li G.G., Aboud A.A., Hopkins C.R., Chamberlin R., Hong C.C., Ess K.C., Bowman A.B. DMH1, a highly selective small molecule BMP inhibitor promotes neurogenesis of hiPSCs: comparison of PAX6 and Sox1 expression during neural induction. ACS Chem. Neurosci. 2012;3:482–491. doi: 10.1021/cn300029t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X., Huang C.T., Chen J., Pankratz M.T., Xi J., Li J., Yang Y., LaVaute T.M., Li X.-J., Ayala M. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H.-F., Liu F.-C. Developmental restriction of the LIM homeodomain transcription factor Islet-1 expression to cholinergic neurons in the rat striatum. Neuroscience. 2001;103:999–1016. doi: 10.1016/s0306-4522(00)00590-x. [DOI] [PubMed] [Google Scholar]
- 55.López-Bendito G., Cautinat A., Sánchez J.A., Bielle F., Flames N., Garratt A.N., Talmage D.A., Role L.W., Charnay P., Marín O. Tangential neuronal migration controls axon guidance: a role for neuregulin-1 in thalamocortical axon navigation. Cell. 2006;125:127–142. doi: 10.1016/j.cell.2006.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowman A.B., Aschner M. Considerations on manganese (Mn) treatments for in vitro studies. Neurotoxicology. 2014;41:141–142. doi: 10.1016/j.neuro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams B.B., Kwakye G.F., Wegrzynowicz M., Li D., Aschner M., Erikson K.M., Bowman A.B. Altered manganese homeostasis and manganese toxicity in a Huntington's disease striatal cell model are not explained by defects in the iron transport system. Toxicol. Sci. 2010;117:169–179. doi: 10.1093/toxsci/kfq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Banin S., Moyal L., Shieh S.-Y., Taya Y., Anderson C., Chessa L., Smorodinsky N., Prives C., Reiss Y., Shiloh Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 59.Canman C.E., Lim D.-S., Cimprich K.A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M.B., Siliciano J.D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 60.Jones R.G., Plas D.R., Kubek S., Buzzai M., Mu J., Xu Y., Birnbaum M.J., Thompson C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 61.She Q.-B., Chen N., Dong Z. ERKs and p38 kinase phosphorylate p53 protein at serine 15 in response to UV radiation. J. Biol. Chem. 2000;275:20444–20449. doi: 10.1074/jbc.M001020200. [DOI] [PubMed] [Google Scholar]
- 62.Chan D.W., Son S.-C., Block W., Ye R., Khanna K.K., Wold M.S., Douglas P., Goodarzi A.A., Pelley J., Taya Y. Purification and Characterization of ATM from Human Placenta a Manganese-Dependent, Wortmannin-Sensitive Serine/Threonine Protein Kinase. J. Biol. Chem. 2000;275:7803–7810. doi: 10.1074/jbc.275.11.7803. [DOI] [PubMed] [Google Scholar]
- 63.Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 64.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 65.Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 66.Kwakye G.F., Li D., Bowman A.B. Novel high-throughput assay to assess cellular manganese levels in a striatal cell line model of Huntington's disease confirms a deficit in manganese accumulation. Neurotoxicology. 2011;32:630–639. doi: 10.1016/j.neuro.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar K.K., Aboud A.A., Patel D.K., Aschner M., Bowman A.B. Optimization of Fluorescence Assay of cellular manganese status for high throughput screening. J. Biochem. Mol. Toxicol. 2013;27:42–49. doi: 10.1002/jbt.21457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waghorn B., Yang Y., Baba A., Matsuda T., Schumacher A., Yanasak N., Hu T.C.C. Assessing manganese efflux using SEA0400 and cardiac T1-mapping manganese-enhanced MRI in a murine model. NMR Biomed. 2009;22:874–881. doi: 10.1002/nbm.1414. [DOI] [PubMed] [Google Scholar]
- 69.Dally H., Hartwig A. Induction and repair inhibition of oxidative DNA damage by nickel (II) and cadmium (II) in mammalian cells. Carcinogenesis. 1997;18:1021–1026. doi: 10.1093/carcin/18.5.1021. [DOI] [PubMed] [Google Scholar]
- 70.Kumar K.K., Lowe J.E.W., Aboud A.A., Neely M.D., Redha R., Bauer J.A., Odak M., Weaver C.D., Meiler J., Aschner M., et al. Cellular manganese content is developmentally regulated in human dopaminergic neurons. Sci. Rep. 2014;4:1–8. doi: 10.1038/srep06801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferlazzo M.L., Sonzogni L., Granzotto A., Bodgi L., Lartin O., Devic C., Vogin G., Pereira S., Foray N. Mutations of the Huntington's Disease Protein Impact on the ATM-Dependent Signaling and Repair Pathways of the Radiation-Induced DNA Double-Strand Breaks: Corrective Effect of Statins and Bisphosphonates. Mol. Neurobiol. 2013;49:1–12. doi: 10.1007/s12035-013-8591-7. [DOI] [PubMed] [Google Scholar]
- 72.Jung-Il C., Dong-Wook K., Nayeon L., Young-Joo J., Iksoo J., Jihye K., Jumi K., Yunjo S., Dong-Seok L., Kang S.S. Quantitative proteomic analysis of induced pluripotent stem cells derived from a human Huntington's disease patient. Biochem. J. 2012;446:359–371. doi: 10.1042/BJ20111495. [DOI] [PubMed] [Google Scholar]
- 73.Ehrnhoefer D.E., Skotte N.H., Ladha S., Nguyen Y.T., Qiu X., Deng Y., Huynh K.T., Engemann S., Nielsen S.M., Becanovic K. p53 increases caspase-6 expression and activation in muscle tissue expressing mutant huntingtin. Hum. Mol. Genet. 2014;23:717–729. doi: 10.1093/hmg/ddt458. [DOI] [PubMed] [Google Scholar]
- 74.Chattopadhyay B., Baksi K., Mukhopadhyay S., Bhattacharyya N.P. Modulation of age at onset of Huntington disease patients by variations in TP53 and human caspase activated DNase (hCAD) genes. Neurosci. Lett. 2005;374:81–86. doi: 10.1016/j.neulet.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 75.Helton E.S., Chen X. p53 modulation of the DNA damage response. J. Cell. Biochem. 2007;100:883–896. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- 76.Kalia K., Jiang W., Zheng W. Manganese accumulates primarily in nuclei of cultured brain cells. Neurotoxicology. 2008;29:466–470. doi: 10.1016/j.neuro.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morello M., Canini A., Mattioli P., Sorge R., Alimonti A., Bocca B., Forte G., Martorana A., Bernardi G., Sancesario G. Sub-cellular localization of manganese in the basal ganglia of normal and manganese-treated rats: an electron spectroscopy imaging and electron energy-loss spectroscopy study. Neurotoxicology. 2008;29:60–72. doi: 10.1016/j.neuro.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 78.Chiang M.-C., Chen H.-M., Lee Y.-H., Chang H.-H., Wu Y.-C., Soong B.-W., Chen C.-M., Wu Y.-R., Liu C.-S., Niu D.-M. Dysregulation of C/EBPŒ± by mutant Huntingtin causes the urea cycle deficiency in Huntington's disease. Hum. Mol. Genet. 2007;16:483–498. doi: 10.1093/hmg/ddl481. [DOI] [PubMed] [Google Scholar]
- 79.Lievens J.-C., Woodman B., Mahal A., Spasic-Boscovic O., Samuel D., Kerkerian-Le Goff L., Bates G. Impaired glutamate uptake in the R6 Huntington's disease transgenic mice. Neurobiol. Dis. 2001;8:807–821. doi: 10.1006/nbdi.2001.0430. [DOI] [PubMed] [Google Scholar]
- 80.Podolsky S., Leopold N., Sax D. Increased frequency of diabetes mellitus in patients with Huntington's chorea. Lancet. 1972;299:1356–1359. doi: 10.1016/s0140-6736(72)91092-6. [DOI] [PubMed] [Google Scholar]
- 81.Müller F.-J., Schuldt B.M., Williams R., Mason D., Altun G., Papapetrou E.P., Danner S., Goldmann J.E., Herbst A., Schmidt N.O. A bioinformatic assay for pluripotency in human cells. Nat. Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.