Abstract
Autophagy is required for the long-term maintenance of antigen-specific memory B cells. However, whether autophagy is also important for the initial formation of memory B cells remains unclear. Here we show that newly generated memory B cells do not display active autophagy, but are capable of forming antibody secreting cells after re-challenge with antigens. Increases in autophagy took place over time after the initial formation of memory B cells. The expression of transcription factors involved in autophagy, but not changes in epigenetic regulation by DNA methylation, was required for autophagy gene expression and the development of active autophagy in memory B cells. This indicates that autophagy is not critical for the initial generation of memory B cells, but is required for their long-term persistence. Our results suggest that promoting autophagy to improve antibody-dependent immunological memory is more effective during memory B cell maintenance stage.
Introduction
Exposure to pathogens leads to the activation and significant expansion of antigen-specific lymphocytes. This is followed by a contraction phase when most of these expanded lymphocytes undergo programmed cell death after the clearance of the pathogens (1–3). However, a small number of these antigen-specific lymphocytes develop into memory cells (4, 5). The persistence of antigen-specific memory cells is crucial for the maintenance of immunological memory against the original pathogens (6). Memory B cells are a heterogeneous population of quiescent antigen-experienced long-lived B cells (7–12). In T cell-dependent antigen responses, the interaction of B cells with T cells leads to the formation of germinal centers (GC), where B cells undergo isotype switching and somatic hypermutations in the immunoglobulin gene (11, 13). These antigen-specific GC B cells can give rise to memory B cells or plasma cells (11, 13–15). After re-encountering the antigens, memory B cells rapidly proliferate and differentiate into antibody secreting plasma cells (ASCs) to produce high-affinity antibodies that neutralize antigens (8, 11, 13).
In order to maintain immunological memory, the antigen-experienced memory lymphocytes need to inhibit cell death for their long-term survival. Mechanisms underlying long-term survival of memory B cells have not been fully elucidated. It has been shown that the presence of antigens is not required for the persistence of memory B cells (16). Intrinsic mechanisms may play a major role in the protection of long-term survival of memory B cells. Autophagy is an important mechanism to maintain cell survival. It is a well conserved process from yeast to mammals by which the cells sequester cytoplasmic components into double-membraned autophagosomes, leading to the degradation of enclosed materials upon fusion with lysosomes (17, 18). Autophagy helps to provide energy and metabolic intermediates to sustain cell viability during the deprivation of nutrients or growth factors (17, 19, 20). In addition, autophagy is important for quality control of cellular proteins and organelles to promote cell survival (21). Autophagy may be especially important for sustaining the survival of long-lived cell types, such as neurons (22, 23).
We have detected active autophagy and reduced cell death in memory B cells (24). Autophagy deficiency in B cells leads to a significant reduction of memory B cells and antibody-dependent immunological memory in mice. Interestingly, however, memory B cells appears in normal numbers initially after immunization in autophagy-deficient mice (24). However, it remains to be determined whether autophagy is important for the initial formation of memory B cells.
Memory B cells express increased levels of autophagy genes compared to naïve and GC B cells (24). However, the mechanisms for the increases in autophagy in memory B cells remain to be determined. Autophagy can be regulated at the epigenetic level by DNA methylation at the promoter regions of autophagy genes (25, 26). Moreover, formation of memory T cells is characterized by changes in DNA methylation of genes important for T cell functions (27). We therefore investigated the potential involvement for epigenetic and transcriptional regulation of autophagy genes in memory B cells. We found that the expression of several key autophagy genes was independent of epigenetic regulation by DNA methylation, but was controlled by the levels of transcription factors required for autophagy gene expression. Autophagy genes were not induced during the initial formation of memory B cells, but their levels were increased in these cells over time after immunization. Our data suggest that promoting autophagy during the memory B cell maintenance phase is likely to be effective in improving B cell memory.
Materials and Methods
Mice and immunization
Mice with B cell-specific deletion of Atg7 (B/Atg7−/−) were generated by crossing Atg7flox mice (28) with CD19-cre knock-in mice (The Jackson Laboratory) as described (24). Sex and age-matched 6- to 10-week old mice on the C57BL/6 background were immunized with 100 μg NP-KLH (Biosearch Technologies) precipitated with 100 μl Imject Alum (Thermo Scientific) intraperitoneally. The mice were housed in a specific pathogen-free facility at Baylor College of Medicine, and experiments were performed according to federal and institutional guidelines and with the approval of the Institutional Animal Care and Use Committee.
Flow cytometry
Splenocytes of immunized mice were stained with PE-conjugated antibodies to mouse CD11b, IgM, IgD, Gr-1 and CD138, APC-anti-IgG1, BD Horizon BV421-anti-PD-L2 (BD Bioscience), PerCP-Cy5.5-anti-B220 (eBioscience), FITC-anti-CD38, Brilliant Violet 605-anti-CD80 (Biolegend) and biotin-NIP-BSA (Biosearch), followed by staining with PE-Cy7-straptavidin (eBioscience). Alternatively, the cells were stained with PE-conjugated antibodies to CD11b, IgM, IgD, Gr-1 and CD138, APC-anti-IgG1, PerCP-Cy5.5-anti-B220, biotin-NIP-BSA plus PE-Cy7-straptavidin as above and Pacific blue-anti-CD38 (Biolegend), together with FITC-conjugated antibodies to CD38, CD138, I-Ab or isotype controls (BD Bioscience). The cells were analyzed using a LSRII flow cytometer (BD Bioscience).
Real time RT-PCR
B220+IgMlowIgD+CD23+IgG- naïve B cells were sorted from unimmunized mouse spleen by flow cytometry. B220+IgG1+NP+CD38- GC B cells were sorted from mouse spleen two weeks after immunization with NP-KLH. B220+IgG1+NP+CD38+ memory B cells were sorted from mouse spleen at different time after immunization. The purity of sorted cells is between 96% and 99%. RNA extracted from the cells was used to prepare cDNA with the High Capacity cDNA Reverse Transcription Kit (Life Technologies). Real-time PCR was performed using Taqman Universal PCR Master Mix with specific primers for autophagy genes or 18S rRNA from the TaqMan Gene Expression Assay Kit (AB Applied Biosystem) in the ABI PRISM 7000 Sequence Detection System. The Taqman specific primers for autophagy genes from AB Applied Biosystem include Mm00504340_m1 (Atg5), Mm00512209_m1 (Atg7), Mm01265461_m1 (Beclin 1), Mm00553733_m1 (Atg14), Mm00437238_m1 (ULK1), Mm00458725_g1 (MAP1LC3A), Mm00490672_m1 (FOXO1) and Mm01185722_m1 (FOXO3).
Determination of DNA methylation by bisulfite sequencing
Genomic DNA of GC and memory B cells was subjected to bisulfite conversion using the Epitect Bisulfite Conversion Kit (Qiagen) according to manufacturer’s protocol. Primers were designed to amplify CpG islands in the promoter regions of different genes using the Methprimer software (29). The promoter region of different genes were amplified by PCR from bisulfite converted DNA and cloned into pBluescript, followed by DNA sequencing to determine non-methylated (with C-T conversion) and methylated CpG (no C to T conversion) sites. The following primers were used: Beclin 1: forward 5’-TAGTTGAATTCGAAGTTTATTTGTATGGTGTTGTTGG-3’; reverse 5’-TTAGTCTCGAGAATCCTCTTAATATCATCCCACTCC-3’; Atg14, forward 5’-TAGTTGAATTCGTTTTTTAGTGGGAAGGGATTTT-3’, reverse 5’-TTAGTCTCGAGTCAAAAAAACAAAAACAATAAAACC-3’; Ulk1, forward 5’-TAGTTGAATTCAGTTTTGATATATTTTAGTTTTAAGTT-3’, reverse 5’-TTAGTCTCGAGACATTAACATACCCAATCTACTCC-3’; Hif-1α, forward 5’-TAGTTGAATTCGTTATTTGGGAAGGAGGGATTT-3’, reverse, 5’-TTAGTCTCGAGTCTCTACACCTTCAATAAAAAACTAC-3’; Foxo1, forward 5’-TAGTTGAATTCTTTGTAGGTGTGTATAGGTAGGGTG-3’, reverse 5’-TTAGTCTCGAGAATACTCCAAACAAAACCCAAAC -3’.
Cell death analyses
B220+IgG1+NP+CD38+ memory B cells or B220+IgG1+NP+CD38- GC B cells were cultured in RPMI 1640 complete medium for 0 or 4 h in 96-well plates (104 cells/well). Live cells with exclusion of propidium iodide (PI) staining were determined by flow cytometry (24). Percentage of cell loss of live cells was calculated as follows: (Bcontrol - Bcultured)/Bcontrol × 100%, with Bcontrol and Bcultured represent B cells without or with culture in vitro.
Analyses of somatic hypermutation of VH186.2 gene in NP-specific memory B cells
B220+IgG1+NP+CD38+ memory B cells were sorted from the spleens of WT or B/Atg7−/− mice 2 or 8 weeks after immunization with NP-KLH. Total RNA was extracted using Direct-zol RNA Miniprep Kit and treated with DNase I (Zymo Research), followed by reverse transcription using Superscript Vilo cDNA Synthesis Kit (Invitrogen). Rearranged V186.2DJH-Cγ1 DNA coding for the anti-NP V region was amplified similar to previously established protocols (30) by PCR with Phusion Hi-Fidelity DNA polymerase (New England Biolabs) with the following primer pair: forward, CATGCTCTTCTTGGCAGCAACAGC (for VH186.2); and reverse, GTGCACACCGCTGGACAGGGATCC (for Cg1). Second round of PCR was performed using the following nested primers: CAGGTCCAACTGCAGCAG and CCATGGAGTTAGTTTGGGCAG. PCR products were purified with Qiaquick Gel-extraction Kit (Qiagen) and cloned into the pCRII-Blunt-TOPO vector (Invitrogen), followed by sequencing with the M13 primer. Somatic mutations over a 276 bp region containng framework 1 (FR1), complementarity-determining region 1 (CDR1), FR2, CDR2 and FR3 were analyzed by comparing with the germline sequence (31).
Adoptive transfer of memory B cells
B220+IgG1+NP+CD38+ memory B cells were sorted from the pooled spleens of 20 to 50 immunized WT or B/Atg7−/− mice at different time after immunization. The cells were adoptively transferred intravenously (104/mouse) into wild type recipient mice with 105 CD4+ T cells from immunized wild type mice. Some groups were treated with rapamycin (LC Laboratories; 75 μg/kg body weight intraperitoneally) daily after the transfer of the cells. One day after the transfer, B/Atg7−/− recipients were immunized with NP-KLH. ASCs in the spleen were quantitated by ELISPOT at day 4 after NP-KLH rechallenge. Antibody titers in the sera were measured by ELISA.
Enzyme-linked immunosorbent assay (ELISA)
Sera with serial dilutions were added to 96-well plates coated with 5 μg/ml NP5-BSA (Biosearch Technologies) and incubated at room temperature for 2 h, followed by incubation with HRP-conjugated secondary antibodies against mouse IgG1 (Southern Biotechnology). The plates were developed with TMB peroxidase substrate kit (Bio-Rad Laboratories, Hercules, CA) and optical densities at 450 nm were measured. A mixture of sera from wild type mice immunized with NP-KLH was used to establish standard curves in each plate and antibody levels were shown as relative titers.
ELISPOT
MultiScreen 96-well Filtration plates (Millipore) were coated with 20 μg/ml NP5-BSA. Splenocytes or bone marrow cells (1–5×105/well) were then added to the plates and incubated at 37 °C for 5 h. The cells were lysed with H2O and the wells were probed with HRP-conjugated goat anti-mouse IgG1 (Southern Biotechnology), followed by development with 3-amino-9-ethylcarbzole (Sigma).
Immunocytochemistry
B220+IgG1+CD38+NP+ memory B cells were added to slides by cytospin. The cells were fixed, incubated with rabbit anti-LC3 (Abgent), followed by staining with Alexa Fluor-conjugated anti-rabbit IgG (Molecular Probes). The nucleus was counter-stained with DAPI. The cells were then analyzed using a SoftWorx Image deconvolution microscope (Applied Precision).
Transfection of memory B cells
Sorted NP-specific memory B cells (105 in 10 μl) were suspended in T buffer of the Neon Transfection System (Life Technologies) with 100 nM siRNA targeting FOXO3 (Cell Signaling Technology) or control siRNA. Electroporation was carried out using the Neon Transfection System (1 pulse at 2150 volts for a pulse width of 20 mini seconds). Virtually all cells were transfected using this condition with fluorescently FAM-labeled siRNA (Life Technologies). The transfected cells were used for adoptive transfer as above, followed by immunization and analyses of antibody production. Alternatively, the cells were cultured in vitro in 20 U/ml IL-2, 10 ng/ml IL-10, 10 ng/ml IL-15 and 10 μM CpG for 2 days, followed by real-time PCR analyses of autophagy genes.
Statistical analyses
Data were presented as the mean ± SD and P values were determined by two-tailed Student’s t-test or by Mann Whitney test (for somatic mutations) using the GraphPad Prism software.
Results
Active autophagy in memory B cells is regulated at the transcriptional level
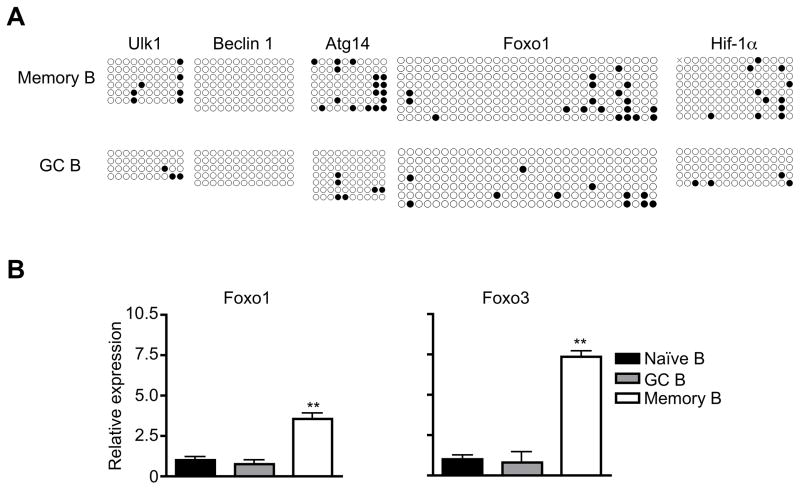
The development from GC B cells into memory B cells is accompanied by increased expression of autophagy genes and the presence of active autophagy (24). Autophagy can be regulated at the epigenetic level by DNA methylation at the promoter regions of autophagy gene (25, 26). We therefore examined whether DNA methylation might be changed at the promoter regions of several core autophagy genes important for the initiation of autophagy, including Beclin 1, Atg14 and ULK1 (32–34). Sodium bisulfite causes deamination of unmethylated cytosine but not 5-methylcytosine and convert deaminated unmethylated cytosine to uracil, which can then be amplified by PCR for sequencing (35). We subjected genomic DNA from GC and memory B cells to bisulfite conversion and sequencing analyses of autophagy-related genes. Promoter regions of Beclin 1, Atg14 and ULK1, as well as transcription factors Foxo1 and HIF-1α that are involved in regulating autophagy gene expression (36, 37), were then amplified by PCR and cloned into plasmids for sequencing analyses. We found no methylation among 13 CpG sites in the promoter of Beclin 1 in memory B cells or GC B cells (Fig. 1A). We also detected a generally low level of CpG methylation at promoter regions of ULK1, Atg14 in GC and memory B cells (Fig. 1A). The level of DNA methylation for Foxo1 and HIF-1α was also low in these cells (Fig. 1A). These data suggest that the increased expression of Beclin 1, Atg14, and ULK1 in memory B cells is not regulated at the epigenetic level by DNA methylation.
Figure 1.
Transcriptional regulation of autophagy genes in memory B cells. (A) DNA methylation at the promoter regions of autophagy genes in GC and memory B cells. Each row represents one clone of the PCR product of the promoter region with each circle represents one CpG site. Filled circle, methylated CpG. Open circle: unmethylated CpG. (B) The expression of transcription factors in GC and memory B cells. **P<0.01 (n=3).
Interestingly, we found increases in the expression of transcription factors, including FOXO1 and FOXO3 that are implicated in the expression of autophagy genes Atg4, Atg5, Atg14, Beclin 1, ULK1 and VPS34 (36, 38–44), in memory B cells compared to naïve or GC B cells (Fig. 1B). These data suggest that the induction of transcription factors, but not epigenetic changes in DNA methylation, is the major mechanism that regulates autophagy gene expression in memory B cells.
Antigen-specific memory B cells can be formed independently of autophagy
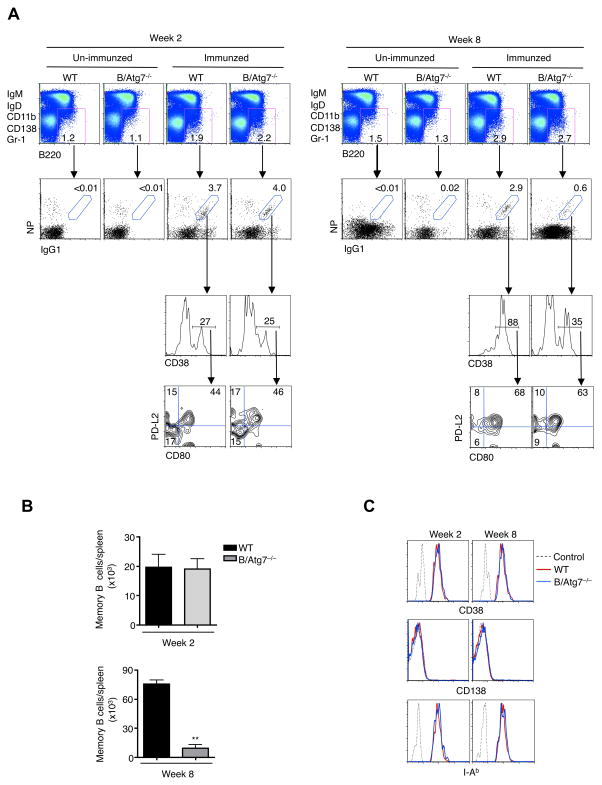
Although the long-term survival of memory B cells depends on autophagy (24), it is not known whether the initial formation of these cells requires autophagy. We therefore examined memory B cells at different time points after immunization with NP-KLH. Consistent with our previous observations (24), we found that B220+CD38+IgG1+NP+ memory B cells were generated in normal numbers in B/Atg7−/− mice two weeks after immunization (Fig. 2A). It has been shown that memory B cells contain the PD-L2-CD80- population with high self renewal potential, as well as the PD-L2+CD80+ subtype with increased capacity in forming antibody secreting cells (45). We found that these subtypes of memory B cells were similarly represented in wild type and B/Atg7−/− mice at 2 weeks after immunization (Fig. 2B). By 8 weeks after immunization, memory B cells were markedly lower in B/Atg7−/− mice, with reduction similarly observed in both the PD-L2-CD80- and PD-L2+CD80+ populations (Fig. 2B, C). These results indicate that autophagy is dispensable for the initial formation of memory B cells but is required for their long-term maintenance.
Figure 2.
Generation of memory B cells after immunization. (A) Wild type and B/Atg7−/− mice were immunized with NP-KLH. At week 2 and week 8 after immunization, NP-specific memory B cells in the spleen of immunized mice and un-immunized controls were examined by flow cytometry. (B) Counts of NP-specific memory B cells in the spleens of mice immunized in (A). **P<0.01 (n=5). (C) Analyses of levels of CD38, CD138 and I-Ab on NP-specific memory B cells by flow cytometry. Dashed line: isotype control.
The initially formed memory B cells in B/Atg7−/− mice can convert into ASCs
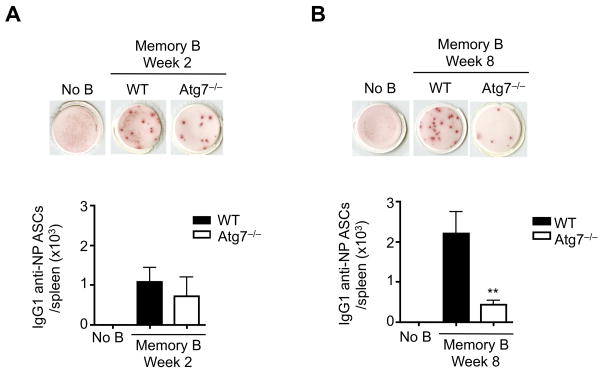
We next determined whether the newly generated memory B cells from B/Atg7−/− mice can develop into ASCs. After adoptive transfer, memory B cells from both wild type and B/Atg7−/− mice formed ASCs after re-exposure to antigens as detected by ELISPOT (Fig. 3). Compared to wild type memory B cells at 8 weeks after immunization, Atg7−/− memory B cells from this time point showed reduced capacity in forming ASCs (Fig. 3). This suggests that the newly formed memory B cells do not require autophagy for their functions in vivo.
Figure 3.
The ability for initially formed memory B cells in generating antibody secreting cells. IgG1 NP-specific memory B cells were sorted from wild type or B/Atg7−/− mice at week 2 (A) and week 8 (B) after immunization. The cells were adoptively transferred into recipient mice (5 mice/group), followed by challenge with NP-KLH one day later. Antibody secreting cells in the spleen were determined by ELISPOT. **P<0.01 (n=5).
Newly formed memory B cells show reduced cell death compared to GC B cells independent of autophagy
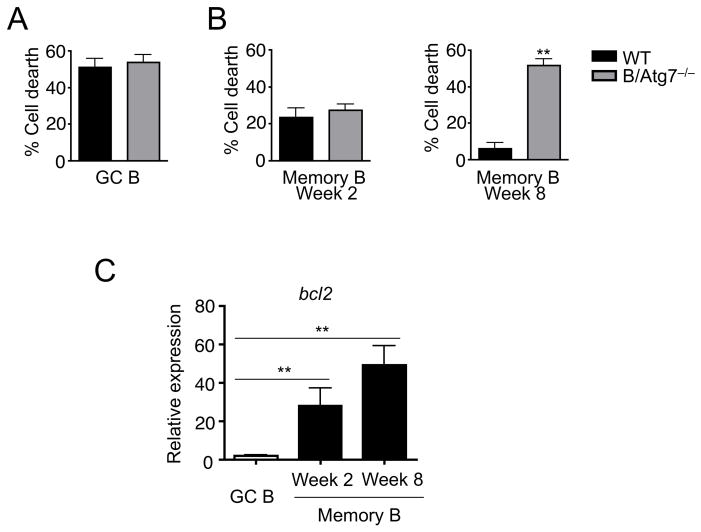
Consistent with our previous findings (24), spontaneous cell death was not affected by autophagy in GC B cells (Fig. 4A), while autophagy-deficient memory B cells isolated 2 months after immunization show significantly increased cell death during in vitro culture (Fig. 4B). Interestingly, wild type memory B cells isolated 2 weeks after immunization displayed reduced spontaneous cell death compared to GC B cells (Fig. 4B). However, autophagy-deficient memory B cells at this stage did not display significantly increased cell death after in vitro culture (Fig. 4B). This indicates that these initially formed memory B cells do not depend on autophagy for their survival.
Figure 4.
Reduced cell death in memory B cells 2 weeks after immunization. (A) GC B cells purified from mice 2 weeks after immunization with NP-KLH were cultured in the absence of cytokines for 4 h. Percentages of cells were quantitated. (B) NP-specific memory B cells from mice at 2 or 8 weeks after immunization with NP-KLH were cultured in the absence of cytokines for 4 h. Percentages of cells were quantitated. **P<0.01 (n=5). (C) Real-time RT-PCR for bcl-2 in GC cells, or in memory B cells at 2 or 8 weeks after immunization. **P<0.01 (n=3).
In addition to the expression of CD38 on the cell surface during GC to memory B cell transition (14), memory B cells showed significant up-regulation of Bcl-2 compared to GC B cells (46, 47). We observed that the level of Bcl-2 was indeed higher in memory B cells 2 weeks after immunization (Fig. 4C). These observations support the conclusion that these early memory B cells have gained anti-apoptotic properties. However, autophagy does not appear to be important to maintain their survival at this early stage.
We have previously found that germinal center responses and the affinities of NP-specific antibodies are not changed in B/Atg7−/− mice (24). To further determine whether autophagy deficiency affects affinity maturation, we performed sequence analyses of somatic mutations in the VH186.2 gene (30, 48) of IgG1+ NP-specific memory B cells from wild type and B/Atg7−/− mice at 2 or 8 weeks after immunization. We found that memory B cells in B/Atg7−/− and wild type mice showed similar mutation frequencies in the VH186.2 gene (Fig. 5). This supports the conclusion that autophagy deficiency does not affect affinity maturation of immunoglobulin genes coding for antigen-specific antibodies.
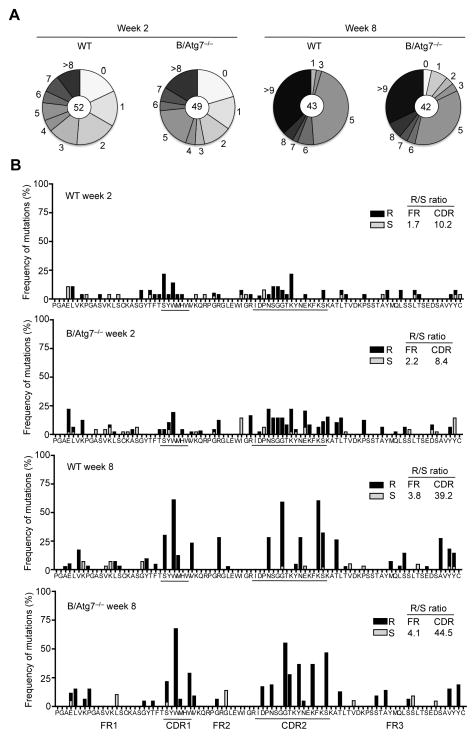
Figure 5.
Somatic mutations of VH186.2 gene in memory B cells. IgG1+ NP-specific memory B cells were sorted from wild type or B/Atg7−/− mice at week 2 and week 8 after immunization (as in Fig. 3). cDNAs of rearranged VH186.2-Cγ1 sequences were amplified and cloned into the pCRII-Blunt-TOPO plasmid for sequencing. (A) Pie charts represent the proportion of sequences that carry one, two, three, etc. nucleotide mutations over a 276 bp region in VH186.2 that covers FR1, CDR1, FR2, CDR2 and FR3. Total numbers of sequences analyzed are shown in the inner circle. (B) Frequency of mutations that resulted in amino acid replacement (R) or are silent (S) are shown in FR1, CDR1, FR2, CDR2 and FR3. Average frequency (%) of nucleotide mutation per base pair in the entire VH186.2 region sequenced: week 2, 1.37 ± 1.47 (WT) and 1.51 ± 1.54 (B/Atg7−/−), P=0.66; week8, 2.79 ± 1.87 (WT) and 2.97 ± 1.54 (B/Atg7−/−), P=0.68.
Autophagy genes are up-regulated in memory B cells after their initial formation
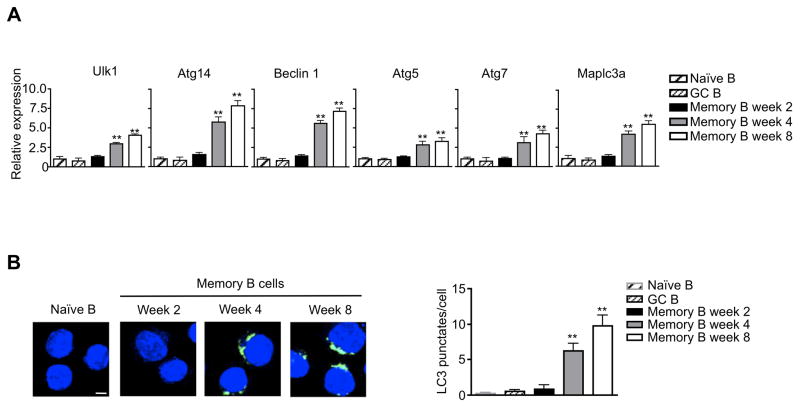
Atg7 deficiency did not affect cell death in early memory B cells at two weeks after immunization (Fig. 4B). It is possible that memory B cells have not developed active autophagy at this early time point. Indeed, we found that autophagy genes were not increased in memory B cells isolated at 2 weeks after immunization (Fig. 6A). However, memory B cells at 4 or 8 weeks after immunization showed increased expression of autophagy genes (Fig. 6A). We also determined the autophagy levels in memory B cells by immunocytochemistry staining. We did not detect autophagosomes in memory B cells at two weeks after immunization (Fig. 6B). However, memory B cells isolated at 4 or 8 weeks after immunization showed significantly higher levels of LC3 punctate staining (Fig. 6B). This indicates that memory B cells develop autophagy after their initial formation.
Figure 6.
Autophagy in memory B cells. (A) Real-time RT-PCR for autophagy genes in naïve or GC B cells, or in memory B cells at 2, 4 or 8 weeks after immunization. Comparison to naïve B cells: **P<0.01 (n=3). (B) Immunocytochemistry staining for LC3 (green). Nuclei were stained with DAPI (blue). Scale bar: 5 μM. The number of LC3 punctates per cell were counted. Comparison to naïve B cells: **P<0.01 (n=10).
Increased expression of transcription factors for autophagy genes in memory B cells
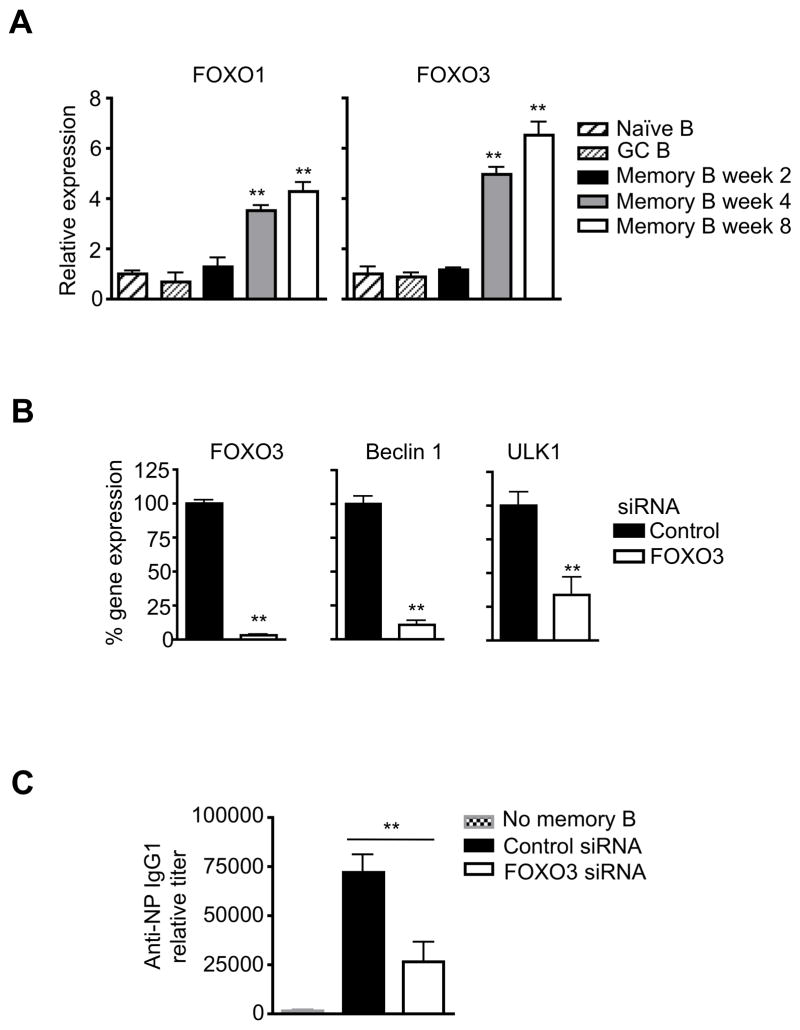
Next, we examined the potential mechanism for transcriptional regulation of autophagy genes in memory B cells. We found that several genes implicated in the activation of autophagy gene expression, including FOXO1 and FOXO3 (44), were increased in memory B cells at 4 or 8 weeks after immunization (Fig. 7A). This suggests that transcriptional control plays an important role in the expression of autophagy genes in memory B cells. Consistently, the expression of Beclin 1 and ULK1 that are important for the initiation of autophagy, was significantly decreased in memory B cells after silencing of FOXO3 (Fig. 7B). This suggests that FOXO3 is indeed important for increased autophagy gene expression in memory B cells.
Figure 7.
Expression of transcription factors for autophagy genes. (A) Real-time RT-PCR for FOXO1 and FOXO3 in naïve B cells or in memory B cells at 2, 4 or 8 weeks after immunization. Comparison to naïve B cells: **P<0.01 (n=3). (B) Memory B cells sorted at week 8 after immunization were transfected with siRNA targeting FOXO3. The cells were cultured for 2 days. RNA was extracted for real-time PCR for FOXO3, Beclin 1 and ULK1. **P<0.01 (n=3). (C) NP-specific memory B cells transfected with FOXO3 or control siRNA were transferred into recipient mice, followed by immunization with NP-KLH and analyses of NP-specific antibodies. **P<0.01 (n=5).
To determine whether transcriptional regulation of autophagy genes is indeed important for memory B cell functions, we test the effects of silencing of FOXO3 on memory B cell after adoptive transfer. We found that NP-specific memory B cells have reduced capacity in forming antibody secreting cells in recipient mice after re-challenge with the antigen (Fig. 7C). This supports the conclusion that transcriptional control of autophagy genes is important for the maintenance of memory B cell functions.
Activation of autophagy increases the survival of memory B cells
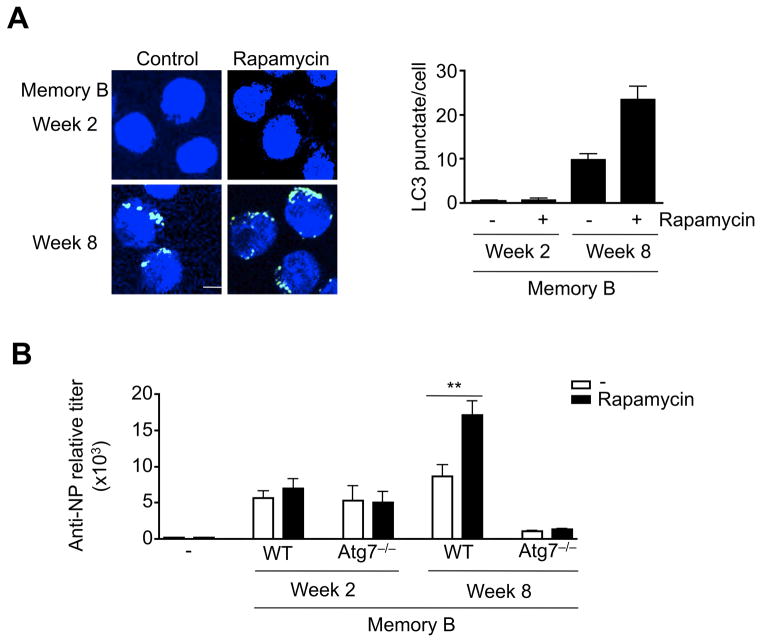
We have previously observed that treatment with rapamycin increased autophagy in memory B cells and improved their ability in forming antibody secreting cells after adoptive transfer (24). Although rapamycin increased autophagy in memory B cells from mice 8 weeks after immunization, it had no significant effect on memory B cells form mice 2 weeks after immunization (Fig. 8A). This is potentially due to the lack of autophagy machinery in memory B cells at this initial stage with the lack of expression of key autophagy genes.
Figure 8.
Activation of autophagy in memory B cells. (A) Memory B cells sorted at week 2 or 8 after immunization were treated with rapamycin. The cells were used for immunocytochemistry staining for LC3 (green). Nuclei were stained with DAPI (blue). Scale bar: 5 μM. The number of LC3 punctates per cells was counted. **P<0.01 (n=10). (B) Memory B cells were sorted from wild type or B/Atg7−/− mice at week 2 or 8 after immunization and transferred into recipient mice and re-challenged with NP-KLH. The mice were injected with rapamycin or PBS. Anti-NP IgG1 in the sera was measured at day 5 after adoptive transfer. **P<0.01 (2-week memory B cell transfer: n=5 for wild type and Atg7−/−; 8-week memory B cell transfer: n=5 for wild type and n=3 for Atg7−/−).
We then determined whether rapamycin could promote autophagy in vivo to improve the functions of memory B cells in forming antibody secreting cells. As expected, wild type memory B cells obtained 8 weeks after immunization showed increased ability in forming antibody secreting cells in the presence of rapamycin (Fig. 8B). In contrast, Atg7−/− memory B cells obtained 8 weeks after immunization did not show such an effect after rapamycin treatment (Fig. 8B). This suggests that the effect of rapamycin on memory B cells is mediated through autophagy. However, rapamycin did not have effect on memory B cells obtained 2 weeks after immunization from wild type or B/Atg7−/− mice (Fig. 8B). It is potentially more efficient to stimulate autophagy to improve memory B cell function during memory B cell maintenance stage when autophagy is active in these cells.
Discussion
Autophagy is essential for the long-term maintenance of memory B cells. Our current study indicates that autophagy is not required for the initial formation of memory B cells. Newly generated memory B cells displayed increased expression of Bcl-2 but not autophagy genes. Several transcription factors for the expression autophagy genes were increased in memory B cells over several weeks after their initial formation. We found no significant DNA methylation at promoter regions of several key autophagy genes in either GC or memory B cells, suggesting that epigenetic regulation at the DNA methylation levels is not major mechanism that regulate autophagy gene expression in these cells. Rather, the expression of transcription factors, such as FOXO1 and FOXO3, likely promotes autophagy gene expression. The ability for these newly formed memory B cells to develop into antibody secreting cells was independent of autophagy. Between 2 and 8 weeks after immunization, the expression of autophagy genes in memory B cells increased. These data suggest that after their initiation formation, memory B cells continue with a maturation process to further up-regulate Bcl-2 and induce the expression of autophagy genes.
How can newly formed memory B cells survive independent of autophagy is not known. They showed up-regulation of Bcl-2, but the level was still lower than that in memory B cells 8 weeks after immunization. It is possible that cytokines in the microenvironment, as well as survival signals provided in their development niche by other cell types, such helper T cells, may help to promote the survival of newly generated memory B cells. After leaving their initial development environment, the cells will need to gain intrinsic survival protection more independently from the environment and the presence of antigens. Up-regulation of autophagy therefore provides an important mechanism to increase their intrinsic ability in protecting themselves when antigen stimulation and T cell help are absent.
Transcriptional regulation of autophagy genes is a major mechanism for the regulation of autophagy (44). We found that transcription factors FOXO1 and FOXO3 were increased in memory B cells over time after immunization. Consistently, silencing of FOXO3 significantly suppressed the expression of autophagy gene in memory B cells. Increased expression of these transcription factor are likely to be important for the induction of autophagy genes in memory B cells. It will be interesting to determine whether these key transcription factors are potential pharmacological targets for the stimulation of autophagy in memory B cells.
Autophagy has been shown to play an important role in the survival of antibody-secreting plasma cells involving the protection against stress in the endoplasmic reticulum caused by antibody secretion (49, 50). Autophagy is therefore an important mechanism to protect different steps of antibody-mediated immune responses. It has also been shown that autophagy is important for T cell development by maintaining the function of the mitochondrion and endoplasmic reticulum (51, 52). In the CD8+ T cell compartment, it has been shown that autophagy deficiency does not affect effector T cell responses to antigen but causes severe defects in CD8+ memory T cells (53, 54). Therefore, the importance for autophagy in immunological memory lies in its functions in different compartments of the immune system. We observed that the expression of autophagy genes in memory B cells increased over time after the initial formation of these cells. It was not efficient to promote autophagy in memory B cells at the initial formation stages. This is probably due to the lack of autophagy machinery at this stage. Therefore, boosting autophagy would be most effective at memory B cell maintenance stage when autophagy genes are induced, but not at the initial memory B cell forming stage. Our study suggests it is effective to promote autophagy during the maintenance phase of immunological memory in the protection against infection.
Acknowledgments
This work was supported by grants from the Lupus Research Institute and the NIH, and by NIH T32 training grant to S.K. (GM08231-22).
We thank Dr. Zhenming Xu for advice on somatic mutation analyses, and Dr. Masaaki Komatsu for providing the Atg7flox mice.
Abbreviations
- GC
germinal center
- ASCs
antibody secreting cells
- FR
framework region
- CDR
complementarity-determining region
References
- 1.Lenardo M, Chan KM, Hornung F, McFarland H, Siegel R, Wang J, Zheng L. Mature T lymphocyte apoptosis--immune regulation in a dynamic and unpredictable antigenic environment. Annu Rev Immunol. 1999;17:221–253. doi: 10.1146/annurev.immunol.17.1.221. [DOI] [PubMed] [Google Scholar]
- 2.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer PH, Arnold R, Lavrik IN. Life and death in peripheral T cells. Nat Rev Immunol. 2007;7:532–542. doi: 10.1038/nri2115. [DOI] [PubMed] [Google Scholar]
- 4.Lanzavecchia A, Sallusto F. Progressive differentiation and selection of the fittest in the immune response. Nat Rev Immunol. 2002;2:982–987. doi: 10.1038/nri959. [DOI] [PubMed] [Google Scholar]
- 5.Kalia V, Sarkar S, Gourley TS, Rouse BT, Ahmed R. Differentiation of memory B and T cells. Curr Opin Immunol. 2006;18:255–264. doi: 10.1016/j.coi.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McHeyzer-Williams M, Okitsu S, Wang N, McHeyzer-Williams L. Molecular programming of B cell memory. Nat Rev Immunol. 2012;12:24–34. doi: 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurosaki T, Aiba Y, Kometani K, Moriyama S, Takahashi Y. Unique properties of memory B cells of different isotypes. Immunol Rev. 2010;237:104–116. doi: 10.1111/j.1600-065X.2010.00939.x. [DOI] [PubMed] [Google Scholar]
- 9.Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, Ravindran R, Stewart S, Alam M, Kwissa M, Villinger F, Murthy N, Steel J, Jacob J, Hogan RJ, Garcia-Sastre A, Compans R, Pulendran B. Programming the magnitude and persistence of antibody responses with innate immunity. Nature. 2011;470:543–547. doi: 10.1038/nature09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pape KA, Taylor JJ, Maul RW, Gearhart PJ, Jenkins MK. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331:1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012;247:52–63. doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 12.Berkowska MA, Driessen GJ, Bikos V, Grosserichter-Wagener C, Stamatopoulos K, Cerutti A, He B, Biermann K, Lange JF, van der Burg M, van Dongen JJ, van Zelm MC. Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood. 2011;118:2150–2158. doi: 10.1182/blood-2011-04-345579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 14.Ridderstad A, Tarlinton DM. Kinetics of establishing the memory B cell population as revealed by CD38 expression. J Immunol. 1998;160:4688–4695. [PubMed] [Google Scholar]
- 15.Vikstrom I, Carotta S, Luthje K, Peperzak V, Jost PJ, Glaser S, Busslinger M, Bouillet P, Strasser A, Nutt SL, Tarlinton DM. Mcl-1 is essential for germinal center formation and B cell memory. Science. 2010;330:1095–1099. doi: 10.1126/science.1191793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruyama M, Lam KP, Rajewsky K. Memory B-cell persistence is independent of persisting immunizing antigen. Nature. 2000;407:636–642. doi: 10.1038/35036600. [DOI] [PubMed] [Google Scholar]
- 17.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 18.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 20.Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio-Carrion A, Waterhouse NJ, Li CW, Mari B, Barbry P, Newmeyer DD, Beere HM, Green DR. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 21.Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 23.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 24.Chen M, Hong MJ, Sun H, Wang L, Shi X, Gilbert BE, Corry DB, Kheradmand F, Wang J. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat Med. 2014;20:503–510. doi: 10.1038/nm.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Narvajas Artal-Martinez A, Gomez TS, Zhang JS, Mann AO, Taoda Y, Gorman JA, Herreros-Villanueva M, Gress TM, Ellenrieder V, Bujanda L, Kim DH, Kozikowski AP, Koenig A, Billadeau DD. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33:3983–3993. doi: 10.1128/MCB.00813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla S, Patric IR, Patil V, Shwetha SD, Hegde AS, Chandramouli BA, Arivazhagan A, Santosh V, Somasundaram K. Methylation silencing of ULK2, an autophagy gene, is essential for astrocyte transformation and tumor growth. J Biol Chem. 2014;289:22306–22318. doi: 10.1074/jbc.M114.567032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 30.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJ, III, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 33.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashimoto K, Kokubun S, Itoi E, Roach HI. Improved quantification of DNA methylation using methylation-sensitive restriction enzymes and real-time PCR. Epigenetics. 2007;2:86–91. doi: 10.4161/epi.2.2.4203. [DOI] [PubMed] [Google Scholar]
- 36.Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Xu P, Das M, Reilly J, Davis RJ. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–322. doi: 10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 41.Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez AM, Csibi A, Raibon A, Cornille K, Gay S, Bernardi H, Candau R. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem. 2012;113:695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 43.Schips TG, Wietelmann A, Hohn K, Schimanski S, Walther P, Braun T, Wirth T, Maier HJ. FoxO3 induces reversible cardiac atrophy and autophagy in a transgenic mouse model. Cardiovasc Res. 2011;91:587–597. doi: 10.1093/cvr/cvr144. [DOI] [PubMed] [Google Scholar]
- 44.Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 45.Zuccarino-Catania GV, Sadanand S, Weisel FJ, Tomayko MM, Meng H, Kleinstein SH, Good-Jacobson KL, Shlomchik MJ. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat Immunol. 2014;15:631–637. doi: 10.1038/ni.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lebecque S, de Bouteiller O, Arpin C, Banchereau J, Liu YJ. Germinal center founder cells display propensity for apoptosis before onset of somatic mutation. J Exp Med. 1997;185:563–571. doi: 10.1084/jem.185.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi Y, Ohta H, Takemori T. Fas is required for clonal selection in germinal centers and the subsequent establishment of the memory B cell repertoire. Immunity. 2001;14:181–192. doi: 10.1016/s1074-7613(01)00100-5. [DOI] [PubMed] [Google Scholar]
- 48.Yang Z, Sullivan BM, Allen CD. Fluorescent in vivo detection reveals that IgE(+) B cells are restrained by an intrinsic cell fate predisposition. Immunity. 2012;36:857–872. doi: 10.1016/j.immuni.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 49.Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, Fagioli C, Merlini A, Mariani E, Pasqualetto E, Orfanelli U, Ponzoni M, Sitia R, Casola S, Cenci S. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- 50.Conway KL, Kuballa P, Khor B, Zhang M, Shi HN, Virgin HW, Xavier RJ. ATG5 regulates plasma cell differentiation. Autophagy. 2013;9:528–537. doi: 10.4161/auto.23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 52.Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186:1564–1574. doi: 10.4049/jimmunol.1001822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, Watson AS, Cerundolo V, Townsend AR, Klenerman P, Simon AK. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife. 2014;3 doi: 10.7554/eLife.03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X, Araki K, Li S, Han JH, Ye L, Tan WG, Konieczny BT, Bruinsma MW, Martinez J, Pearce EL, Green DR, Jones DP, Virgin HW, Ahmed R. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol. 2014;15:1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]