Abstract
The 2q37 and 17q12-q22 loci are linked to an increased prostate cancer (PrCa) risk. No candidate gene has been localized at 2q37 and the HOXB13 variant G84E only partially explains the linkage to 17q21-q22 observed in Finland. We screened these regions by targeted DNA sequencing to search for cancer-associated variants. Altogether, four novel susceptibility alleles were identified. Two ZNF652 (17q21.3) variants, rs116890317 and rs79670217, increased the risk of both sporadic and hereditary PrCa (rs116890317: OR = 3.3 – 7.8, P = 0.003 – 3.3 × 10−5; rs79670217: OR = 1.6 – 1.9, P = 0.002 – 0.009). The HDAC4 (2q37.2) variant rs73000144 (OR = 14.6, P = 0.018) and the EFCAB13 (17q21.3) variant rs118004742 (OR = 1.8, P = 0.048) were overrepresented in patients with familial PrCa. To map the variants within 2q37 and 17q11.2-q22 that may regulate PrCa-associated genes, we combined DNA sequencing results with transcriptome data obtained by RNA sequencing. This expression quantitative trait locus (eQTL) analysis identified 272 SNPs possibly regulating six genes that were differentially expressed between cases and controls. In a modified approach, pre-filtered PrCa-associated SNPs were exploited and interestingly, a novel eQTL targeting ZNF652 was identified. The novel variants identified in this study could be utilized for PrCa risk assessment, and they further validate the suggested role of ZNF652 as a PrCa candidate gene. The regulatory regions discovered by eQTL mapping increase our understanding of the relationship between regulation of gene expression and susceptibility to PrCa and provide a valuable starting point for future functional research.
Keywords: prostate cancer risk, genetic predisposition, susceptibility loci, 2q37, 17q11.2-q22
Introduction
A large proportion of familial prostate cancer (PrCa) cases can be explained by genetic risk factors.1 Despite extensive research, the identification of these factors has proven challenging. In Finland, mutations in hereditary prostate cancer (HPC) risk genes are relatively rare, with the exception of the HOXB13 G84E mutation,2 which is present in 8.4% of familial PrCa cases and has been significantly associated with an increased PrCa risk in unselected cases.3
The involvement of chromosomal regions 2q37 and 17q12-q22 with PrCa has been previously reported in numerous linkage4–6 and genome-wide association studies (GWAS).7, 8 Cropp et al.9 performed a genome-wide linkage scan of 69 Finnish high-risk HPC families and in the dominant model, the loci on 2q37.3 and 17q21-q22 exhibited the strongest linkage signals. No known PrCa candidate gene resides on 2q37.3, and as demonstrated in our earlier study, the HOXB13 G84E mutation only partially explains the observed linkage to 17q21-q22.3
Here, we performed targeted re-sequencing that covered the linkage peaks on 2q37 and 17q11.2-q22. The sequence data were filtered to identify the variants within genes predicted to be involved in PrCa predisposition. These variants were validated in Finnish HPC families and in unselected PrCa patients by Sequenom genotyping, and several novel variants were discovered that were significantly associated with PrCa. To study the impact of SNPs on the regulation of gene expression within the two linked regions, we performed transcriptome sequencing followed by expression quantitative trait loci (eQTL) mapping. eQTLs are known to modify the penetrance of rare deleterious variants and therefore likely contribute to genetic predisposition to complex diseases. New information was obtained on several genes as well as their regulatory elements that generated fresh insights into PrCa susceptibility, especially in HPC.
Materials and Methods
All of the subjects were of Finnish origin. The samples were collected with written and signed informed consent. The cancer diagnoses were confirmed using medical records and the annual update from the Finnish Cancer Registry. The project was approved by the local research ethics committee at Pirkanmaa Hospital District and by the National Supervisory Authority for Welfare and Health.
Targeted re-sequencing of 2q37 and 17q11.2-q22
Based on the linkage analysis results from Cropp et al.,9 63 PrCa patients and five unaffected individuals belonging to 21 Finnish high-risk HPC families10 were selected for targeted re-sequencing of the 2q37 and 17q11.2-q22 regions (Table S1). Each family had at least three first- or second-degree relatives diagnosed with PrCa. Paired-end next generation sequencing was performed at the Technology Centre, Institute for Molecular Medicine Finland (FIMM), University of Helsinki. The sequenced fragments spanned approximately 6.8 Mb for chromosome 2q and 21.6 Mb for 17q. The target regions were captured using SeqCap EZ Choice array probes (Roche NimbleGen, Inc., Madison, WI, USA) and were sequenced on a Genome Analyzer IIx (Illumina, Inc., San Diego, CA, USA) following the manufacturer’s protocol. The read alignment and variant calling were performed according to FIMM’s Variant-Calling Pipeline (VCP).11
Bioinformatics workflow for variant characterization
A schematic overview of our bioinformatics workflow is shown in Figure 1. Only those variants that were present in all the affected family members were selected for subsequent analysis. The variants were annotated using Ensembl V65 gene set retrieved from the UCSC Genome Browser.12 The phenotypic effects of the variants were studied with three in silico pathogenicity prediction programs. MutationTaster13 classifies single nucleotide variants (SNVs) and small insertion/deletion polymorphisms (indels) as polymorphic or pathogenic. PolyPhen-214 and PON-P15 only predict the effects of non-synonymous SNVs that result in amino acid replacement. PolyPhen-2 classifies the variants as benign, possibly pathogenic or probably pathogenic, whereas PON-P defines them as neutral, unclassified or pathogenic. Variants categorized as pathogenic by at least one tolerance predictor were defined as pathogenic. In addition, minor allele frequencies (MAF) were obtained from the dbSNP database and information on known PrCa-associated genes was retrieved from the COSMIC16 and DDPC17 databases. Pathway data were gathered from Pathway Commons,18 KEGG19 and WikiPathways20 and Gene Ontology data were retrieved from Ensembl BioMart v.65.21 Higher priority was assigned to rare variants (MAF <0.05), variants located in genes previously linked to PrCa, and variants located in genes functionally similar to PrCa-associated genes.
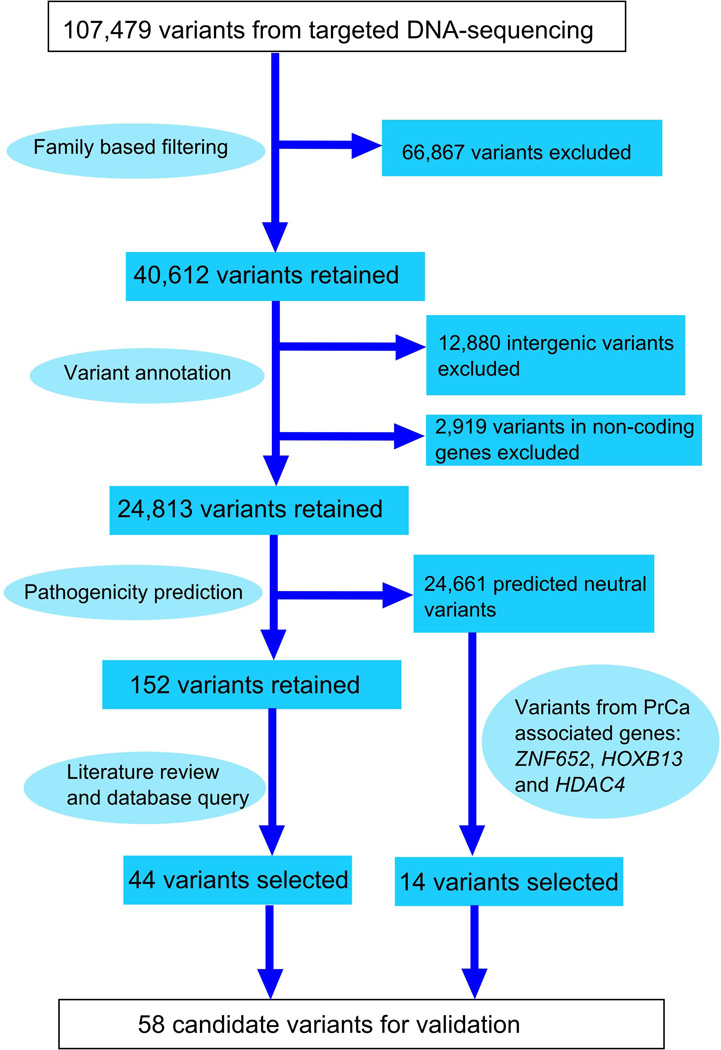
Figure 1. A flowchart describing the variant characterization pipeline.
The targeted re-sequencing of 2q37 and 17q11.2-q22 from 68 Finnish HPC family members produced a total of 107,479 unique sequence variants. Family-based filtering excluded 66,867 variants that did not co-segregate with affection status. Annotation enabled the selection of 24,813 variants that were located within protein-coding genes. Pathogenicity predictions were performed in silico using MutationTaster, PolyPhen-2 and PON-P. As a result, the number of candidate variants was reduced to 152. The final filtering step exploited diverse information on genes and variants as well as gene ontology and pathway data stored in several public databases. In addition, select HDAC4, ZNF652 and HOXB13 variants, which were predicted to be non-pathogenic, were included in the validation because these genes have been associated with PrCa in previous studies.
Validation of predicted PrCa-associated variants with Sequenom
After filtering, 58 variants in 35 target genes (listed in Tables S2–S4) were selected for validation which was performed on germline DNA from 2216 subjects, including 1293 cases and 923 population controls. The majority of the cases (1105 individuals) represented unselected PrCa patients from the Pirkanmaa Hospital District, Tampere, Finland. In addition, 188 index cases from Finnish HPC families10 were included in the study. The control DNA samples from anonymous male blood donors were provided by the Finnish Red Cross Blood Transfusion Service. Genotyping was performed at the Technology Centre, FIMM using the Sequenom MassARRAY system and iPLEX Gold assays (Sequenom, Inc., San Diego, CA, USA). Genotyping reactions were performed with 20 ng of dried genomic DNA according to manufacturer’s recommendations and with their reagents. The genotypes were called using TyperAnalyzer software (Sequenom). For quality control (QC) reasons, the genotype calls were also checked manually. Genotyping quality was examined using a detailed QC procedure that included success rate checks, duplicated samples and water controls.
Statistical and bioinformatic analyses of the validated variants
Association and Hardy-Weinberg Equilibrium (HWE) tests were performed using PLINK.22 The P value threshold for the HWE test was set to 0.05. Samples with low genotyping frequencies (<0.80) were excluded from the association analysis. The statistical significance of the association was evaluated using a two-sided Fisher’s exact test. Odds ratios (OR) were calculated using PLINK with option --fisher. No further model adjustments for confounding factors were made. ENCODE information23 for non-coding variants was retrieved from the Regulome database (RegulomeDB).24 The linkage disequilibrium (LD) analysis of the statistically significant variants is described in Supplementary Methods.
Genotyping of the top four candidate variants in Finnish HPC families
Four variants were chosen for segregation analysis in Finnish HPC families based on a strong association with PrCa, a high OR value and/or predicted pathogenicity. The co-segregation of rs116890317 and rs79670217 in ZNF652 (RefSeq NM_001145365), rs73000144 in HDAC4 (RefSeq NM_006037) and rs118004742 in EFCAB13 (RefSeq NM_152347) with affection status was determined in 41 families whose index cases were mutation-positive in the Sequenom validation. For these families, DNA samples were available from 243 PrCa cases and 204 healthy family members. The variants were genotyped in two to 17 (median: seven) individuals per family by Sanger sequencing.
RNA extraction and sequencing
Peripheral blood samples collected in PAXgene® Blood RNA Tubes (PreAnalytiX GmbH, Switzerland) were available from 84 PrCa patients and 15 healthy male relatives belonging to 31 Finnish HPC families. These included 11 families from the targeted re-sequencing step (Table S1) and additional 20 high-risk families10. Total RNA was purified with MagMAX™ for Stabilized Blood Tubes RNA Isolation Kit (Ambion®/Life Technologies, Carlsbad, CA, USA) and with a PAXgene Blood miRNA Kit (PreAnalytiX GmbH). RNA integrity and quality were analyzed using the Agilent 2100 Bioanalyzer and the Agilent RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA). The massively parallel paired-end RNA sequencing was performed at Beijing Genomics Institute (BGI Hong Kong Co., Ltd., Tai Po, Hong Kong) using an Illumina HiSeq2000 sequencing platform (Illumina Inc.).
RNA sequencing data analysis
On average, RNA sequencing produced 45 million reads per sample. The QC check was performed using fastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). The reads were aligned with Tophat225 using GRCh37/hg19 as the reference genome. The read counts for the genes were determined using HTSeq (http://www-huber.embl.de/users/anders/HTSeq/). The raw read counts were transformed into comparable expression values via normalization using the DESeq package for R26 and the genes with very low or no expression (normalized read counts of < 20) were removed. A differential gene expression analysis was then performed using a two-sided Mann-Whitney test with a P value cut-off of 0.05.
eQTL mapping and data analysis
The eQTL analysis was based on the RNA-seq data and on the SNP genotypes obtained from targeted DNA sequencing. This data existed for 19 samples at 2q37 and for 17 samples at 17q11.2-q22. In total, 54,919 SNPs (average 6,865 per gene, see Table S5 for details) were tested for association with their candidate target genes. Only genes with differential expression (DE) patterns between health status groups were included in the eQTL analysis, to increase the probability that found SNP-gene associations also link PrCa with a certain SNP genotype. The eQTL mapping was applied on 2q37 and 17q11.2-q22 to identify cis-regulated genes. SNPs associated in cis were defined as variants located within 1 Mb up- or downstream of the gene under study. The significance level for SNP-gene associations was set to P ≤ 0.005. A multiple testing adjustment was omitted because of the large number of tested SNPs and the nature of the permutation type tests, acknowledging that this resulted in compromised resolution.
A modified cis-eQTL approach was also utilized, wherein a large genotype dataset from the iCOGS study27 was used to pre-identify possible PrCa-associated SNPs for 2,824 unselected Finnish PrCa patients and 2,440 controls. Here, Fisher’s exact test with a modest significance level of 0.005 was used to study the association. Significant iCOGS variants that were also observed in the targeted DNA sequencing data were then selected for eQTL analysis, which was restricted to the fine-mapped regions. Additional details for the eQTL analysis are presented in Supplementary Methods.
RegulomeDB was used to annotate and assess the regulatory potential of the detected eQTLs.24 The ENCODE datasets23 were retrieved from the UCSC Genome Browser website for visualization purposes using the Table Browser tool.12 As a general indicator of regulatory potential, we used the dataset that contained enriched DNase hypersensitive sites in 125 cell types. To highlight the regulatory potential of eQTLs in PrCa tissue, we used the LNCaP DNase (wgEncodeAwgDnaseUwdukeLncapUniPk) and LNCaP (Andr) DNase (wgEncodeAwgDnaseUwDukeLncapandrogenUniPk) datasets containing DNase hypersensitive sites in LNCaP cells under normal and androgen-induced conditions, respectively. Transcription factor (TF) binding site data were gathered from the Txn Fac ChIP V3 dataset, which contains ChIP-seq experimental data on 91 cell types and 189 TFs.
Results
Targeted DNA sequencing data analysis
The percentage of mapped reads was 95.0% and 95.7% for the samples sequenced for 2q37 and 17q11.2-q22, respectively. The target coverage was 99.8% for 2q37 and 99.5% for 17q11.2-q22. Correspondingly, the percentage of bases having coverage of 20× or more was 79.9% and 63.4%. The total number of unique variants across all samples discovered by the utilized VCP was 107,479 (Figure 1). Among the 41 predicted pathogenic variants in 2q37, there were 20 missense SNVs, 16 non-coding SNVs and five indels. Of all 111 predicted pathogenic variants in 17q11.2-q22, two variants were nonsense SNVs, 49 were missense SNVs, 36 were non-coding SNVs and 24 were indels.
PrCa-associated variants identified by Sequenom validation
Following prioritization, a total of 58 variants were selected for validation in a larger sample set (Table S2). In the QC analysis, four variants failed the HWE test (P < 0.05), and 20 samples were omitted due to low genotyping frequencies (< 0.80). In the case-control association analysis, a total of 13 variants in seven different genes were statistically significantly associated with PrCa (P < 0.05; Tables 1, 2, S3 and S4). Three variants were located in the ZNF652 gene at 17q21.3, and the HDAC4 (2q37.2), HOXB3 (17q21.3), ACACA (17q21) and MYEOV2 (2q37.3) genes harbored two variants each. A single variant was identified in the HOXB13 and EFCAB13 genes at 17q21.3. Only three of these 13 PrCa-associated variants were located within exons, whereas the majority, 10 variants, resided in non-coding regions.
Table 1.
Variants significantly associated with prostate cancer based on a comparison of familial cases (n=186) and controls (n=914).
| SNP Id | Function | Gene | Chr | Mina / Majb | F_Ac / F_Ud (%) | P value | OR (95% CI) | Pathogenicity predictione |
|---|---|---|---|---|---|---|---|---|
| rs116890317 | intronic | ZNF652 | 17 | A / T | 2.96 / 0.39 | 3.3 × 10−5 | 7.8 (3.0 – 20.3) | polymorphism/-/- |
| rs79670217 | intronic | ZNF652 | 17 | G / T | 6.65 / 3.56 | 0.009 | 1.9 (1.2 – 3.1) | polymorphism/-/- |
| rs10554930 | intronic | HOXB3 | 17 | −ACA / ACA | 27.5 / 21.3 | 0.010 | 1.4 (1.1 – 1.8) | pathogenic/-/- |
| rs35384813 | 5’-UTR | HOXB3 | 17 | +T / - | 26.7 / 20.8 | 0.013 | 1.4 (1.1 – 1.8) | pathogenic/-/- |
| rs73000144 | missense | HDAC4 | 2 | T / C | 0.80 / 0.06 | 0.018 | 14.6 (1.5 – 140.2) | polymorphism/benign/neutral |
| rs13411615* | near gene 5’ | MYEOV2 | 2 | C / A | 52.1 / 45.6 | 0.023 | 1.3 (1.0 – 1.6) | polymorphism/-/- |
| rs9899142 | intronic | HOXB13 | 17 | T / C | 11.2 / 15.6 | 0.031 | 0.7 (0.5 – 1.0) | polymorphism/-/- |
| rs118004742 | nonsense | EFCAB13 | 17 | G / T | 4.79 / 2.73 | 0.048 | 1.8 (1.0 – 3.1) | pathogenic/-/- |
| rs142044482 | 3’-UTR | ZNF652 | 17 | +A / - | 2.94 / 1.59 | 0.087 | 1.9 (0.9–3.8) | polymorphism/-/- |
| rs140611363* | near gene 5’ | ACACA | 17 | −A / A | 28.8 / 31.1 | 0.421 | 0.9 (0.7–1.1) | pathogenic/-/- |
| rs72828246* | near gene 5’ | ACACA | 17 | G / A | 28.8 / 30.9 | 0.459 | 0.9 (0.7–1.2) | pathogenic/benign/neutral |
| rs13406410* | near gene 5’ | MYEOV2 | 2 | C / T | 47.6 / 46.8 | 0.817 | 1.0 (0.8–1.3) | pathogenic/-/- |
| rs61752234 | synonymous | HDAC4 | 2 | C / T | 7.22 / 6.83 | 0.823 | 1.1 (0.7–1.6) | polymorphism/-/- |
Min = minor allele
Maj = major allele
F_A = frequency of the minor allele in cases
F_U = frequency of the minor allele in controls
Pathogenicity prediction results from: Mutation Taster / PolyPhen-2 / Pon-P
Variants are in linkage disequilibrium.
Chr = chromosome, OR = odds ratio, CI = confidence interval
Bold signifies P < 0.05.
Table 2.
Variants significantly associated with prostate cancer based on a comparison of unselected cases (n=1096) and controls (n=914).
| SNP Id | Function | Gene | Chr | Mina / Majb | F_Ac / F_Ud (%) | P value | OR (95% CI) | Pathogenicity predictione |
|---|---|---|---|---|---|---|---|---|
| rs79670217 | intronic | ZNF652 | 17 | G / T | 5.66 / 3.56 | 0.002 | 1.6 (1.2 – 2.2) | polymorphism/-/- |
| rs116890317 | intronic | ZNF652 | 17 | A / T | 1.27 / 0.39 | 0.003 | 3.3 (1.4 – 7.5) | polymorphism/-/- |
| rs13406410* | near gene 5’ | MYEOV2 | 2 | C / T | 51.5 / 46.8 | 0.006 | 1.2 (1.1 – 1.4) | pathogenic/-/- |
| rs61752234 | synonymous | HDAC4 | 2 | C / T | 4.85 / 6.83 | 0.008 | 0.7 (0.5 – 0.9) | polymorphism/-/- |
| rs142044482 | 3’-UTR | ZNF652 | 17 | +A / - | 0.68 / 1.59 | 0.009 | 0.4 (0.2 – 0.8) | polymorphism/-/- |
| rs140611363* | near gene 5’ | ACACA | 17 | −A / A | 27.9 / 31.1 | 0.032 | 0.9 (0.7 – 1.0) | pathogenic/-/- |
| rs10554930 | intronic | HOXB3 | 17 | −ACA / ACA | 24.1 / 21.3 | 0.034 | 1.2 (1.0 – 1.4) | pathogenic/-/- |
| rs13411615* | near gene 5’ | MYEOV2 | 2 | C / A | 49.0 / 45.6 | 0.037 | 1.1 (1.0 – 1.3) | polymorphism/-/- |
| rs72828246* | near gene 5’ | ACACA | 17 | G / A | 28.0 / 30.9 | 0.044 | 0.9 (0.8 – 1.0) | pathogenic/benign/neutral |
| rs35384813 | 5’-UTR | HOXB3 | 17 | +T / - | 23.2 / 20.8 | 0.073 | 1.1 (1.0–1.3) | pathogenic/-/- |
| rs73000144 | missense | HDAC4 | 2 | T / C | 0.33 / 0.06 | 0.078 | 5.9 (0.7–47.9) | polymorphism/benign/neutral |
| rs118004742 | nonsense | EFCAB13 | 17 | G / T | 3.0 / 2.7 | 0.637 | 1.1 (0.8–1.6) | pathogenic/-/- |
| rs9899142 | intronic | HOXB13 | 17 | T / C | 16.1 / 15.6 | 0.665 | 1.0 (0.9–1.2) | polymorphism/-/- |
Min = minor allele
Maj = major allele
F_A = frequency of the minor allele in cases
F_U = frequency of the minor allele in ontrols
Pathogenicity prediction results from: Mutation Taster / PolyPhen-2 / Pon-P
Variants are in linkage disequilibrium.
Chr = chromosome, OR = odds ratio, CI = confidence interval
Bold signifies P < 0.05.
Four of the variants with a statistically significant association with PrCa were present in both the familial and the unselected sample sets. These were rs116890317 and rs79670217 in ZNF652, rs10554930 in HOXB3, and rs13411615 in MYEOV2. The two ZNF652 variants had the strongest association with an increased PrCa risk. Rs116890317 had the most significant association with the familial cases (OR = 7.8, 95% CI 3.0 – 20.3, P = 3.3 × 10−5) and the same variant conferred the highest risk of 3.3 (95% CI 1.4 – 7.5, P = 0.003) among the unselected cases. Rs79670217 had the most significant association with PrCa in the unselected sample set (P = 0.002) and was the second most significant variant in the familial PrCa patients (OR = 1.9, 95% CI 1.2 – 3.1, P = 0.009; Tables 1 and 2).
The highest OR of 14.6 (95% CI 1.5 – 140.2, P = 0.018) was observed for the HDAC4 variant rs73000144 (c.958C>T, p.Val320Ile) among the familial samples (Table 1). Only three familial PrCa patients (1.6%), seven unselected patients (0.6%) and one control individual (0.1%) carried the minor allele in a heterozygous state, and none of the genotyped individuals were homozygous. Rs73000144 was predicted to be benign or neutral by all three in silico pathogenicity prediction algorithms (Table S2).
The rs118004742 nonsense mutation (c.1638T>G, p.Tyr546Ter) in the EFCAB13 gene was predicted to be pathogenic by MutationTaster (Table S2). Three familial cases (1.6%) were homozygous for the minor allele. There were 12 heterozygotes among the familial index cases (6.5%) and 66 among the unselected cases (6.0%). A statistically significant association between rs118004742 and PrCa was only observed for the familial patients (Table 1). The OR of 1.8 (95% CI 1.0 – 3.1) suggested an increased risk of HPC. Rs118004742 carriers in the unselected sample set did not have an increased cancer risk (OR = 1.1, 95% CI 0.8 – 1.6, P = 0.637; Table S4).
Two common non-coding variants in the HOXB3 gene, rs10554930 and rs35384813, had a moderate effect on PrCa risk, with OR values ranging from 1.2 to 1.4 (Tables 1 and 2). MutationTaster predicted both of these variants to be pathogenic (Table S2). For five variants, the odds ratios were < 1.0, indicating a modulatory role in PrCa predisposition. These variants were located near or within the ZNF652, HDAC4, HOXB13 and ACACA genes (Tables 1 and 2). According to the RegulomeDB, three of the 13 statistically significant variants were likely to affect protein binding: rs9899142 in HOXB13 (Regulome score of 1f), rs13406410 in MYEOV2 and rs72828246 in ACACA (both having Regulome score of 2b).
In case-case comparisons, none of the identified variants were significantly associated with Gleason score, average age or the serum prostate specific antigen (PSA) level at diagnosis (data not shown). The LD analysis (Figure S1) revealed that none of our 13 statistically significant variants (Tables 1 and 2) were in linkage disequilibrium with previously reported PrCa-associated variants27 (see Supplementary Results for details).
Segregation analysis of the top four candidate variants
Altogether, 41 familial index cases out of 188 genotyped by Sequenom carried at least one of the top four candidate variants. Segregation analysis was performed for these 41 HPC families. Rs116890317, rs79670217 and rs118004742 were more common among PrCa patients than healthy family members and provided evidence for co-segregation with affection status in 20 families (Tables S6, S7 and S8). However, in 15 of these families, unaffected male mutation carriers were also observed. In seven families, all of the unaffected male carriers were young enough (< 55 years) to develop PrCa later in life. Rs116890317 segregated completely with affection status in one family (Figure S2A), as did rs79670217 (Figure S2B). Complete segregation of rs118004742 was observed in three families (Table S8). The HDAC4 variant rs73000144 was detected in three families, and approximately one-third of the family members were identified as carriers, irrespective of their health status (Table S9).
Multiple variants were observed in 16 individuals from 14 families. Two families harbored rs116890317, rs79670217 and rs118004742, whereas one family was positive for rs79670217, rs73000144 and rs118004742. In the remaining families, the most common combination detected was rs79670217 together with rs118004742 (six families). Evidence for segregation with affection status was obtained for a maximum of one variant per family.
eQTL mapping results
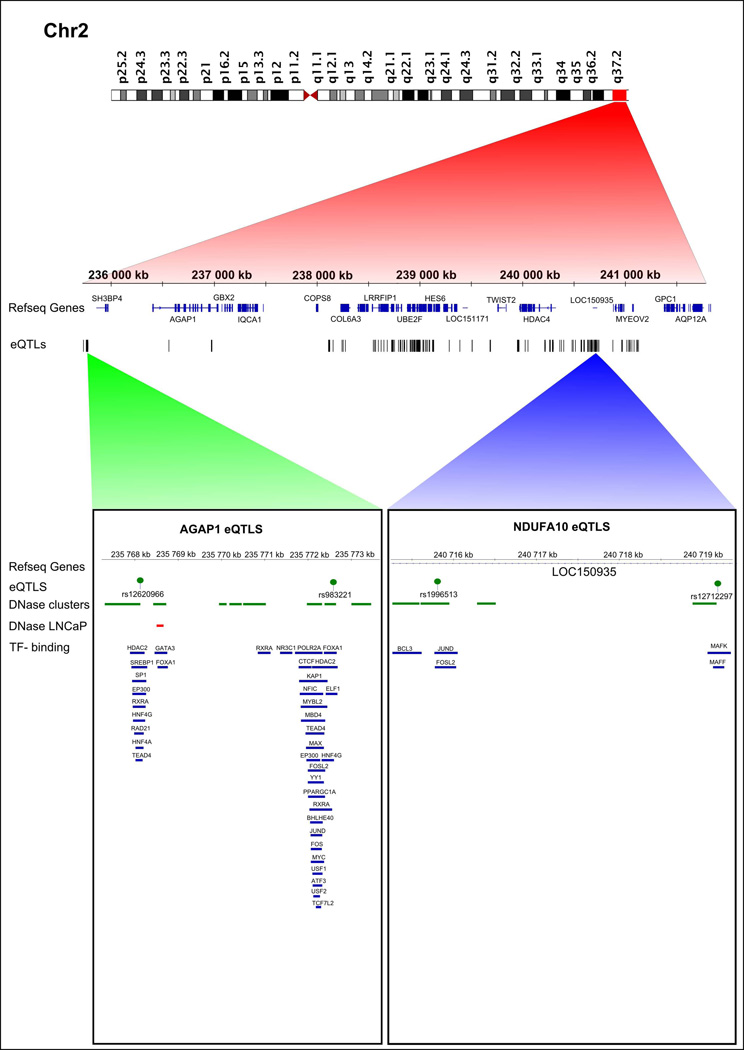
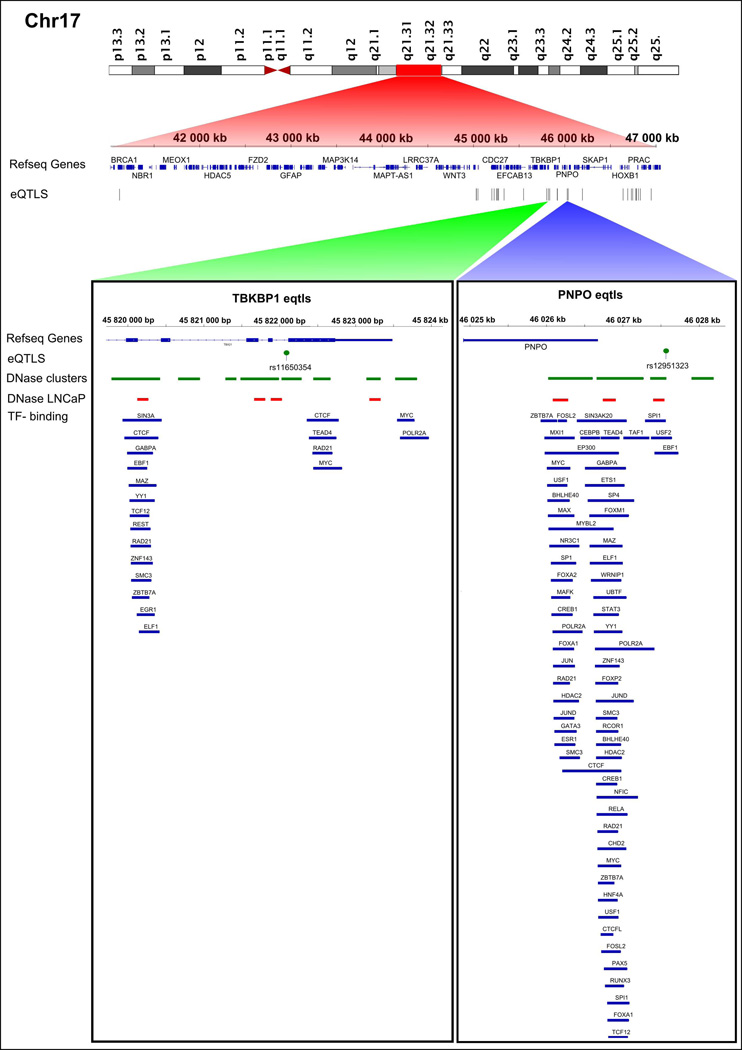
Differential gene expression analysis revealed three genes (out of 173 tested) located at 2q37 and five genes (out of 761 tested) at 17q11.2-q22 whose expression levels differed significantly between cases and controls (P < 0.05). In the targeted cis-eQTL analysis, SNPs within 2 Mb windows were tested for association with each of these eight DE genes (Table S5). Altogether, 272 candidate regulatory SNPs were identified for six DE genes only (Table S10). A vast majority, 237 candidate SNPs potentially regulate the expression of AGAP1, SCLY and NDUFA10 at 2q37 (Figure 2). The remaining 35 candidate SNPs possibly regulate TBKBP1, PNPO and NAGS at 17q11.2-q22 (Figure 3). Based on the ENCODE data, the strongest evidence for regulatory potential was found for rs11650354 on chromosome 17, which targets the TBKBP1 gene. This known eQTL overlaps with an open chromatin region (Mcf7 and Gm12892 cell lines) and its role in the regulation of TBKBP1 expression has been confirmed in a previous study.28 Rs12620966 targeting AGAP1 on chromosome 2 overlaps with several TF binding sites discovered by ChIP-seq (HepG2 cell line), position weight matrix (PWM) matching and digital DNaseI footprinting studies (Table S10). None of the coding variants that were identified by targeted DNA sequencing and validated by Sequenom were statistically significant eQTLs (data not shown).
Figure 2. Cis-eQTLs targeting differentially expressed genes on chromosome 2.
All statistically significant eQTLs are indicated with a track of black bars. Selected eQTLs, rs12620966 and rs983221 (targeting AGAP1) and rs1996513 and rs12712297 (targeting NDUFA10) are illustrated in more detail. DNaseI hypersensitive sites from the DNase cluster and LNCaP datasets are indicated with green and red rectangles, respectively. Blue rectangles denote TF binding sites.
Figure 3. Cis-eQTLs targeting differentially expressed genes on chromosome 17.
All statistically significant eQTLs are indicated with a track of black bars. Selected eQTLs, rs11650354 (targeting TBKBP1) and rs12951323 (targeting PNPO) are illustrated in more detail. DNaseI hypersensitive sites from the DNase cluster and LNCaP datasets are indicated with green and red rectangles, respectively. Blue rectangles denote TF binding sites.
The modified cis-eQTL analysis was based on 12 SNPs at 2q37 and 22 SNPs at 17q11.2-q22 that were shared between the iCOGS dataset and our set of variants obtained by targeted re-sequencing. The regulatory potential of these 34 SNPs was evaluated for 144 genes at 2q37 and for 160 genes at 17q11.2-q22. The modified eQTL approach identified only one PrCa-associated candidate eQTL on chromosome 2 and 36 candidate eQTLs on chromosome 17. Selected examples of these eQTLs and their target genes are shown in Table S11. The ENCODE data from RegulomeDB indicated the strongest evidence of regulatory potential for two variants on chromosome 17, rs4796751 and rs4796616, which target the DHX58, MLX and JUP genes. Both variants have previously been reported as eQTLs targeting MGC20781 and NT5C3L29 and they overlap with open chromatin regions (in 16 and 17 cell lines, respectively). Rs4796616 is also located within a TF binding site (U2OS cell line). Two additional chromosome 17 variants, rs4793943 and rs16941107 were defined as likely to affect gene expression. These variants target the ZNF652 and ARL17B genes, respectively, and overlap with open chromatin regions (in 6 and 42 cell lines, respectively) as well as several TF binding sites (Table S11). Of particular interest was the chromosome 17 variant rs4793976 targeting the SPOP gene. Although no data for this eQTL was available in the RegulomeDB, the importance of SPOP in PrCa predisposition has been recognized.30
Discussion
Prior studies have identified a strong relationship between PrCa and linkage to chromosomal regions 2q37 and 17q11.2-q22. Inspired by the lack of candidate genes and mutations, we re-sequenced the linkage peaks and confirmed the sequencing results by validating select variants. As the number of variants provided by the VCP was high, their prioritization for validation was critical.
The variants that were statistically significantly associated with PrCa were clustered in two genes on chromosome 2q37, HDAC4 and MYEOV2, and in five genes on chromosome 17q11.2-q22, ZNF652, HOXB3, HOXB13, EFCAB13 and ACACA (Tables 1 and 2). Interestingly, four of these genes, HDAC4, ZNF652, HOXB3 and HOXB13 encode TFs. Transcriptional regulation plays an essential role in maintaining normal gene control, and mutations in genes coding for TFs have been identified in PrCa. Examples of commonly occurring alterations include the fusion of TMPRSS2 with ERG, and mutations in genes coding for the forkhead-box family of TFs.31
The ZNF652 gene at 17q21.3 codes for a DNA-binding transcriptional repressor protein with seven zinc finger motifs.32 Highest expression levels have been detected in normal breast, prostate and pancreas, whereas in primary tumors and cancer cell lines, ZNF652 expression is generally lower.32 However, in PrCa, the co-expression of high levels of ZNF652 and the androgen receptor (AR) has been shown to increase the risk of PSA relapse.33 In addition, the recently characterized ZNF652 DNA binding site was found in the promoters of several genes that are involved in PrCa development and progression.34 ZNF652 also interacts with CBFA2T3, a putative breast cancer tumor suppressor, which has been shown to enhance the repressor activity of ZNF652.32
To date, only a single PrCa-associated risk variant has been identified in the ZNF652 gene. Rs7210100 has been reported to predispose men of African descent to PrCa. The risk allele is present at an extremely low frequency (<1%) in non-African populations.35 A possible European-specific risk variant, rs11650494, is located in a lincRNA just downstream of the ZNF652 gene and was recently described by the PRACTICAL Consortium.27 The present study identified two novel ZNF652 gene variants, rs116890317 and rs79670217, which were significantly associated with PrCa in both familial and unselected cases. The risk association was particularly apparent in patients with a positive family history of the disease. Correspondingly, both variants showed evidence for at least partial co-segregation with affection status in a substantial portion of Finnish HPC families. Like rs7210100, these two novel variants are located in the first intron of the gene, suggesting that they may play a role in regulating ZNF652 by affecting splicing events and/or tissue-specific expression.
The HDAC4 gene at 2q37.2 encodes a well-characterized transcriptional repressor. HDAC4 has been reported to accumulate in the nucleus in hormone-refractory PrCa36 and to bind to and inhibit the activity of AR by SUMOylation.37 Here, we determined that the exonic HDAC4 variant rs73000144 (c.958C>T) was significantly associated with familial PrCa (OR = 14.6, 95% CI 1.5 – 140.2, P = 0.018). The variant also had a high OR (= 5.8, 95% CI 0.7 – 47.9) among the unselected cases (Table S4), suggesting an increased cancer risk, but this result was not statistically significant (P = 0.078). The pathogenicity of rs73000144 is uncertain. The resulting amino acid change, a substitution of isoleucine for valine (p.Val320Ile) is conservative and was not considered pathogenic by any of the in silico predictors used (Table S2). The strikingly high OR for the familial sample set, together with the observation that this variant was detected in only three out of 186 index cases from the Finnish HPC families, suggested that rs73000144 may be a private mutation. The importance of private mutations has been emphasized in many diseases, some of which are associated with specific ethnic groups.
The protein encoded by the EFCAB13 (EF-hand calcium binding domain 13) gene at 17q21.3 contains a particular helix-loop-helix domain, the EF-hand, which is required for calcium ion binding. EF-hands are often found in calcium sensor and calcium signal modulator proteins. Ca2+ binding triggers a conformational change in the EF-hand motif, which leads to the activation or inactivation of target proteins. Currently, there is no evidence linking EFCAB13 with PrCa. The nonsense mutation rs118004742 in the EFCAB13 gene introduces a premature stop codon, leading to a significant truncation of the nascent protein. Truncating mutations are generally considered deleterious and, as expected, rs118004742 was predicted pathogenic by MutationTaster (Table S2). The variant segregated completely with affection status in three Finnish mutation-positive HPC families and showed evidence for partial co-segregation in four additional families. In these seven families, the variant was observed in all of the patients but in only half of the genotyped unaffected men (Table S8). It is possible that rs118004742 contributes to hereditary, but not sporadic, disease. Once a more detailed characterization of the EFCAB13 protein function is available, it will be possible to assess the indicative role of EFCAB13 as a PrCa risk gene more accurately.
Considering the importance of the HOXB13 variant G84E2 in familial PrCa predisposition, we compared the families that were positive for the top four SNPs with the existing G84E genotyping data.3 Interestingly, ten of the 11 families that were positive for the ZNF652 variant rs116890317 also harbored G84E. In these ten families, 12/21 (57%) of PrCa patients carried both the rs116890317 variant and the HOXB13 variant G84E. Co-segregation of the ZNF652 variant rs79670217 (Table S7) and G84E was detected in 6/42 (14%) of affected individuals, and among the 31 PrCa patients carrying the EFCAB13 variant rs118004742 (Table S8), G84E was identified in only 2 (6%) patients. In addition, one of the three PrCa patients carrying the HDAC4 variant rs73000144 also carried G84E. The co-occurrence of the ZNF652 variant rs116890317 with the HOXB13 variant G84E suggests possible interaction between these two genomic regions and is an interesting issue for future research.
The HOXB3 gene belongs to the same evolutionarily conserved HOXB gene family at 17q21-q22 as HOXB13. Recently, HOXB3 overexpression was observed in primary PrCa tissues, predicting poor survival.38 In our study, two possibly pathogenic HOXB3 variants were associated with a moderately increased PrCa risk, rs10554930 in both datasets and rs35384813 in the familial sample set only (Tables 1 and 2). Rs10554930 is intronic, located ~730 bp upstream of the HOXB3 transcription start site (TSS), whereas rs35384813 is in the 5’-UTR of the gene. Most variants affecting the expression level of a particular gene are located near the TSS of that gene29 making it possible that these two variants participate in the regulation of HOXB3 gene expression.
The ENCODE data supported a possible regulatory role for three of the statistically significant non-coding variants validated by Sequenom. The intronic HOXB13 variant rs9899142 likely affects the binding of ZNF263, a transcriptional repressor that participates in cell structure maintenance and proliferation.39 This variant is also a known cis-eQTL that regulates the expression of the SKAP1 gene which has been associated with PrCa-specific mortality.40 The SNPs rs13406410 and rs72828246 are located near the 5’ ends of the MYEOV2 and ACACA genes, respectively. Both of these variants likely affect the binding of E2F1. This TF plays a central role in DNA damage-induced apoptosis and DNA repair.41 Recently, a strong correlation between E2F1 and increased expression of NuSAP, a protein that binds DNA to the mitotic spindle, was observed in recurrent PrCa.42 The minor alleles of rs9899142, rs13406410 and rs72828246 had a low OR and were present at a high frequency in both cases and controls. Nevertheless, according to the common disease – common variant hypothesis, it is possible that the major alleles, rather than the minor alleles, explain a proportion of PrCa susceptibility.
The eQTL mapping enabled us to identify genomic regions that were likely to be regulated by variants in the 2q37 and 17q11.2-q22 loci. A drawback of the eQTL analysis was the use of peripheral blood for RNA-sequencing. However, fresh PrCa tissue is rarely available and, due to the multifocal nature of PrCa, the quality of prostate biopsies may be compromised. Post-mortem material, on the other hand, represents expression profiles typical for end-stage disease, whereas our aim was to identify inherited mutations predisposing their carriers to PrCa. Therefore, we consider blood to be a valid starting point for expression profiling of the early changes in PrCa. It will be exciting to see whether future studies confirm our results in another, independent sample set, preferably a collection of PrCa tissue samples.
The traditional eQTL analysis identified six DE genes that were putatively regulated by eQTLs in cis (Figures 2 and 3, Table S10). None of these genes has previously been associated with PrCa. The protein encoded by the AGAP1 gene is involved in membrane trafficking and cytoskeleton dynamics.43 SCLY and PNPO participate in metabolic processes, SCLY in the decomposition of L-selenocysteine44 and PNPO in the biosynthesis of vitamin B6. The adaptor protein encoded by TBKBP1 plays a role in the TNF-alpha/NF-kappa B signal transduction pathway.45 NDUFA10 and NAGS are mitochondrial enzymes. NDUFA10, a member of the respiratory chain complex I, is responsible for electron transport.46 NAGS catalyzes the formation of N-acetylglutamate, an activator of urea cycle enzyme CPSI.47
In the modified eQTL analysis, several cis-acting variants that were associated with altered gene expression were identified (Table S11). The most interesting finding was the association of rs4793943 with ZNF652 expression. This interaction may alter the TF function of ZNF652, thereby modulating susceptibility to PrCa. Data from RegulomeDB suggest that rs4793943 may have a more generalized role in transcriptional regulation. It is located within the binding site of ZNF26339 and it overlaps with HOXA9 and HOXB13 binding motifs. Both of these TFs have been connected with PrCa initiation and progression.2, 48 Furthermore, our data provided suggestive evidence that rs4793976 is an eQTL regulating the expression of SPOP (Table S11). SPOP, a putative tumor suppressor gene, is frequently mutated in localized and advanced prostate tumors.30 SPOP mutations are regarded as driver lesions in prostate carcinogenesis31 and the loss of SPOP expression may contribute to PrCa development.49
While interpreting the eQTL results, it is important to recall that the significant DE genes and SNP-gene associations could be identified merely by chance. The number of observed significant test results lies in the same magnitude as the number of expected significant test results, if the null hypothesis would hold for all performed tests. However, the risk of an excess of false positive results was accepted in favor of minimizing the risk of obtaining too many false negative results. Although several of the SNP-gene connections detected in this study achieved statistical significance, this does not necessarily indicate biological significance. Neither is the mechanism of interaction between the individual eQTLs and their target genes currently known. Further validation with independent datasets is required to confirm the significance of the SNP-gene associations identified here.
In conclusion, the present study demonstrated that next-generation sequencing is a valid and reliable approach for identifying novel disease-associated variants and mutations, especially those rare enough to escape the resolution of GWAS. In contrast to imputation and related prediction-based methods, next-generation sequencing methods provide true genotype data with a minimal error rate. The integrated analysis of rare and common variants with gene expression data generated unique knowledge of PrCa-associated variants with effects at the transcriptional level. This study provided a broader view of the causative factors in PrCa, implicating that regulatory variants co-operating with coding variants can modulate the inherited risk for the disease. The findings reported here encourage further research to elucidate the regulatory networks that control PrCa initiation and development.
Supplementary Material
What’s new?
The single nucleotide polymorphisms (SNPs) identified by genome-wide association studies explain only a fraction of the familial clustering of prostate cancer (PrCa). In this study, we have exploited next-generation sequencing approaches to uncover less common alleles contributing to PrCa risk. Several novel PrCa-associated variants were identified by targeted re-sequencing of two genomic regions, 2q37 and 17q11.2-q22. RNA sequencing of the selected regions followed by eQTL analysis revealed new relationships between regulatory SNPs and PrCa predisposition.
Acknowledgements
The authors wish to thank all the patients and families who participated in this study. The authors also thank Ms. Riitta Vaalavuo and Ms. Riina Kylätie for technical assistance. The genotyping of variants with Sequenom was performed by the Technology Centre, Institute of Molecular Medicine (FIMM), University of Helsinki, Finland. This work was supported by the Academy of Finland [251074]; The Finnish Cancer Organisations; the Sigrid Juselius Foundation; and the Competitive State Research Financing of the Expert Responsibility Area of Tampere University Hospital [X51003]. The PRACTICAL consortium was supported by the European Commission's Seventh Framework Programme [HEALTH-F2-2009-223175]; Cancer Research UK [C5047/A7357, C1287/A10118, C5047/A3354, C5047/A10692, C16913/A6135]; and The National Institutes of Health [Cancer Post-Cancer GWAS initiative grant No. 1 U19 CA 148537-01].
Abbreviations
- AR
Androgen Receptor
- ChIP-seq
Chromatin Immunoprecipitation Combined with Massively Parallel DNA Sequencing
- CI
Confidence Interval
- DB
Database
- DE
Differentially Expressed (gene)
- eQTL
Expression Quantitative Trait Locus
- GWAS
Genome Wide Association Study
- HPC
Hereditary Prostate Cancer
- HWE
Hardy-Weinberg Equilibrium
- Indel
Insertion/Deletion Polymorphism
- LD
Linkage Disequilibrium
- LincRNA
Large Intergenic Non-Coding RNA
- LNCaP
Androgen-Sensitive Human Prostate Adenocarcinoma Cell Line Derived From Lymph Node Metastasis
- MAF
Minor Allele Frequency
- OR
Odds Ratio
- PrCa
Prostate Cancer
- PSA
Prostate Specific Antigen
- PWM
Position Weight Matrix
- RNA-seq
Massively Parallel RNA Sequencing
- SNP
Single Nucleotide Polymorphism
- SNV
Single Nucleotide Variant
- TF
Transcription Factor
- TSS
Transcription Start Site
- UTR
Untranslated Region
- VCP
Variant-Calling Pipeline
- QC
Quality Control
§ The PRACTICAL consortium
Rosalind Eeles1,2, Doug Easton3, Kenneth Muir4, Graham Giles5,6, Fredrik Wiklund7, Henrik Grönberg7, Christopher Haiman8, Johanna Schleutker9,10, Maren Weischer11, Ruth C. Travis12, David Neal13, Paul Pharoah14, Kay-Tee Khaw15, Janet L. Stanford16,17, William J. Blot18, Stephen Thibodeau19, Christiane Maier20,21, Adam S. Kibel22,23, Cezary Cybulski24, Lisa Cannon-Albright25, Hermann Brenner26, Jong Park27, Radka Kaneva28, Jyotnsa Batra29, Manuel R. Teixeira30, Zsofia Kote-Jarai1,Ali Amin Al Olama3, Sara Benlloch3
1 The Institute of Cancer Research, 15 Cotswold Road, Sutton, Surrey, SM2 5NG, UK,2 Royal Marsden NHS Foundation Trust, Fulham and Sutton, London and Surrey, UK,3 Centre for Cancer Genetic Epidemiology, Department of Public Health and Primary Care, University of Cambridge, Strangeways Laboratory, Worts Causeway, Cambridge, UK,4 University of Warwick, Coventry, UK,5 Cancer Epidemiology Centre, The Cancer Council Victoria, 1 Rathdowne street, Carlton Victoria, Australia,6 Centre for Molecular, Environmental, Genetic and Analytic Epidemiology, The University of Melbourne, Victoria, Australia,7 Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm, Sweden,8 Department of Preventive Medicine, Keck School of Medicine, University of Southern California/Norris Comprehensive Cancer Center, Los Angeles, California, USA,9 Department of Medical Biochemistry and Genetics, University of Turku, Turku, Finland,10 BioMediTech, University of Tampere and FimLab Laboratories, Tampere, Finland,11 Department of Clinical Biochemistry, Herlev Hospital, Copenhagen University Hospital, Herlev Ringvej 75, DK-2730 Herlev, Denmark,12 Cancer Epidemiology Unit, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, UK,13 Surgical Oncology (Uro-Oncology: S4), University of Cambridge, Box 279, Addenbrooke’s Hospital, Hills Road, Cambridge, UK and Cancer Research UK Cambridge Research Institute, Li Ka Shing Centre, Cambridge, UK,14 Centre for Cancer Genetic Epidemiology, Department of Oncology, University of Cambridge, Strangeways Laboratory, Worts Causeway, Cambridge, UK,15 Cambridge Institute of Public Health, University of Cambridge, Forvie Site, Robinson Way, Cambridge CB2 0SR,16 Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA,17 Department of Epidemiology, School of Public Health, University of Washington, Seattle, Washington, USA,18 International Epidemiology Institute, 1455 Research Blvd., Suite 550, Rockville, MD 20850,19 Mayo Clinic, Rochester, Minnesota, USA,20 Department of Urology, University Hospital Ulm, Germany,21 Institute of Human Genetics University Hospital Ulm, Germany,22 Brigham and Women's Hospital/Dana-Farber Cancer Institute, 45 Francis Street- ASB II-3, Boston, MA 02115,23 Washington University, St Louis, Missouri,24 International Hereditary Cancer Center, Department of Genetics and Pathology, Pomeranian Medical University, Szczecin, Poland,25 Division of Genetic Epidemiology, Department of Medicine, University of Utah School of Medicine,26 Division of Clinical Epidemiology and Aging Research, German Cancer Research Center, Heidelberg Germany,27 Division of Cancer Prevention and Control, H. Lee Moffitt Cancer Center, 12902 Magnolia Dr., Tampa, Florida, USA,28 Molecular Medicine Center and Department of Medical Chemistry and Biochemistry, Medical University - Sofia, 2 Zdrave St, 1431, Sofia, Bulgaria,29 Australian Prostate Cancer Research Centre-Qld, Institute of Health and Biomedical Innovation and Schools of Life Science and Public Health, Queensland University of Technology, Brisbane, Australia,30 Department of Genetics, Portuguese Oncology Institute, Porto, Portugal and Biomedical Sciences Institute (ICBAS), Porto University, Porto, Portugal
Footnotes
The authors have declared that no conflicts of interest exist.
References
- 1.Baker SG, Lichtenstein P, Kaprio J, Holm N. Genetic susceptibility to prostate, breast, and colorectal cancer among Nordic twins. Biometrics. 2005;61:55–63. doi: 10.1111/j.0006-341X.2005.030924.x. [DOI] [PubMed] [Google Scholar]
- 2.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, Wiley KE, Isaacs SD, Johng D, Wang Y, Bizon C, Yan G, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laitinen VH, Wahlfors T, Saaristo L, Rantapero T, Pelttari LM, Kilpivaara O, Laasanen SL, Kallioniemi A, Nevanlinna H, Aaltonen L, Vessella RL, Auvinen A, et al. HOXB13 G84E mutation in Finland: population-based analysis of prostate, breast, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2013;22:452–460. doi: 10.1158/1055-9965.EPI-12-1000-T. [DOI] [PubMed] [Google Scholar]
- 4.Xu J, Dimitrov L, Chang BL, Adams TS, Turner AR, Meyers DA, Eeles RA, Easton DF, Foulkes WD, Simard J, Giles GG, Hopper JL, et al. A combined genomewide linkage scan of 1,233 families for prostate cancer-susceptibility genes conducted by the international consortium for prostate cancer genetics. Am J Hum Genet. 2005;77:219–229. doi: 10.1086/432377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lange EM, Robbins CM, Gillanders EM, Zheng SL, Xu J, Wang Y, White KA, Chang BL, Ho LA, Trent JM, Carpten JD, Isaacs WB, et al. Fine-mapping the putative chromosome 17q21-22 prostate cancer susceptibility gene to a 10 cM region based on linkage analysis. Hum Genet. 2007;121:49–55. doi: 10.1007/s00439-006-0274-2. [DOI] [PubMed] [Google Scholar]
- 6.Pierce BL, Friedrichsen-Karyadi DM, McIntosh L, Deutsch K, Hood L, Ostrander EA, Austin MA, Stanford JL. Genomic scan of 12 hereditary prostate cancer families having an occurrence of pancreas cancer. Prostate. 2007;67:410–415. doi: 10.1002/pros.20527. [DOI] [PubMed] [Google Scholar]
- 7.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 8.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 9.Cropp CD, Simpson CL, Wahlfors T, Ha N, George A, Jones MS, Harper U, Ponciano-Jackson D, Green TA, Tammela TL, Bailey-Wilson J, Schleutker J. Genome-wide linkage scan for prostate cancer susceptibility in Finland: evidence for a novel locus on 2q37.3 and confirmation of signal on 17q21-q22. Int J Cancer. 2011;129:2400–2407. doi: 10.1002/ijc.25906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schleutker J, Matikainen M, Smith J, Koivisto P, Baffoe-Bonnie A, Kainu T, Gillanders E, Sankila R, Pukkala E, Carpten J, Stephan D, Tammela T, et al. A genetic epidemiological study of hereditary prostate cancer (HPC) in Finland: frequent HPCX linkage in families with late-onset disease. Clin Cancer Res. 2000;6:4810–4815. [PubMed] [Google Scholar]
- 11.Sulonen AM, Ellonen P, Almusa H, Lepisto M, Eldfors S, Hannula S, Miettinen T, Tyynismaa H, Salo P, Heckman C, Joensuu H, Raivio T, et al. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011;12:R94. doi: 10.1186/gb-2011-12-9-r94. 2011-12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Research. 2010 doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 14.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olatubosun A, Valiaho J, Harkonen J, Thusberg J, Vihinen M. PON-P: Integrated predictor for pathogenicity of missense variants. Hum Mutat. 2012 doi: 10.1002/humu.22102. [DOI] [PubMed] [Google Scholar]
- 16.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maqungo M, Kaur M, Kwofie SK, Radovanovic A, Schaefer U, Schmeier S, Oppon E, Christoffels A, Bajic VB. DDPC: Dragon Database of Genes associated with Prostate Cancer. Nucleic Acids Research. 2010 doi: 10.1093/nar/gkq849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. 2010-11-10-r106. Epub 2010 Oct 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45:385–391. 391e1–391e2. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, Eleftheriadis M, Sinning CR, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, Montgomery S, Tavare S, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbieri CE, Bangma CH, Bjartell A, Catto JW, Culig Z, Gronberg H, Luo J, Visakorpi T, Rubin MA. The mutational landscape of prostate cancer. Eur Urol. 2013;64:567–576. doi: 10.1016/j.eururo.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar R, Manning J, Spendlove HE, Kremmidiotis G, McKirdy R, Lee J, Millband DN, Cheney KM, Stampfer MR, Dwivedi PP, Morris HA, Callen DF. ZNF652, a novel zinc finger protein, interacts with the putative breast tumor suppressor CBFA2T3 to repress transcription. Mol Cancer Res. 2006;4:655–665. doi: 10.1158/1541-7786.MCR-05-0249. [DOI] [PubMed] [Google Scholar]
- 33.Callen DF, Ricciardelli C, Butler M, Stapleton A, Stahl J, Kench JG, Horsfall DJ, Tilley WD, Schulz R, Nesland JM, Neilsen PM, Kumar R, et al. Co-expression of the androgen receptor and the transcription factor ZNF652 is related to prostate cancer outcome. Oncol Rep. 2010;23:1045–1052. doi: 10.3892/or_00000731. [DOI] [PubMed] [Google Scholar]
- 34.Kumar R, Selth LA, Schulz RB, Tay BS, Neilsen PM, Callen DF. Genome-wide mapping of ZNF652 promoter binding sites in breast cancer cells. J Cell Biochem. 2011;112:2742–2747. doi: 10.1002/jcb.23214. [DOI] [PubMed] [Google Scholar]
- 35.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, Rybicki BA, Isaacs WB, Ingles SA, Stanford JL, Diver WR, Witte JS, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halkidou K, Cook S, Leung HY, Neal DE, Robson CN. Nuclear accumulation of histone deacetylase 4 (HDAC4) coincides with the loss of androgen sensitivity in hormone refractory cancer of the prostate. Eur Urol. 2004;45:382–389. doi: 10.1016/j.eururo.2003.10.005. author reply 389. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Tse AK, Li P, Ma Q, Xiang S, Nicosia SV, Seto E, Zhang X, Bai W. Inhibition of androgen receptor activity by histone deacetylase 4 through receptor SUMOylation. Oncogene. 2011;30:2207–2218. doi: 10.1038/onc.2010.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Zhu S, Jiang N, Shang Z, Quan C, Niu Y. HoxB3 promotes prostate cancer cell progression by transactivating CDCA3. Cancer Lett. 2013;330:217–224. doi: 10.1016/j.canlet.2012.11.051. [DOI] [PubMed] [Google Scholar]
- 39.Frietze S, Lan X, Jin VX, Farnham PJ. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J Biol Chem. 2010;285:1393–1403. doi: 10.1074/jbc.M109.063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang CN, Huang SP, Pao JB, Chang TY, Lan YH, Lu TL, Lee HZ, Juang SH, Wu PP, Pu YS, Hsieh CJ, Bao BY. Genetic polymorphisms in androgen receptor-binding sites predict survival in prostate cancer patients receiving androgen-deprivation therapy. Ann Oncol. 2012;23:707–713. doi: 10.1093/annonc/mdr264. [DOI] [PubMed] [Google Scholar]
- 41.Biswas AK, Johnson DG. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012;72:13–17. doi: 10.1158/0008-5472.CAN-11-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gulzar ZG, McKenney JK, Brooks JD. Increased expression of NuSAP in recurrent prostate cancer is mediated by E2F1. Oncogene. 2013;32:70–77. doi: 10.1038/onc.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nie Z, Stanley KT, Stauffer S, Jacques KM, Hirsch DS, Takei J, Randazzo PA. AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J Biol Chem. 2002;277:48965–48975. doi: 10.1074/jbc.M202969200. [DOI] [PubMed] [Google Scholar]
- 44.Mihara H, Kurihara T, Watanabe T, Yoshimura T, Esaki N. cDNA cloning, purification, and characterization of mouse liver selenocysteine lyase. Candidate for selenium delivery protein in selenoprotein synthesis. J Biol Chem. 2000;275:6195–6200. doi: 10.1074/jbc.275.9.6195. [DOI] [PubMed] [Google Scholar]
- 45.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 46.Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 47.Caldovic L, Morizono H, Gracia Panglao M, Gallegos R, Yu X, Shi D, Malamy MH, Allewell NM, Tuchman M. Cloning and expression of the human N-acetylglutamate synthase gene. Biochem Biophys Res Commun. 2002;299:581–586. doi: 10.1016/s0006-291x(02)02696-7. [DOI] [PubMed] [Google Scholar]
- 48.Chen JL, Li J, Kiriluk KJ, Rosen AM, Paner GP, Antic T, Lussier YA, Vander Griend DJ. Deregulation of a Hox protein regulatory network spanning prostate cancer initiation and progression. Clin Cancer Res. 2012;18:4291–4302. doi: 10.1158/1078-0432.CCR-12-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim MS, Je EM, Oh JE, Yoo NJ, Lee SH. Mutational and expressional analyses of SPOP, a candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. APMIS. 2013;121:626–633. doi: 10.1111/apm.12030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.