Abstract
Increased expression of the immuno-suppressive cytokines, transforming growth factor-β1 (TGFβ1) and interleukin-10 (IL-10), is a hallmark of the advanced stages of cutaneous T cell lymphoma (CTCL), where it has been associated with suppressed immunity, increased susceptibility to infections, and diminished antitumor responses. Yet, little is known about the transcriptional regulation of TGFβ1 and IL-10 in CTCL, and about their function in regulating the CTCL cell responses. Here, we show that TGFβ1 and IL-10 expression in CTCL cells is regulated by NFκB, and suppressed by bortezomib (BZ), which has shown promising results in the treatment of CTCL. However, while the TGFβ1 expression is IκBα-dependent and is regulated by the canonical pathway, the IL-10 expression is IκBα-independent, and its inhibition by BZ is associated with increased promoter recruitment of p52 that characterizes the non-canonical pathway. TGFβ1 suppression decreases CTCL cell viability and increases apoptosis, and adding exogenous TGFβ1 increases viability of BZ-treated CTCL cells, indicating TGFβ1 pro-survival function in CTCL cells. In addition, TGFβ1 suppression increases expression of the pro-inflammatory cytokines IL-8 and IL-17 in CTCL cells, suggesting that TGFβ1 also regulates the IL-8 and IL-17 expression. Importantly, our results demonstrate that BZ inhibits expression of the chemokine receptor CXCR4 in CTCL cells, resulting in their decreased migration, and that the CTCL cell migration is mediated by TGFβ1. These findings provide the first insights into the BZ-regulated TGFβ1 and IL-10 expression in CTCL cells, and indicate that TGFβ1 has a key role in regulating CTCL survival, inflammatory gene expression, and migration.
Keywords: Bortezomib, cutaneous T-cell lymphoma, CXCL12, CXCR4, IL-10, NFκB, TGFβ1
Introduction
Enhanced TGFβ1 and IL-10 expression by T regulatory (Treg) cells is a hallmark of the advanced stages of CTCL, especially mycosis fungoides (MF) and the leukemic variant, Sézary syndrome (SS) (1–5). The increased levels of IL-10 and TGFβ1 have been associated with suppressed immunity, increased susceptibility to infections, and diminished antitumor responses in CTCL patients (6–10). In addition, the advanced stages of CTCL are associated with the increased expression of the chemokine receptor CXCR4 and its ligand CXCL12, also known as stromal cell-derived factor 1 (SDF-1); both CXCR4 and CXCL12 have been linked with the increased CTCL cell proliferation, skin recruitment and accumulation (11–14).
At the transcriptional level, both IL-10 and TGFβ1 expression is regulated by the transcription factor NFκB (15–21). However, compared to anti-apoptotic and pro-inflammatory genes, the transcriptional regulation of immunosuppressive genes by NFκB is much less understood. Majority of genes involved in inflammatory response are regulated by NFκB canonical pathway that is IκBα-dependent, and involves predominantly p65 and p50 subunits. In contrast, lymphocyte development is regulated by non-canonical pathway, which is IκBα-independent, and activates complexes containing mostly RelB and p52 (22, 23). Binding of the individual NFκB dimers, and other transcription factors and co-regulators to different κB sites provides the transcriptional selectivity of NFκB responses (24–26). The NFκB DNA binding activity is constitutively increased in CTCL cells, and has been associated with the high CTCL cell survival and resistance to chemotherapy (27–32). There is no effective strategy to prolong survival of patients with advanced stages of CTCL; thus, novel therapeutic strategies are needed (33–38).
Bortezomib (BZ, Velcade, PS-341) is the first FDA approved proteasome inhibitor that has shown remarkable anti-tumor activity in the treatment of patients with multiple myeloma (39, 40). BZ has shown promising results also in patients with relapsed or refractory CTCL (41–44). However, the precise molecular mechanisms are not fully understood (45, 46). BZ has been originally developed as an inhibitor of the IκBα-dependent canonical pathway, resulting in the inhibition of NFκB-dependent anti-apoptotic genes in cancer cells (39, 40). However, we have recently shown that the proteasome inhibition also induces nuclear translocation and accumulation of IκBα, which has a promoter specific effect on the regulation of NFκB-dependent genes, depending on the subunit composition of NFκB proteins recruited to the different κB binding sites (47–50).
In this study, we report that both TGFβ1 and IL-10 expression in CTCL cells is suppressed by the BZ-mediated proteasome inhibition. However, while the TGFβ1 expression is regulated by the IκBα-dependent canonical pathway, the IL-10 expression is IκBα-independent, and its inhibition is associated with the increased promoter recruitment of p52 that characterizes the non-canonical pathway. In addition, TGFβ1 suppression induces apoptosis, and IL-8 and IL-17 expression in CTCL cells, indicating that TGFβ1 regulates survival and pro-inflammatory gene expression in CTCL. Importantly, our results indicate that TGFβ1 mediates the CXCL12/CXCR4-dependent CTCL cell migration, and suggest that BZ inhibits the CTCL migration, at least partly, by inhibiting TGFβ1.
Materials and Methods
Antibodies and reagents
Purified antibodies against human IκBα (sc-371), NFκB p65 (sc-372X), p50 (sc-7178X), cRel (sc-71X), RelB (sc-226X), p52 (sc-848X), CXCR4 (sc-9046), TβRI (sc-398), TβRII (sc-17792), and lamin B (sc-6216) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Recombinant human TGFβ1 (240-B-002) and CXCL12 (350-NS-010) proteins, TGFβ neutralizing antibody (MAB1835), and the corresponding isotype control antibody (MAB002) were from R&D Systems (Minneapolis, MN, USA). Purified polyclonal antibody against lactate dehydrogenase (LDH; 20-LG22) was from Fitzgerald Industries International (Concord, MA, USA), and actin antibody was from Sigma (St Louis, MO, USA). Horseradish peroxidase (HRP)-conjugated anti-rabbit, anti-mouse and anti-goat secondary antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Bortezomib was from ChemieTek (Indianapolis, IN, USA). All other reagents were molecular biology grade and were from Sigma (St Louis, MO).
Cell culture
CTCL cell lines, Hut-78 (ATCC® TIB-161), H9 (ATCC® HTB-176), and HH (ATCC® CRL-2105) cells, derived from peripheral blood of patients with SS and non-MF/SS aggressive CTCL were obtained from the American Type Culture Collection (ATCC; Rockville, MD, USA), and used between the passage numbers 10 and 15. Peripheral blood mononuclear cells (PBMC) from healthy human volunteers were purchased from Zen-Bio (SER-PBMC-200; Research Triangle Park, NC, USA). Cells were maintained at 37°C in RPMI 1640 medium, supplemented with 10% heat inactivated fetal bovine serum (FBS) and 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin, in a humidified atmosphere with 5% CO2. Prior to cell treatment, cells were seeded (5×105 cells/ml) in 6-well plates and grown 24 hours at 37°C with 5% CO2. Bortezomib was dissolved in DMSO and stored at −80°C. An equivalent volume of DMSO was used in all experiments as a solvent control. Cell viability was measured by using Trypan Blue exclusion.
Transfections
Human IκBα (sc-29360), TGFβ1 (sc-44146), TβRI (sc-40222), TβRII (sc-36657), and non-silencing (sc-37007) small interfering RNAs (siRNAs) were obtained from Santa Cruz Biotechnology. Prior to transfections, cells were seeded (5×105 cells/ml) into a 6-well plate and incubated in a humidified 5% CO2 atmosphere at 37°C in antibiotic-free RPMI medium supplemented with 10% FBS for 24 hours. For each transfection, 50 nmol (final concentration) of siRNA was used. Cells were transfected with TransIT-siQUEST transfection reagent (Mirus Bio, Madison, WI, USA) as described (50). After transfection, fresh RPMI medium supplemented with FBS and antibiotics was added and cells were incubated for 48 hours.
Preparation of cell extracts and western blotting
Whole cell extracts (WCE), and cytoplasmic (CE) and nuclear extracts (NE) were prepared as described previously (47–50), and separated on 12% SDS gels. Contamination of nuclear and cytoplasmic fractions by cytoplasmic and nuclear proteins, respectively, was determined by western analysis using LDH and lamin B as specific markers as described (47–50). To determine equal protein loading, membranes were stripped and re-probed with anti-actin antibody as described (50).
Real time RT-PCR
Total RNA was isolated by using RNeasy mini-kit (Qiagen, Valencia, CA, USA). The iScript one-step RT-PCR kit with SYBR Green (BioRad, Hercules, CA, USA) was used as a supermix and 20 ng of RNA was used as template on a Bio-Rad MyIQ Single Color Real-Time PCR Detection System (BioRad). The primers used for quantification of TGFβ1, IL-10, IL-8, IL-17, CXCR4, and actin mRNA were purchased from SA Biosciences (Frederick, MD, USA).
Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed as described (51). Briefly, proteins and DNA were cross-linked by formaldehyde, cells were washed and sonicated. The lysates were centrifuged (15,000 g, 10 min, 4 °C), and the supernatant extracts were diluted with ChIP dilution buffer and pre-cleared with Protein A/G Agarose (Santa Cruz, CA) for 2 hours at 4 °C. Immunoprecipitation was performed overnight at 4 °C, with p65, p50, cRel, RelB or p52 antibodies. Following immunoprecipitation, the samples were incubated with Protein A/G Agarose (1 h, 4 °C), and the immune complexes were collected by centrifugation (150 g, 5 min, 4 °C), washed, and extracted with 1% SDS–0.1 M NaHCO3. After reversing the cross-linking, proteins were digested with proteinase K, and the samples were extracted with phenol/chloroform, followed by precipitation with ethanol. The pellets were resuspended in nuclease-free water and subjected to real time PCR. Immunoprecipitated DNA was analyzed by real-time PCR (25 μl reaction mixture) using the iQ SYBR Green Supermix and the Bio-Rad MyIQ Single Color Real-Time PCR Detection System (Bio-Rad). Each immunoprecipitation was performed at least three times using different chromatin samples, and the occupancy was calculated by using the human IGX1A negative control primers (SA Biosciences, Frederick, MD, USA), which detect specific genomic ORF-free DNA sequence that does not contain binding site for any known transcription factors. The results were calculated as fold difference in occupancy of the particular protein at the particular locus in comparison with the IGX1A locus. Primers for TGFβ1 and IL-10 were as follows: TGFβ1: Forward, 5′-ATCTCCCTCCCACCTCCCT-3′ and reverse, 5′-TCCTGCTCGTCTCAGACTCTG-3′; IL-10-κB1: forward, 5′-CTATGGAATTGAGGCTC-TTGC-3′ and reverse, 5′-GCAGAAGCAGTTAGAGGTGC-3′; IL-10-κB2: forward, 5′-CAGCCAATGTGGAATTCCC-3′ and reverse, 5′-GTGGCCAAGGCTCAAACAC-3′; IL-10-κB3: forward, 5′-TCCCACTCACTTCACGCTG-3′ and reverse, 5′-AGATGGAGGGCATTTCCAGT-3′.
Apoptosis assay
Apoptosis was evaluated with a cell death detection ELISA kit that quantifies release of nucleosomes into the cytoplasm (Cell Death Detection ELISAPLUS, Roche, Indianapolis, IN) as described (47). The assay was performed at the indicated time points as per the manufacturer’s instructions.
ELISA
TGFβ1, IL-10, and CXCL12 cytokine release was measured in cell culture supernatants by commercially available ELISA kits (R&D, Minneapolis, MN) as previously described (50).
Transwell migration assay
Migration assays were performed using 8-μm pore polyester Transwell inserts (Corning, MA) equilibrated in PBS buffer. Cells were pre-cultured 12 hours in serum-free RPMI medium, counted, re-suspended (5×104) in 100 μl of serum-free RPMI medium containing 0, 10 or 100 nM BZ, or 300 ng/ml of TGFβ neutralizing antibody or IgG1 isotype control, and added to the upper chamber. RPMI medium (650 μl) containing 10% FBS was added to the lower chamber with or without 50 ng/ml of CXCL12 (12). Migration was allowed to proceed for 6 hours at 37°C in 5% CO2 incubator. The inserts were then removed, washed with PBS, fixed 10 minutes in 100% methanol, and stained for 30 minutes by using 2% crystal violet. After cells were stained, the inserts were washed with water and allowed to dry before counting the migrated cells. Migrated cells were counted in five random fields. Results are expressed as migration index, which represents the ratio between the numbers of migrated treated and untreated (UT) cells.
Statistical analysis
The results represent at least three independent experiments. Numerical results are presented as means +/− SE. Data were analyzed by using an InStat software package (GraphPAD, San Diego, CA). Statistical significance was evaluated by using Mann-Whitney U test with Bonferroni correction for multiple comparisons, and p<0.05 was considered significant.
Results
Proteasome inhibition down-regulates TGFβ1 and IL-10 expression in CTCL cells
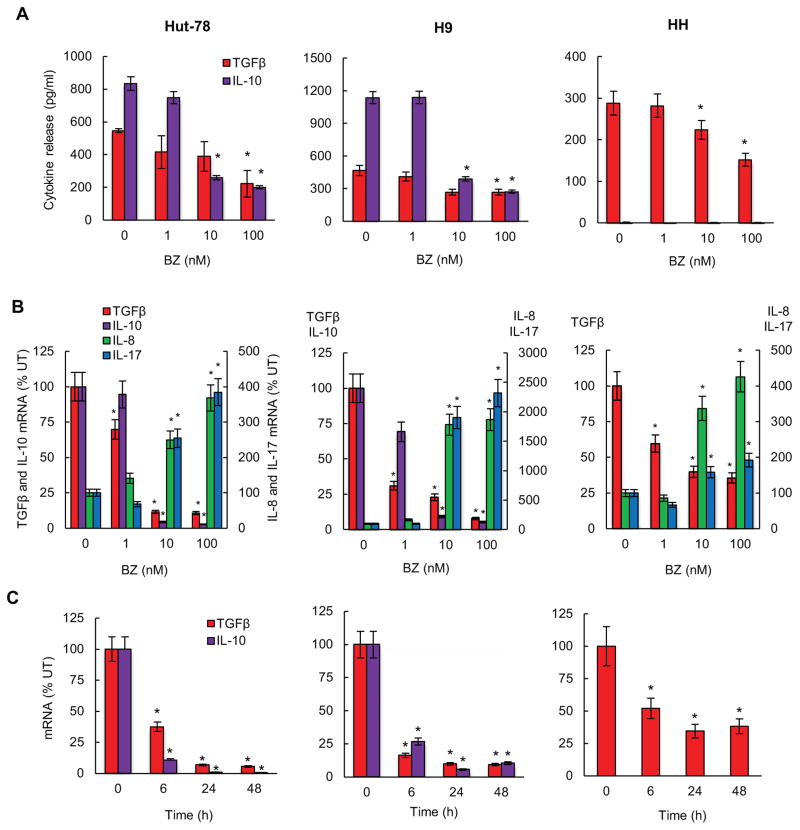
Since we have previously shown that proteasome inhibition has a promoter-specific effect on the expression of NFκB-dependent genes (47, 50), we wanted to determine whether BZ regulates expression of the immunosuppressive cytokines TGFβ1 and IL-10 in CTCL cells. To this end, we first measured TGFβ1 and IL-10 release from CTCL Hut-78 (left panels), H9 (middle panels) and HH cells (right panels) incubated 24 hours with increasing BZ concentrations. All three CTCL cell types release considerable amounts of TGFβ1, and 100 nM BZ, which approximately corresponds to the clinically used BZ concentrations (52), significantly inhibits the TGFβ1 release from all three CTCL cells (Fig. 1A). In contrast, IL-10 is released only by Hut-78 and H9 cells, but not HH cells, and 10 and 100 nM BZ concentrations significantly inhibit the IL-10 release (Fig. 1A).
Figure 1. Proteasome inhibition suppresses TGFβ1 and IL-10 expression in CTCL cells.
(A) ELISA assay of TGFβ1 and IL-10 release measured in cell culture supernatants of Hut-78 (left panels), H9 (middle panels), and HH (right panels) cells incubated 24 hours with increasing BZ concentrations. (B) Real time RT-PCR analysis of TGFβ1, IL-10, IL-8 and IL-17 mRNA levels in Hut-78, H9, and HH cells treated 24 hours with increasing BZ concentrations. (C) Real time RT-PCR of TGFβ1 and IL-10 mRNA levels in Hut-78, H9, and HH cells treated 0, 6, 24, and 48 hours with 10 nM BZ. The values in Figs. 1A-C represent the mean +/−SE of four experiments. Asterisks denote a statistically significant (p<0.05) change compared to control untreated (UT) cells.
10 and 100 nM BZ also greatly reduced the mRNA levels of TGFβ1 and IL-10 in all CTCL cells (Fig. 1B). To ensure that the decreased expression of TGFβ1 and IL-10 in BZ-treated cells was not caused by the BZ-induced apoptosis (47), we have analyzed, as a control, expression of the NFκB-dependent pro-inflammatory genes IL-8 and IL-17. In contrast to the decreased mRNA levels of TGFβ1 and IL-10, the expression of IL-8 and IL-17 was significantly increased in CTCL cells incubated with 10 and 100 nM BZ (Fig. 1B), demonstrating specificity of BZ effect on the expression of NFκB-dependent genes. The BZ inhibition of TGFβ1 and IL-10 gene expression in all three CTCL cell types was time dependent (Fig. 1C).
TGFβ1 inhibition is regulated by IκBα, while IL-10 inhibition is IκBα-independent
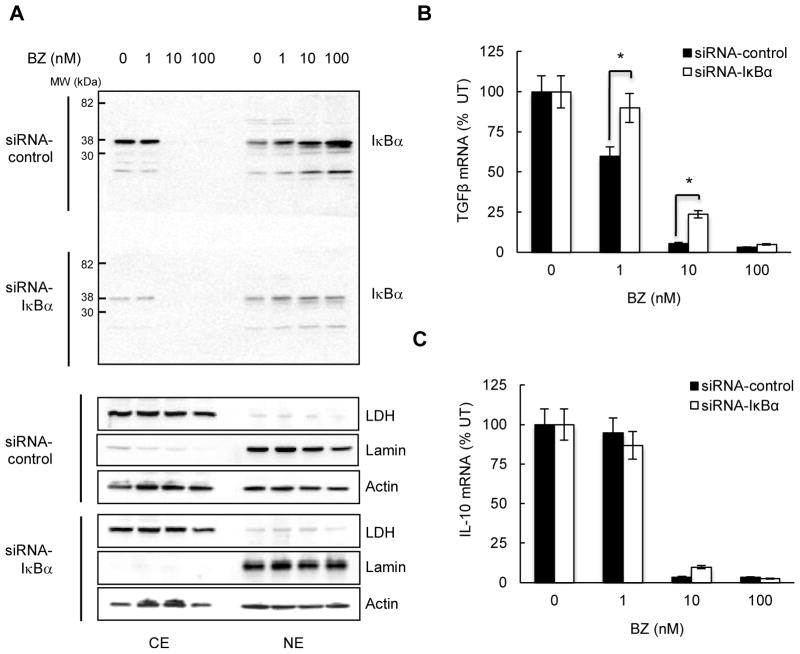
Our previous studies have demonstrated that proteasome inhibition induces nuclear translocation and accumulation of IκBα, which has a promoter-specific effect on the inhibition of NFκB-dependent genes (47–50). Thus, we wanted to determine whether the BZ-induced inhibition of TGFβ1 and IL-10 expression is IκBα-dependent. Hut-78 cells were transfected with control and IκBα specific siRNA, treated with BZ, and analyzed for IκBα, TGFβ1 and IL-10 expression. In agreement with our previous observations (47–50), 10 and 100 nM BZ induced the nuclear translocation and accumulation of IκBα in cells transfected with control siRNA (Fig. 2A, top panel). Transfection with IκBα siRNA reduced both the cytoplasmic and the nuclear IκBα levels in BZ-treated cells (Fig. 2A, top panel, bottom gel, exposed together with the control siRNA gel). However, while the TGFβ1 mRNA levels were increased in cells transfected with IκBα siRNA (Fig. 2B), the IL-10 mRNA levels were not affected by IκBα suppression (Fig. 2C). These data indicated that the BZ-mediated TGFβ1 inhibition is IκBα-dependent, while the IL-10 inhibition is not regulated by IκBα.
Figure 2. TGFβ1 inhibition is mediated by IκBα, while IL-10 inhibition is IκBα-independent.
(A) Western blotting of cytoplasmic (CE) and nuclear (NE) extracts prepared from Hut-78 cells transfected with control non-silencing (top gel) and IκBα specific (bottom gel) siRNA, incubated 24 hours with increasing BZ concentrations, and analyzed by using IκBα antibody. Both gels were transferred to a membrane and exposed together at the same time. The purity of cytoplasmic and nuclear fractions was monitored by using lactate dehydrogenase (LDH) and lamin B antibodies. To confirm equal protein loading, the membranes were stripped and re-probed with actin antibody. (B) TGFβ1 and (C) IL-10 mRNA levels in Hut-78 cells transfected with control and IκBα siRNA, and treated 24 hours with increasing BZ concentrations. The values represent the mean +/−SE of four experiments; the asterisks denote a statistically significant (p<0.05) change compared to cells transfected with control siRNA.
TGFβ1 is regulated by NFκB canonical pathway, while IL-10 is regulated by non-canonical pathway in CTCL cells
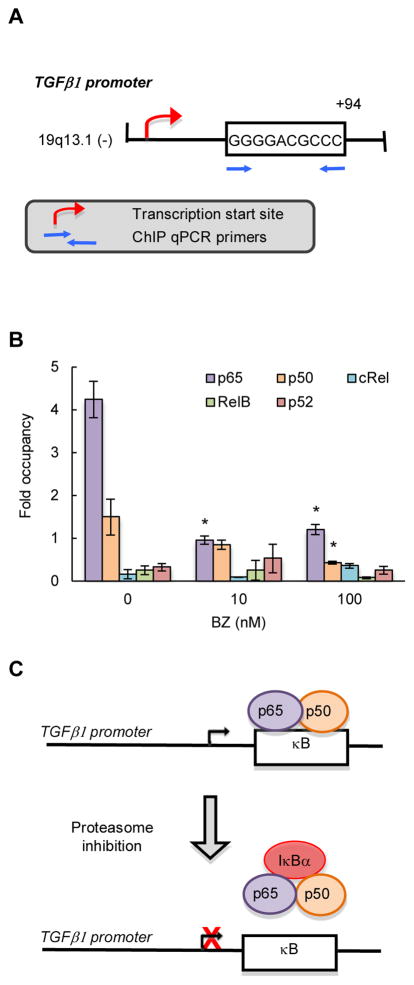
The dependence of TGFβ1 expression on IκBα has suggested that TGFβ1 is regulated by the canonical NFκB pathway. Using TFSEARCH program, we have identified NFκB-binding site (GGGGACGCCC) located +94 from the transcription start site in human TGFβ1 promoter (Fig. 3A). Even though we have recently shown that p65, p50 and RelB NFκB subunits are localized in the nucleus in CTCL cells (50), chromatin immunoprecipitation (ChIP) analysis using p65, p50, RelB, cRel and p52 antibodies has demonstrated that only p65 and p50 are recruited to the TGFβ1 promoter in Hut-78 cells (Fig. 3B). Bortezomib significantly decreased the p65/p50 recruitment (Fig. 3B), correlating with the BZ-reduced TGFβ1 expression (Fig. 1). Together with the data demonstrating that the TGFβ1 expression is IκBα-dependent (Fig. 2B), these results have indicated that the TGFβ1 expression in CTCL cells is mediated by the classical canonical pathway (Fig. 3C).
Figure 3. TGFβ1 is regulated by NFκB canonical pathway in CTCL cells.
(A) Schematic illustration of NFκB binding site in human TGFβ1 promoter, and the ChIP primers used in the ChIP assay. (B) Recruitment of NFκB p65, p50, cRel, RelB and p52 subunits to TGFβ1 promoter in Hut-78 cells treated 24 hours with 0, 10 and 100 nM BZ was analyzed by ChIP and quantified by real time PCR. The data are presented as the difference in occupancy of each protein between the TGFβ1 promoter and the IGX1A (SA Biosciences) negative control locus, and represent the mean +/−SE of four experiments. Asterisks denote a statistically significant (p<0.05) change compared to control (no BZ) cells. (C) Model of the regulation of TGFβ1 transcription in CTCL cells.
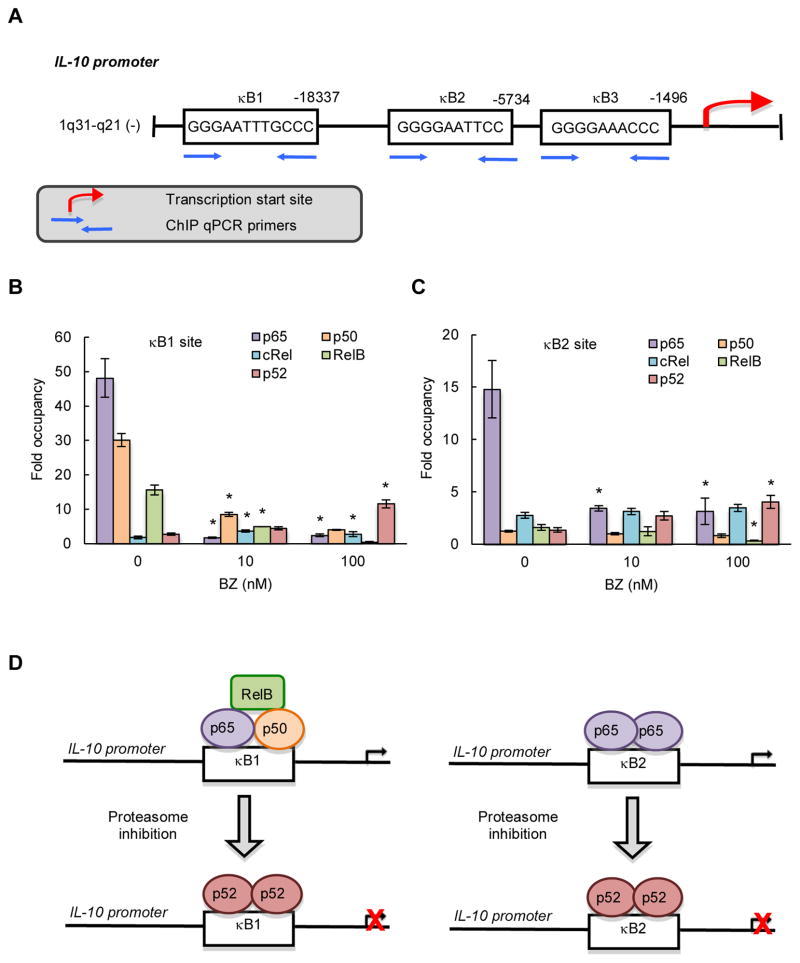
The human IL-10 promoter contains three putative NFκB binding sites: κB1 (GGGAA-TTTGCCC; −18,337), κB2 (GGGGAATTCC; −5,734), and κB3 (GGGGAAACCC; −1,496) (Fig. 4A). p65, p50 and RelB were all heavily recruited to the κB1 site, and this recruitment was decreased by BZ. Interestingly, BZ concomitantly increased the p52 recruitment (Fig. 4B); this is in an agreement with our recent data demonstrating that proteasome inhibition induces p52 accumulation in the nucleus in CTCL cells (50). The κB2 site was occupied predominantly by p65 (Fig. 4C), indicating that the κB2 site is regulated by p65 homodimers (Fig. 4D). Proteasome inhibition by BZ that decreased the IL-10 expression (Fig. 1), significantly reduced p65 recruitment to the κB2 site, but increased the recruitment of p52 (Figs. 4C, D). We did not find any significant recruitment of p65, p50, RelB, cRel or p52 to the κB3 site (data not shown). These results have indicated that the IL-10 expression is regulated by the IκBα-independent non-canonical pathway, and that the BZ-mediated inhibition of IL-10 expression is associated with increased recruitment of p52 NFκB that functions as a repressor of IL-10 transcription in CTCL cells (Fig. 4D).
Figure 4. IL-10 is regulated by NFκB non-canonical pathway in CTCL cells.
(A) Schematic illustration of NFκB binding sites in human IL-10 promoter, and the ChIP primers used in the ChIP assay. Recruitment of NFκB p65, p50, cRel, RelB and p52 subunits to the IL-10 κB1 (B) and κB2 (C) sites in Hut-78 cells treated 24 hours with 0, 10 and 100 nM BZ was analyzed by ChIP and quantified by real time PCR. The data are presented as the difference in occupancy of each protein between the κB1 and κB2 sites and the IGX1A negative control locus, and represent the mean +/−SE of four experiments. Asterisks denote a statistically significant (p<0.05) change compared to control (no BZ) cells. (D) Model of the regulation of IL-10 transcription by the exchange of NFκB subunits in CTCL cells.
TGFβ1 increases CTCL cell viability, while inhibiting pro-inflammatory gene expression
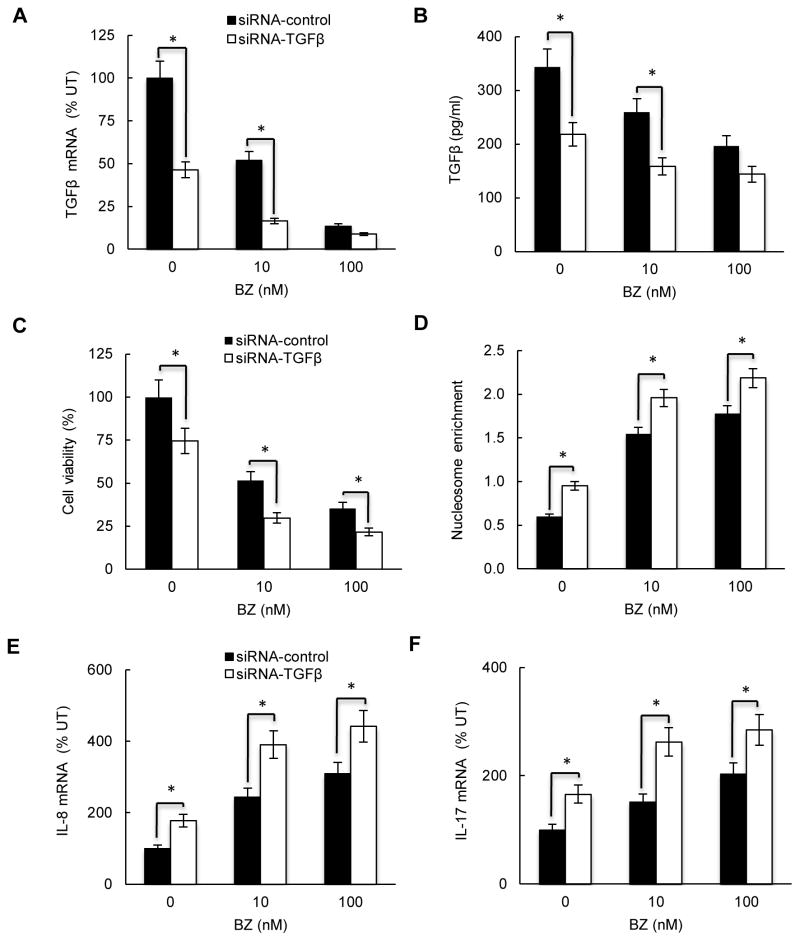
In most cell types, TGFβ1 has growth inhibitory and pro-apoptotic effects; however, in some tumors, TGFβ1 has tumor-promoting effects that include increased cell proliferation, angiogenesis, and metastasis (53, 54). To investigate the TGFβ1 effect on survival and pro-inflammatory gene expression in CTCL cells, we suppressed TGFβ1 in HH cells that express TGFβ1 in the absence of IL-10 (Fig. 1). HH cells were transfected with control or TGFβ1 specific siRNA, and incubated 24 hours with 0, 10 and 100 nM BZ. As expected, suppression of TGFβ1 with siRNA resulted in significantly decreased TGFβ1 mRNA levels (Fig. 5A) and TGFβ1 release (Fig. 5B). Importantly, TGFβ1 suppression significantly decreased the viability of untreated and BZ-treated HH cells (Fig. 5C), indicating that TGFβ1 induces survival in CTCL cells, and suggesting that the BZ-induced apoptosis is mediated, at least partly, by the BZ-reduced TGFβ1 levels. To test whether the BZ-induced apoptosis is mediated by the reduced TGFβ1 levels, we evaluated apoptosis by the quantitative cytoplasmic nucleosome release assay. As shown in Fig. 5D, suppression of TGFβ1 by siRNA significantly increased the nucleosome release in untreated and BZ-treated HH cells, indicating that TGFβ1 mediates the spontaneous and BZ-induced apoptosis in CTCL cells. To investigate whether TGFβ1 contributes to the BZ increased IL-8 and IL-17 expression in CTCL cells (shown in Fig. 1B), we analyzed IL-8 and IL-17 mRNA levels in HH cells transfected with TGFβ1 and control siRNA. As shown in Figs. 5E and F, cells transfected with TGFβ1 siRNA exhibited significantly higher IL-8 and IL-17 mRNA levels than cells transfected with control siRNA, indicating that TGFβ1 suppresses IL-8 and IL-17 expression in CTCL cells.
Figure 5. TGFβ1 suppression decreases viability, and increases pro-inflammatory gene expression in CTCL cells.
HH cells were transfected with control (full columns) or TGFβ1 specific siRNA (empty columns), treated 24 hours with 0, 10 and 100 nM BZ, and analyzed for TGFβ1 mRNA expression by real time RT-PCR (A), and for TGFβ1 release by ELISA (B). (C) Cell viability measured by Trypan Blue exclusion, and (D) apoptosis analyzed by the cytoplasmic nucleosome enrichment assay in HH cells transfected with control or TGFβ1 specific siRNA. Real time RT-PCR analysis of IL-8 (E) and IL-17 (F) mRNA levels in HH cells transfected with control (full columns) or TGFβ1 specific siRNA (empty columns) and treated 24 hours with 0, 10 and 100 nM BZ. The values in Figs. 5A-F represent the mean +/−SE of four experiments. Asterisks denote a statistically significant (p<0.05) change compared to cells transfected with the corresponding control siRNA.
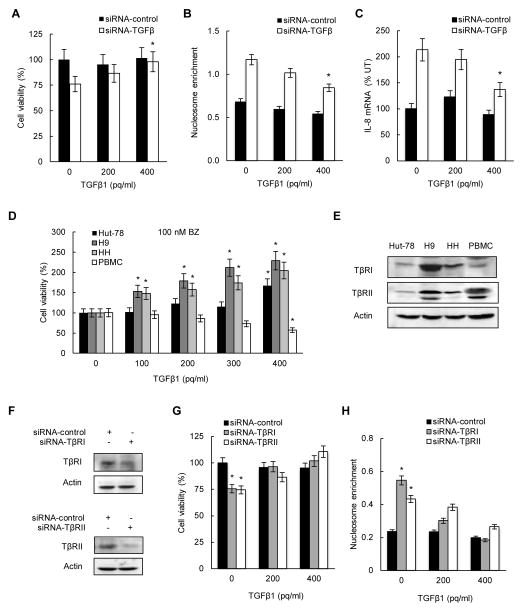
The fact that TGFβ1 suppression decreased viability, and increased apoptosis, and IL-8 and IL-17 expression also in BZ-untreated cells, indicated that TGFβ1 increases CTCL cell viability, and inhibits apoptosis and inflammatory gene expression independently of bortezomib. To test this hypothesis, we have analyzed cell viability, apoptosis, and IL-8 expression in HH cells transfected with control siRNA (producing about 350 pg/ml of TGFβ1; Fig. 5B) and TGFβ1 siRNA (producing about 200 pg/ml of TGFβ1; Fig. 5B), and incubated with 0, 200 and 400 pg/ml of exogenously added recombinant TGFβ1 protein. As shown in Fig. 6A, while addition of TGFβ1 did not have any significant effect on the viability of cells transfected with control siRNA, it increased the viability of TGFβ1 siRNA transfected cells; addition of 400 pg/ml of TGFβ1 reversed the decreased viability in TGFβ1 siRNA transfected cells. Similarly, exogenously added TGFβ1 decreased apoptosis (Fig. 6B) and IL-8 mRNA expression (Fig. 6C) in TGFβ1 siRNA transfected cells, while it did not have any significant effect on cells transfected with control siRNA.
Figure 6. Exogenous TGFβ1 increases viability of BZ-treated CTCL cells.
HH cells were transfected with control (full columns) or TGFβ1 specific (empty columns) siRNA, incubated 24 hours with 0, 200 and 400 pg/ml of exogenously added recombinant TGFβ1 protein, and analyzed for cell viability (A), cytoplasmic nucleosome enrichment (B), and IL-8 mRNA levels (C). (D) Viability of Hut-78, H9, HH, and PBMC cells incubated 24 hours with increasing amounts of exogenously added TGFβ1 protein, in the presence of 100 nM BZ. The values in Figs. 6A–D represent the mean +/−SE of four experiments; asterisks denote a statistically significant (p<0.05) change compared to corresponding cells incubated without exogenously added TGFβ1. (E) Western blotting of whole cell extracts (WCE) prepared from untreated Hut-78, H9, HH, and PBMC cells, and analyzed by using TβRI, TβRII, and control actin antibodies. (F) Western analysis of TβRI and TβRII protein levels in WCE of HH cells transfected with TβRI, TβRII, or control siRNA. (G) Cell viability and (H) nucleosome enrichment analyzed in HH cells transfected with TβRI, TβRII, or control siRNA, and incubated 24 hours with 0, 200 and 400 pg/ml of exogenously added TGFβ1. The values in Figs. 6G and H represent the mean +/−SE of four experiments. Asterisks denote a statistically significant change compared to cells transfected with the corresponding control siRNA.
To determine whether TGFβ1 increases viability also in other CTCL cells, we tested effect of exogenously added recombinant TGFβ1 protein on the viability of untreated and 100 nM BZ-treated Hut-78, H9, and HH cells, as well as normal human peripheral blood mononuclear cells (PBMC) used as a control. Addition of recombinant TGFβ1 did not have any significant effect on the viability of untreated cells (data not shown); this is consistent with the results demonstrating that TGFβ1 does not affect viability of HH cells transfected with control siRNA (Fig. 6A). Interestingly, addition of exogenous TGFβ1 significantly increased the viability of all three BZ-treated CTCL cells, but decreased the viability of BZ-treated PBMCs (Fig. 6D), indicating that TGFβ1 has an opposing effect on the viability of BZ-treated CTCL cells and normal PBMCs.
To examine whether TGFβ1 regulates cell viability through its receptors TβRI and TβRII, we have analyzed TβRI and TβRII protein levels in CTCL cells as well as in PBMCs, and tested whether TβRI and TβRII suppression decreases CTCL cell viability, and whether addition of exogenous TGFβ1 reverses this effect. As shown in Fig. 6E, all three CTCL cells as well as PBMCs express both TβRI and TβRII, even though the protein expression of both receptors is lower in Hut-78 and HH cells compared to H9 cells. Importantly, suppression of TβRI and TβRII in HH cells with siRNA (Fig. 6F) significantly decreased cell viability (Fig. 6G) and increased apoptosis (Fig. 6H), and addition of exogenous recombinant TGFβ1 reversed these effects, suggesting that TGFβ1 induces CTCL cell survival through the TβRI/TβRII signaling.
BZ inhibits CXCR4 expression in CTCL cells via TGFβ1-dependent mechanism
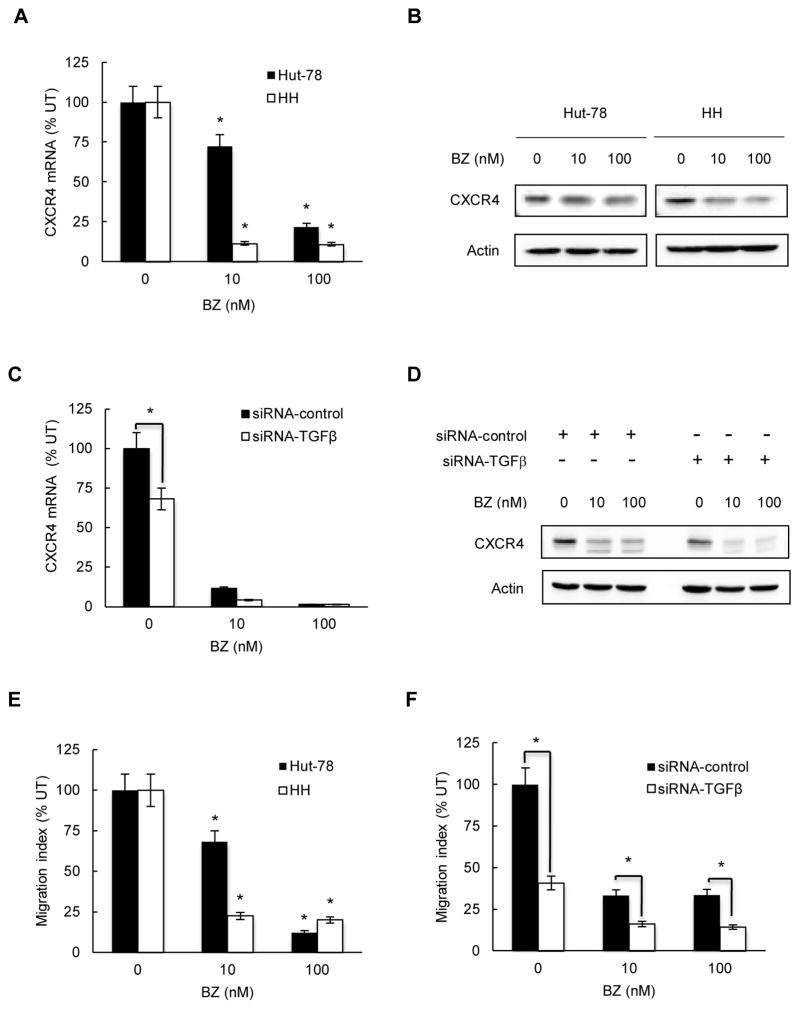
Advanced stages of CTCL and skin accumulation of malignant T cells have been associated with the increased expression of the chemokine receptor CXCR4 (11–14). Since previous studies have shown that TGFβ1 can increase the CXCR4 expression on T cells (55–57), we hypothesized that proteasome inhibition, by reducing the TGFβ1 levels, might inhibit the CXCR4 expression in CTCL cells. First, we analyzed the CXCR4 mRNA levels in CTCL Hut-78 and HH cells incubated 24 hours with 0, 10 and 100 nM BZ. As shown in Fig. 7A, BZ significantly decreased the CXCR4 mRNA levels in both CTCL cells; 10 nM BZ decreased the CXCR4 mRNA expression in Hut-78 cells by about 25%, and in HH cells by 90%. 100 nM BZ decreased CXCR4 mRNA levels in both cell types by about 90%. Bortezomib also substantially reduced the CXCR4 protein levels in Hut-78 and HH cells (Fig. 7B). Again, the CXCR4 protein levels were more affected by 10 nM BZ in HH cells than in Hut-78 cells (Fig. 7B), correlating with the mRNA data (Fig. 7A).
Figure 7. BZ inhibits CXCR4 expression and migration of CTCL cells.
(A) Real time RT-PCR analysis of CXCR4 mRNA levels in Hut-78 (full columns) and HH (empty columns) cells treated 24 hours with 0, 10 and 100 nM BZ. The values represent the mean +/−SE of four experiments. Asterisks denote a statistically significant (p<0.05) inhibition compared to untreated cells. (B) Western analysis of CXCR4 protein levels in WCE prepared from Hut-78 and HH cells treated 24 hours with 0, 10 and 100 nM BZ. Equal protein loading was confirmed by re-probing the membranes with anti-actin antibody. (C) Real time RT-PCR of CXCR4 mRNA levels in HH cells transfected with control (full columns) or TGFβ1 specific (empty columns) siRNA and treated 24 hours with 0, 10 and 100 nM BZ. The values represent the mean +/−SE of four experiments. The asterisk denotes a statistically significant change compared to cells transfected with control siRNA. (D) Western analysis of WCE prepared from HH cells transfected with control or TGFβ1 siRNA, treated 24 hours with 0, 10 and 100 nM BZ, and analyzed by using CXCR4 and actin antibodies. (E) Migration assay of Hut-78 and HH cells incubated 6 hours with 0, 10 and 100 nM BZ. Results are expressed as migration index that represents the ratio between the number of cells that migrated in the presence of BZ and the number of migrated untreated cells. The values represent the mean +/−SE of four experiments. Asterisks denote a statistically significant inhibition compared to untreated cells. (F) Migration assay of HH cells transfected with control (full columns) or TGFβ1 specific (empty columns) siRNA and treated 6 hours with 0, 10 and 100 nM BZ. Results are expressed as migration index representing the ratio between the number of cells migrating in the presence of BZ and the number of untreated migrating cells; the values represent the mean +/−SE of four experiments. Asterisks denote a statistically significant change compared to cells transfected with control siRNA.
To determine whether the CXCR4 expression in CTCL cells is mediated by TGFβ1, we examined the CXCR4 expression in HH cells transfected with control and TGFβ1 siRNA. TGFβ1 suppression significantly reduced both the CXCR4 mRNA (Fig. 7C) and protein levels (Fig. 7D) in HH cells, demonstrating that the CXCR4 expression in CTCL cells is mediated by TGFβ1, and suggesting that TGFβ1 might promote CTCL cell migration.
TGFβ1 and CXCL12 promote CTCL cell migration
Since CXCR4 controls T cell trafficking (58), we hypothesized that suppression of CXCR4 by the BZ-reduced TGFβ1 expression in CTCL cells might decrease their migration potential. First, we tested whether BZ inhibits migration of Hut-78 and HH cells. As shown in Fig. 7E, migration of Hut-78 and HH cells was significantly decreased by 10 and 100 nM BZ compared to untreated cells. Correlating with the CXCR4 expression (Fig. 7A), 10 nM BZ reduced migration of Hut-78 cells by about 30%, and of HH cells, by approximately 80% (Fig. 7E); 100 nM BZ decreased migration of both cell types by 80–90% (Fig. 7E). To examine whether the decreased migration of BZ-treated cells is mediated by the reduced TGFβ1 levels, we suppressed TGFβ1 expression in HH cells by siRNA, and measured their migration after 6-hour treatment with 0, 10 and 100 nM BZ. Suppression of TGFβ1 significantly reduced migration of untreated HH cells (Fig. 7F), strongly indicating that TGFβ1 promotes CTCL cell migration. TGFβ1 suppression decreased the cell migration also in BZ-treated cells (Fig. 7F), suggesting that the BZ-inhibited CTCL cell migration may be mediated, at least partly, by the reduced TGFβ1 levels.
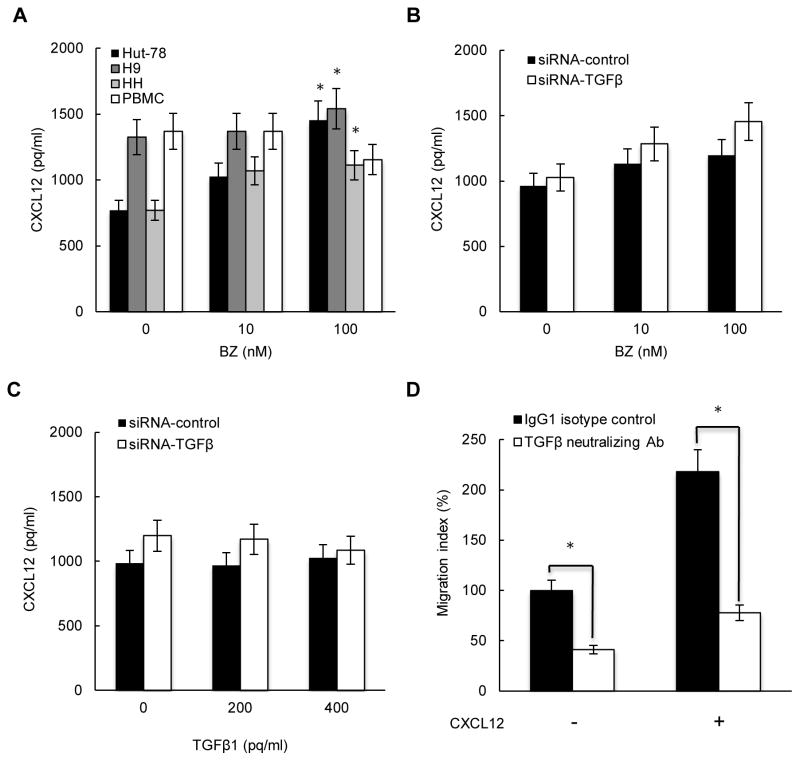
Since CXCL12 is the major ligand for CXCR4, we wanted to determine whether CXCL12 is released by CTCL cells, whether its expression is regulated by BZ and mediated by TGFβ1, and whether it induces migration of CTCL cells. All three CTCL lines as well as normal PBMCs release considerable amounts of CXCL12 (Fig. 8A). Interestingly however, while 100 nM BZ significantly increases CXCL12 release from all three CTCL cells, it reduces CXCL12 release from PBMCs (Fig. 8A). Suppression of endogenous TGFβ1 by siRNA (Fig. 8B) or addition of exogenous TGFβ1 (Fig. 8C) did not have any significant effect on the CXCL12 release from HH cells, indicating that the CXCL12 expression is not dependent on TGFβ1 in CTCL cells.
Figure 8. TGFβ1 and CXCL12 promote CTCL cell migration.
(A) CXCL12 release measured in Hut-78, H9, HH and PBMC cells incubated 24 hours with 0, 10, and 100 nM BZ. Asterisks denote a statistically significant (p<0.05) change compared to untreated cells. (B) CXCL12 release measured in HH cells transfected with control or TGFβ1 specific siRNA, and incubated 24 hours with 0, 10, and 100 nM BZ. (C) CXCL12 release measured in control and TGFβ1 siRNA transfected HH cells, and incubated 24 hours with 0, 200 and 400 pg/ml of exogenously added recombinant TGFβ1. (D) Migration assay of HH cells incubated 6 hours with 300 ng/ml of TGFβ neutralizing antibody (empty columns) or control IgG (full columns) in the absence and presence of 50 ng/ml of CXCL12 added to the lower chamber. The values represent the mean +/−SE of four experiments; asterisks denote a statistically significant (p<0.05) change compared to cells incubated with control antibody.
To determine whether TGFβ1 and CXCL12 induce CTCL migration, we analyzed migration of serum-starved HH cells towards serum and CXCL12 (50 ng/ml), in the presence of 300 ng/ml of TGFβ neutralizing antibody, or isotype control IgG. As shown in Fig. 8D, TGFβ neutralizing antibody significantly reduced the HH cell migration both in the absence and presence of CXCL12 added to the lower chamber. Furthermore, CXCL12 increased the HH cell migration more than two-fold (Fig. 8D). Together, these results strongly indicate that TGFβ1 and CXCL12 induce migration of CTCL cells.
Discussion
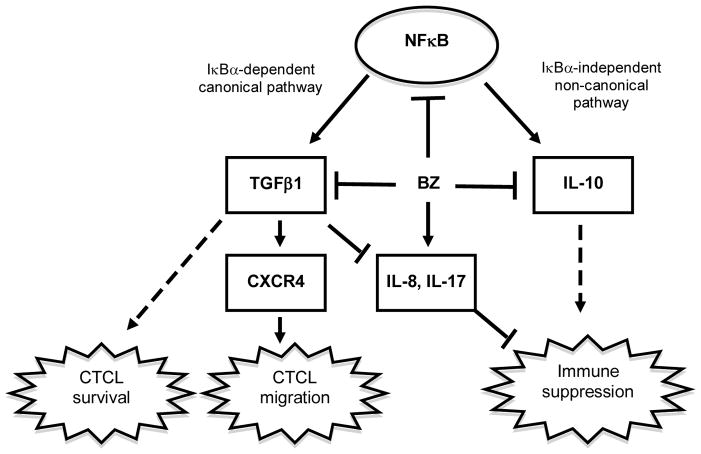
Progression of CTCL has been associated with the increased expression of TGFβ1 and IL-10 that are believed to be responsible for the suppressed immunity, increased susceptibility to infections, and diminished antitumor responses. Yet, little is known about the transcriptional regulation of TGFβ1 and IL-10 in CTCL, and about their function in regulating the CTCL cell responses. Here, we show that both TGFβ1 and IL-10 expression in CTCL cells is regulated by NFκB pathway, and suppressed by BZ-mediated proteasome inhibition. However, while the TGFβ1 expression is IκBα-dependent and is regulated by the canonical pathway, IL-10 expression is IκBα-independent, and its inhibition is associated with the increased promoter recruitment of p52 that characterizes the non-canonical pathway. Interestingly, our data show that TGFβ1 has opposite effect on the viability of BZ-treated cells; while it enhances the viability of CTCL cells, it decreases the viability of normal PBMCs. TGFβ1 suppression increases IL-8 and IL-17 expression in untreated and BZ-treated CTCL cells, indicating that TGFβ1 inhibits the pro-inflammatory gene expression in CTCL cells. Furthermore, our results show that TGFβ1 mediates CXCR4 expression and CXCL12-induced CTCL cell migration, and that BZ inhibits the CXCR4 expression and CTCL cell migration (Fig. 9).
Figure 9. Proposed model of TGFβ1 and IL-10 function and regulation in CTCL cells.
In CTCL cells, proteasome inhibition by BZ inhibits expression of the NFκB-dependent immunosuppressive cytokines IL-10 and TGFβ1. However, while the TGFβ1 expression is regulated by the IκBα-dependent canonical pathway, the IL-10 expression is regulated by the IκBα-independent non-canonical pathway. Suppression of TGFβ1 in CTCL cells induces apoptosis and IL-8 and IL-17 expression, while concomitantly inhibiting CXCR4 expression and CTCL migration.
Bortezomib is the first clinically approved proteasome inhibitor that has been very effective in the treatment of multiple myeloma (MM), and has shown promising results also in patients with relapsed or refractory CTCL (37–42). However, the mechanisms of BZ function in CTCL remain incompletely understood. Previous studies from our laboratory have shown that BZ induces CTCL cell apoptosis by inhibiting expression of the anti-apoptotic genes cIAP1 and cIAP2 and the proto-oncogene Bcl3, while it concomitantly induces release of the pro-inflammatory cytokines IL-8 and IL-17, and increases their release levels to those seen in healthy adults (47, 50). In this study, we show that BZ inhibits TGFβ1 and IL-10 expression, CXCR4 levels, and CTCL cell migration, indicating that this may represent an additional mechanism of BZ function in CTCL.
Our data indicate that the BZ-induced IL-8 and IL-17 expression is mediated, at least partly, by the BZ-inhibited TGFβ1 release, since TGFβ1 suppression enhances the IL-8 and IL-17 expression in BZ-treated CTCL cells. However, since TGFβ1 suppression increases IL-8 and IL-17 mRNA levels also in BZ-untreated cells (Figs. 5E, F), these results indicate that TGFβ1 inhibits the pro-inflammatory gene expression in CTCL cells independently of BZ. The mechanisms by which TGFβ1 inhibits the pro-inflammatory gene expression in CTCL cells are currently under investigation. The increased IL-8 and IL-17 mRNA levels observed after TGFβ1 suppression are compatible with decreased transcription; however, it is also possible that TGFβ1 regulates IL-8 and IL-17 mRNA stability.
Even though TGFβ1 inhibits survival and proliferation in most cells, some tumor cells become unresponsive to the anti-proliferative effect of TGFβ1, and some actually switch to being stimulated by TGFβ1 (53, 54). Interestingly, our data show that TGFβ1 increases viability of BZ-treated CTCL cells, while it decreases viability of BZ-treated PBMCs (Fig. 6D). Intriguingly, however, this opposing effect on cell viability is observed only in BZ-treated cells (Fig. 6D); TGFβ1 has no effect on CTCL or PBMC cell viability when it is added to untreated cells (data not shown). This is consistent with results shown in Figs. 6A–C, demonstrating that exogenous TGFβ1 increases viability and decreases apoptosis and IL-8 expression in HH cells transfected with TGFβ1 siRNA, but not control siRNA. Since both 100 nM BZ and TGFβ1 siRNA suppression decrease the released TGFβ1 levels in HH cells to about 150–200 pg/ml (Figs. 1A, 5B), these results suggest that cells are responsive to exogenously added TGFβ1 only when the endogenous TGFβ1 levels are reduced. Several signaling mechanisms may be responsible for this “saturation” effect; however, the TGFβ1 receptors, TβRI and TβRII seem to be among the most plausible candidates.
Several studies have shown that CTCL patients have decreased expression of TβRI and/or TβRII, and reduced responsiveness to the growth-inhibitory effects of TGFβ1 (59–64). We have detected TβRI and TβRII in all three CTCL lines, even though Hut-78 and HH cells express considerably lower protein levels of both receptors than H9 cells (Fig. 6E). Since transfection of HH cells with TβRI and TβRII siRNA decreased their viability and increased apoptosis, and addition of exogenous TGFβ1 reversed these effects (Figs. 6F–H), these data suggest that the TGFβ1 receptors in HH cells are, at least partially, functional. However, since CTCL cells respond to exogenously added TGFβ1 only when the endogenous TGFβ1 levels are suppressed by TGFβ1 siRNA (Figs. 6A–C), or BZ (Fig. 6D), it is possible that the TGFβ1 receptors in CTCL cells are saturated by the high levels of endogenous TGFβ1, and respond to exogenously added TGFβ1 only when the endogenous levels are decreased. In this respect, BZ might have a previously unrecognized effect on the viability of CTCL cells that is mediated by the reduced TGFβ1 levels. However, since the present study used only CTCL established cell lines, and not primary Sezary cells, it will be important in the future to determine whether the same mechanisms are operational in primary Sezary cells as well. Particularly, it would be interesting to investigate whether primary Sezary cells from BZ-treated patients have functional TGFβ1 receptors and signaling.
Importantly, in addition to the inhibition of TGFβ1 and IL-10, BZ inhibits mRNA and protein expression of the chemokine receptor CXCR4 in CTCL Hut-78 and HH cells (Figs. 7A, B). Since TGFβ1 suppression decreases CXCR4 mRNA and protein levels in CTCL cells (Figs. 7C, D), these data indicate that TGFβ1 induces the CXCR4 expression in CTCL cells, and that the BZ-decreased CXCR4 expression is at least partly mediated by TGFβ1. The proposed BZ inhibition of CXCR4 via TGFβ1 signaling is further supported by studies demonstrating that BZ decreases CXCR4 expression in BZ-resistant mouse multiple myeloma cell lines, while it does not inhibit CXCR4 expression in cells that do not express detectable levels of TGFβ1 (65–67). Since the increased CXCR4 expression has been implicated in promoting skin homing of malignant CTCL cells in patients with Sézary syndrome (12), these findings suggest that the BZ-reduced TGFβ1-CXCR4 signaling may represent yet another mechanism responsible for the BZ function in CTCL. Indeed, we have found that BZ significantly reduces migration of CTCL cells (Fig. 7E), and that TGFβ1 suppression decreases HH cell migration (Fig. 7F), indicating that TGFβ1 induces migration of CTCL cells, and suggesting that BZ inhibits the CTCL cell migration by inhibiting TGFβ1 expression. The role of TGFβ1 in inducing migration of CTCL cells is further supported by our data demonstrating that addition of TGFβ neutralizing antibody significantly decreases migration of HH cells, both in the absence and presence of CXCL12 (Fig. 8D). Our data show that all three CTCL lines release considerable levels of CXCL12. However, in contrast to CXCR4, which is inhibited by BZ and dependent on TGFβ1, the CXCL12 release is increased by 100 nM BZ in CTCL cells (Fig. 8A), and its expression is not regulated by TGFβ1 (Figs. 8B, C). Since CXCL12 is a potent chemoattractant for lymphocytes, these results suggest that BZ suppresses migration of CTCL cells through the decreased autocrine TGFβ1-CXCR4 signaling, while it may concomitantly increase recruitment of non-malignant lymphocytes by the increased CXCL12 release from CTCL cells. It will be interesting to determine in future if this is indeed the case in BZ-treated CTCL patients.
In cancer cells, the immunosuppressive cytokines TGFβ1 and IL-10 are thought to be responsible for the suppression of anti-tumor immune responses and successful tumor escape (68, 69). Our data indicate that in CTCL, TGFβ1 inhibits apoptosis, blocks IL-8 and IL-17 expression, and promotes CXCR4 expression and CTCL cell migration. Thus, understanding the mechanisms regulating TGFβ1 and IL-10 expression in CTCL cells will likely reveal novel pathways that could be targeted for therapy. Our data demonstrate that the TGFβ1 expression in CTCL cells is IκBα-dependent (Fig. 2), and correlates with the p65/p50 occupancy at the TGFβ1 promoter (Fig. 3), indicating that the TGFβ1 expression is regulated by the canonical pathway. In contrast, the IL-10 expression is IκBα-independent (Fig. 2), correlates with p65, p50 and RelB occupancy at the IL-10 promoter, and inversely correlates with p52 recruitment (Fig. 4). The high p65, p50 and RelB occupancy at the IL-10 promoter, and the increased p52 recruitment in BZ-treated cells, are consistent with our recent study demonstrating high levels of p65, p50 and RelB in the nucleus of CTCL cells, and increased p52 nuclear accumulation in response to proteasome inhibition (50). These results indicate that the IL-10 expression is regulated by non-canonical pathway, and suggest that p52 acts as a transcriptional repressor of the IL-10 expression in CTCL cells. Even though p52 lacks the transactivation domain, it can stimulate transcription by forming complexes with p65, or repress transcription by forming inactive p52/p52 homodimers (70–72), or associating with histone deacetylases (HDAC) (73, 74). Our data suggest that the κB1 and κB2 sites of the IL-10 promoter in BZ-treated CTCL cells are occupied by p52 homodimers (Fig. 4); however, it seems plausible that additional recruited transcriptional regulators, such as HDAC (73, 74) or the nuclear onco-protein Bcl3 (50) may contribute to the p52-mediated inhibition of IL-10 expression in CTCL cells.
In conclusion, our study provides a novel insight into the BZ function in CTCL, and demonstrates that in addition to inducing apoptosis, BZ has an important immunomodulatory role by down-regulating expression of the immunosuppressive cytokines TGFβ1 and IL-10, while concomitantly increasing expression of IL-8, IL-17, and CXCL12. Our data show that TGFβ1 regulates the CXCR4 expression and CTCL cell migration, and suggest that it might represent a useful tool to interfere with tumor progression in CTCL and other malignancies characterized by excessive TGFβ1 expression.
Abbreviations used in this paper
- BZ
bortezomib
- ChIP
chromatin immunoprecipitation
- CTCL
cutaneous T cell lymphoma
- LDH
lactate dehydrogenase
Footnotes
This work was supported by NIH grants AI085497 and CA173452 to I. Vancurova.
References
- 1.Berger CL, Edelson R. The life cycle of cutaneous T cell lymphoma reveals opportunities for targeted drug therapy. Curr Cancer Drug Targets. 2004;4:609–619. doi: 10.2174/1568009043332808. [DOI] [PubMed] [Google Scholar]
- 2.Poszepczynska E, Bagot M, Echchakir H, Martinvalet D, Ramez M, Charue D, Boumsell L, Bensussan A. Functional characterization of an IL-7-dependent CD4(+)CD8alphaalpha(+) Th3-type malignant cell line derived from a patient with a cutaneous T-cell lymphoma. Blood. 2000;96:1056–1063. [PubMed] [Google Scholar]
- 3.Berger CL, Tigelaar R, Cohen J, Mariwalla K, Trinh J, Wang N, Edelson RL. Cutaneous T-cell lymphoma: malignant proliferation of T-regulatory cells. Blood. 2005;105:1640–1647. doi: 10.1182/blood-2004-06-2181. [DOI] [PubMed] [Google Scholar]
- 4.Kasprzycka M, Zhang Q, Witkiewicz A, Marzec M, Potoczek M, Liu X, Wang HY, Milone M, Basu S, Mauger J, Choi JK, Abrams JT, Hou JS, Rook AH, Vonderheid E, Woetmann A, Odum N, Wasik MA. Gamma c-signaling cytokines induce a regulatory T cell phenotype in malignant CD4+ T lymphocytes. J Immunol. 2008;181:2506–2512. doi: 10.4049/jimmunol.181.4.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krejsgaard T, Gjerdrum LM, Ralfkiaer E, Lauenborg B, Eriksen KW, Mathiesen AM, Bovin LF, Gniadecki R, Geisler C, Ryder LP, Zhang Q, Wasik MA, Odum N, Woetmann A. Malignant Tregs express low molecular splice forms of FOXP3 in Sézary syndrome. Leukemia. 2008;22:2230–2239. doi: 10.1038/leu.2008.224. [DOI] [PubMed] [Google Scholar]
- 6.Kim EJ, Hess S, Richardson SK, Newton S, Showe LC, Benoit BM, Ubriani R, Vittorio CC, Junkins-Hopkins JM, Wysocka M, Rook AH. Immunopathogenesis and therapy of cutaneous T cell lymphoma. J Clin Invest. 2005;115:798–812. doi: 10.1172/JCI24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh PT, Benoit BM, Wysocka M, Dalton ND, Turka LA, Rook AH. A Role for Regulatory T Cells in Cutaneous T-Cell Lymphoma; Induction of a CD4+CD25+ Foxp3+ T-Cell Phenotype Associated with HTLV-1 Infection. J Invest Dermatol. 2006;126:690–692. doi: 10.1038/sj.jid.5700121. [DOI] [PubMed] [Google Scholar]
- 8.Chong BF, Wilson AJ, Gibson HM, Hafner MS, Luo Y, Hedgcock CJ, Wong HK. Immune function abnormalities in peripheral blood mononuclear cell cytokine expression differentiates stages of cutaneous T-cell lymphoma/mycosis fungoides. Clin Cancer Res. 2008;14:646–653. doi: 10.1158/1078-0432.CCR-07-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abraham RM, Zhang Q, Odum N, Wasik MA. The role of cytokine signaling in the pathogenesis of cutaneous T-cell lymphoma. Cancer Biol Ther. 2011;12:1019–1022. doi: 10.4161/cbt.12.12.18144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krejsgaard T, Odum N, Geisler C, Wasik MA, Woetmann A. Regulatory T cells and immunodeficiency in mycosis fungoides and Sezary syndrome. Leukemia. 2012;26:424–432. doi: 10.1038/leu.2011.237. [DOI] [PubMed] [Google Scholar]
- 11.Kallinich T, Muche JM, Qin S, Sterry W, Audring H, Kroczek RA. Chemokine receptor expression on neoplastic and reactive T cells in the skin at different stages of mycosis fungoides. J Invest Dermatol. 2003;121:1045–1052. doi: 10.1046/j.1523-1747.2003.12555.x. [DOI] [PubMed] [Google Scholar]
- 12.Narducci MG, Scala E, Bresin A, Caprini E, Picchio MC, Remotti D, Ragone G, Nasorri F, Frontani M, Arcelli D, Volinia S, Lombardo GA, Baliva G, Napolitano M, Russo G. Skin homing of Sezary cells involves SDF-1-CXCR4 signaling and down-regulation of CD26/dipeptidylpeptidase IV. Blood. 2006;107:1108–1115. doi: 10.1182/blood-2005-04-1492. [DOI] [PubMed] [Google Scholar]
- 13.Wu XS, Lonsdorf AS, Hwang ST. Cutaneous T-cell lymphoma: roles for chemokines and chemokine receptors. J Invest Dermatol. 2009;129:1115–1119. doi: 10.1038/jid.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daggett RN, Kurata M, Abe S, Onishi I, Miura K, Sawada T, Tanizawa T, Kitagawa M. Expression dynamics of CXCL12 and CXCR4 during the progression of mycosis fungoides. Br J Dermatol. 2014;171:722–731. doi: 10.1111/bjd.13054. [DOI] [PubMed] [Google Scholar]
- 15.Mori N, Prager D. Activation of the interleukin-10 gene in the human T lymphoma line HuT 78: identification and characterization of NFκB binding sites in the regulatory region of the interleukin-10 gene. Eur J Haematol. 1997;59:162–170. doi: 10.1111/j.1600-0609.1997.tb00970.x. [DOI] [PubMed] [Google Scholar]
- 16.Cao S, Zhang X, Edwards JP, Mosser DM. NFκB1 (p50) homodimers differentially regulate pro- and anti-inflammatory cytokines in macrophages. J Biol Chem. 2006;281:26041–26050. doi: 10.1074/jbc.M602222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol Rev. 2008;226:205–218. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 19.Gabryšová L, Howes A, Saraiva M, O’Garra A. The regulation of IL-10 expression. Curr Top Microbiol Immunol. 2014;380:157–190. doi: 10.1007/978-3-662-43492-5_8. [DOI] [PubMed] [Google Scholar]
- 20.Lee KY, Ito K, Hayashi R, Jazrawi EP, Barnes PJ, Adcock IM. NFκB and activator protein 1 response elements and the role of histone modifications in IL-1-induced TGF-β1 gene transcription. J Immunol. 2006;176:603–615. doi: 10.4049/jimmunol.176.1.603. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimura A, Wakabayashi Y, Mori T. Cellular and molecular basis for the regulation of inflammation by TGFβ. J Biochem. 2010;147:781–792. doi: 10.1093/jb/mvq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pomerantz JL, Baltimore D. Two pathways to NFκB. Mol Cell. 2002;10:693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 23.Shih VF, Tsui R, Caldwell A, Hoffmann A. A single NFκB system for both canonical and non-canonical signaling. Cell Res. 2011;21:86–102. doi: 10.1038/cr.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NFκB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 25.Wan F, Lenardo MJ. The nuclear signaling of NFκB: current knowledge, new insights, and future perspectives. Cell Res. 2009;20:24–33. doi: 10.1038/cr.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smale ST. Dimer-specific regulatory mechanisms within the NFκB family of transcription factors. Immunol Rev. 2012;246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 27.Cleere R, Long A, Kelleher D, O’Neill LA. Autocrine regulation of the transcription factor NFκB by TNFα in the human T cell lymphoma line Hut 78. Biochem Soc Trans. 1995;23:113S. doi: 10.1042/bst023113s. [DOI] [PubMed] [Google Scholar]
- 28.O’Connell MA, Cleere R, Long A, O’Neill LA, Kelleher D. Cellular proliferation and activation of NFκB are induced by autocrine production of TNFα in the human T lymphoma line Hut-78. J Biol Chem. 1995;270:7399–7404. doi: 10.1074/jbc.270.13.7399. [DOI] [PubMed] [Google Scholar]
- 29.Giri DK, Aggarwal BB. Constitutive activation of NFκB causes resistance to apoptosis in human cutaneous T cell lymphoma HuT-78 cells. Autocrine role of TNF and reactive oxygen intermediates. J Biol Chem. 1998;273:14008–14014. doi: 10.1074/jbc.273.22.14008. [DOI] [PubMed] [Google Scholar]
- 30.Izban KF, Ergin M, Qin JZ, Martinez RL, Pooley RJ, JR, Saeed S, Alkan S. Constitutive expression of NFκB is a characteristic feature of mycosis fungoides: implications for apoptosis resistance and pathogenesis. Hum Pathol. 2000;31:1482–1490. doi: 10.1053/hupa.2000.20370. [DOI] [PubMed] [Google Scholar]
- 31.Sors A, Jean-Louis F, Pellet C, Laroche L, Dubertret L, Courtois G, Bachelez H, Michel L. Down-regulating constitutive activation of the NFκB canonical pathway overcomes the resistance of cutaneous T-cell lymphoma to apoptosis. Blood. 2006;107:2354–2363. doi: 10.1182/blood-2005-06-2536. [DOI] [PubMed] [Google Scholar]
- 32.Sors A, Jean-Louis F, Bégué E, Parmentier L, Dubertret L, Dreano M, Courtois G, Bachelez H, Michel L. Inhibition of IκB kinase subunit 2 in cutaneous T-cell lymphoma down-regulates NFκB constitutive activation, induces cell death, and potentiates the apoptotic response to antineoplastic chemotherapeutic agents. Clin Cancer Res. 2008;14:901–911. doi: 10.1158/1078-0432.CCR-07-1419. [DOI] [PubMed] [Google Scholar]
- 33.Querfeld C, Rosen ST, Guitart J, Kuzel TM. The spectrum of cutaneous T-cell lymphomas: new insights into biology and therapy. Curr Opin Hematol. 2005;12:273–278. doi: 10.1097/01.moh.0000166498.64515.03. [DOI] [PubMed] [Google Scholar]
- 34.Wong HK, Mishra A, Hake T, Porcu P. Evolving Insights in the Pathogenesis and Therapy of Cutaneous T-cell lymphoma (Mycosis Fungoides and Sezary Syndrome) Br J Haematol. 2011;155:150–166. doi: 10.1111/j.1365-2141.2011.08852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilcox RA. Cutaneous T-cell lymphoma: 2011 update on diagnosis, risk-stratification, and management. Am J Hematol. 2011;86:928–948. doi: 10.1002/ajh.22139. [DOI] [PubMed] [Google Scholar]
- 36.Li JY, Horwitz S, Moskowitz A, Myskowski PL, Pulitzer M, Querfeld C. Management of cutaneous T cell lymphoma: new and emerging targets and treatment options. Cancer Manag Res. 2012;4:75–89. doi: 10.2147/CMAR.S9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bloom T, Kuzel TM, Querfeld C, Guitart J, Rosen ST. Cutaneous T-cell lymphomas: a review of new discoveries and treatments. Curr Treat Options Oncol. 2012;13:102–121. doi: 10.1007/s11864-011-0179-8. [DOI] [PubMed] [Google Scholar]
- 38.Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sézary syndrome): part I. Diagnosis: clinical and histopathologic features and new molecular and biologic markers. J Am Acad Dermatol. 2014;70:205. e1–e16. doi: 10.1016/j.jaad.2013.07.049. [DOI] [PubMed] [Google Scholar]
- 39.Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, Anderson KC. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–3076. [PubMed] [Google Scholar]
- 40.Shah JJ, Orlowski RZ. Proteasome inhibitors in the treatment of multiple myeloma. Leukemia. 2009;23:1964–1979. doi: 10.1038/leu.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zinzani PL, Musuraca G, Tani M, Stefoni V, Marchi E, Fina M, Pellegrini C, Alinari L, Derenzini E, de Vivo A, Sabattini E, Pileri S, Baccarani M. Phase II trial of proteasome inhibitor bortezomib in patients with relapsed or refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25:4293–4297. doi: 10.1200/JCO.2007.11.4207. [DOI] [PubMed] [Google Scholar]
- 42.Horwitz SM. Novel therapies for cutaneous T-cell lymphomas. Clin Lymphoma Myeloma. 2008;S5:S187–192. doi: 10.3816/CLM.2008.s.015. [DOI] [PubMed] [Google Scholar]
- 43.Heider U, Rademacher J, Lamottke B, Mieth M, Moebs M, von Metzler I, Assaf C, Sezer O. Synergistic interaction of the histone deacetylase inhibitor SAHA with the proteasome inhibitor bortezomib in cutaneous T cell lymphoma. Eur J Haematol. 2009;82:440–449. doi: 10.1111/j.1600-0609.2009.01239.x. [DOI] [PubMed] [Google Scholar]
- 44.Kim SJ, Yoon DH, Kang HJ, Kim JS, Park SK, Kim HJ, Lee J, Ryoo BY, Ko YH, Huh J, Yang WI, Kim HK, Min SK, Lee SS, Do IG, Suh C, Kim WS. On behalf of Consortium for Improving Survival of Lymphoma (CISL) investigators. Bortezomib in combination with CHOP as first-line treatment for patients with stage III/IV peripheral T-cell lymphomas: A multicentre, single-arm, phase 2 trial. Eur J Cancer. 2012;48:3223–3231. doi: 10.1016/j.ejca.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 45.Manfè V, Biskup E, Rosbjerg A, Kamstrup M, Skov AG, Lerche CM, Lauenborg BT, Odum N, Gniadecki R. miR-122 regulates p53/Akt signalling and the chemotherapy-induced apoptosis in cutaneous T-cell lymphoma. PLoS One. 2012;7:e29541. doi: 10.1371/journal.pone.0029541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biskup E, Kamstrup MR, Manfé V, Gniadecki R. Proteasome inhibition as a novel mechanism of the proapoptotic activity of γ-secretase inhibitor I in cutaneous T-cell lymphoma. Br J Dermatol. 2013;168:504–512. doi: 10.1111/bjd.12071. [DOI] [PubMed] [Google Scholar]
- 47.Juvekar A, Manna S, Ramaswami S, Chang TP, Vu HY, Ghosh CC, Celiker MY, Vancurova I. Bortezomib induces nuclear translocation of IκBα resulting in gene-specific suppression of NFκB-dependent transcription and induction of apoptosis in CTCL. Mol Cancer Res. 2011;9:183–194. doi: 10.1158/1541-7786.MCR-10-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manna S, Singha B, Phyo SA, Gatla HR, Chang TP, Sanacora S, Ramaswami S, Vancurova I. Proteasome Inhibition by Bortezomib Increases IL-8 Expression in Androgen-Independent Prostate Cancer Cells: The Role of IKKα. J Immunol. 2013;191:2837–2846. doi: 10.4049/jimmunol.1300895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singha B, Gatla HR, Manna S, Chang TP, Sanacora S, Poltoratsky V, Vancura A, Vancurova I. Proteasome inhibition increases recruitment of IκB kinase β (IKKβ), S536P-p65, and transcription factor EGR1 to interleukin-8 (IL-8) promoter, resulting in increased IL-8 production in ovarian cancer cells. J Biol Chem. 2014;289:2687–2700. doi: 10.1074/jbc.M113.502641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang TP, Vancurova I. Bcl3 regulates pro-survival and pro-inflammatory gene expression in cutaneous T-cell lymphoma. Biochim Biophys Acta. 2014;1843:2620–2630. doi: 10.1016/j.bbamcr.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang TP, Kim M, Vancurova I. Analysis of TGFβ1 and IL-10 transcriptional regulation in CTCL cells by chromatin immunoprecipitation. Methods Mol Biol. 2014;1172:329–341. doi: 10.1007/978-1-4939-0928-5_30. [DOI] [PubMed] [Google Scholar]
- 52.Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, Maas J, Pien CS, Prakash S, Elliott PJ. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 1999;59:2615–2622. [PubMed] [Google Scholar]
- 53.Thomas DA, Massagué J. TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8:369–380. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 54.Siegel PM, Massagué J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 55.Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J Exp Med. 1998;187:875. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buckley CD, Amft N, Bradfield PF, Pilling D, Ross E, Arenzana-Seisdedos F, Amara A, Curnow SJ, Lord JM, Scheel-Toellner D, Salmon M. Persistent induction of the chemokine receptor CXCR4 by TGF-β1 on synovial T cells contributes to their accumulation within the rheumatoid synovium. J Immunol. 2000;165:3423–3429. doi: 10.4049/jimmunol.165.6.3423. [DOI] [PubMed] [Google Scholar]
- 57.Franitza S, Kollet O, Brill A, Vaday GG, Petit I, Lapidot T, Alon R, Lider O. TGF-β1 enhances SDF-1alpha-induced chemotaxis and homing of naive T cells by up-regulating CXCR4 expression and downstream cytoskeletal effector molecules. Eur J Immunol. 2002;32:193–202. doi: 10.1002/1521-4141(200201)32:1<193::AID-IMMU193>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 58.Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol. 2001;2:123–128. doi: 10.1038/84219. [DOI] [PubMed] [Google Scholar]
- 59.Dulmage BO, Geskin LJ. Lessons learned from gene expression profiling of cutaneous T-cell lymphoma. Br J Dermatol. 2013;169:1188–1197. doi: 10.1111/bjd.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kadin ME, Cavaille-Coll MW, Gertz R, Massague J, Cheifetz S, George D. Loss of receptors for TGFβ in human T-cell malignancies. Proc Natl Acad Sci USA. 1994;91:6002–6006. doi: 10.1073/pnas.91.13.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Capocasale RJ, Lamb RJ, Vonderheid EC, Fox FE, Rook AH, Nowell PC, Moore JS. Reduced surface expression of TGF receptor type II in mitogen-activated T cells from Sezary patients. Proc Natl Acad Sci USA. 1995;92:5501–5505. doi: 10.1073/pnas.92.12.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knaus PI, Lindemann D, DeCoteau JF, Perlman R, Yankelev H, Hille M, Kadin ME, Lodish HF. A dominant inhibitory mutant of the type II transforming growth factor beta receptor in the malignant progression of a cutaneous T-cell lymphoma. Mol Cell Biol. 1996;16:3480–3489. doi: 10.1128/mcb.16.7.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Doorn R, Dijkman R, Vermeer MH, Out-Luiting JJ, van der Raaij-Helmer EM, Willemze R, Tensen CP. Aberrant expression of the tyrosine kinase receptor EphA4 and the transcription factor twist in Sézary syndrome identified by gene expression analysis. Cancer Res. 2004;64:5578–5586. doi: 10.1158/0008-5472.CAN-04-1253. [DOI] [PubMed] [Google Scholar]
- 64.Chung JS, Shiue LH, Duvic M, Pandya A, Cruz PD, Jr, Ariizumi K. Sézary syndrome cells overexpress syndecan-4 bearing distinct heparan sulfate moieties that suppress T-cell activation by binding DC-HIL and trapping TGF-beta on the cell surface. Blood. 2011;117:3382–3390. doi: 10.1182/blood-2010-08-302034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baughn LB, Di Liberto M, Niesvizky R, Cho HJ, Jayabalan D, Lane J, Liu F, Chen-Kiang S. CDK2 phosphorylation of Smad2 disrupts TGF-β transcriptional regulation in resistant primary bone marrow myeloma cells. J Immunol. 2009;182:1810–1817. doi: 10.4049/jimmunol.0713726. [DOI] [PubMed] [Google Scholar]
- 66.Azab AK, Runnels JM, Pitsillides C, Moreau AS, Azab F, Leleu X, Jia X, Wright R, Ospina B, Carlson AL, Alt C, Burwick N, Roccaro AM, Ngo HT, Farag M, Melhem MR, Sacco A, Munshi NC, Hideshima T, Rollins BJ, Anderson KC, Kung AL, Lin CP, Ghobrial IM. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009;113:4341–4351. doi: 10.1182/blood-2008-10-186668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stessman HA, Mansoor A, Zhan F, Janz S, Linden MR, Baughn LB, Van Ness B. Reduced CXCR4 expression is associated with extramedullary disease in a mouse model of myeloma and predicts poor survival in multiple myeloma patients treated with bortezomib. Leukemia. 2013;27:2075–2077. doi: 10.1038/leu.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whiteside TL. Regulatory T cell subsets in human cancer: are they regulating for or against tumor progression? Cancer Immunol Immunother. 2014;63:67–72. doi: 10.1007/s00262-013-1490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wakefield LM, Hill CS. Beyond TGFβ: roles of other TGFβ superfamily members in cancer. Nat Rev Cancer. 2013;13:328–341. doi: 10.1038/nrc3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang CC, Zhang J, Lombardi L, Neri A, Dalla-Favera R. Mechanism of expression and role in transcriptional control of the proto-oncogene NFκB-2/LYT-10. Oncogene. 1994;9:923–933. [PubMed] [Google Scholar]
- 71.Zhang J, Chang CC, Lombardi L, Dalla-Favera R. Rearranged NFκB2 gene in the HUT78 T-lymphoma cell line codes for a constitutively nuclear factor lacking transcriptional repressor functions. Oncogene. 1994;9:1931–1937. [PubMed] [Google Scholar]
- 72.Perkins ND, Schmid RM, Duckett CS, Leung K, Rice NR, Nabel GK. Distinct combinations of NFκB subunits determine the specificity of transcriptional activation. Proc Natl Acad Sci. 1992;89:1529–1533. doi: 10.1073/pnas.89.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rocha S, Martin AM, Meek DW, Perkins ND. p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NFκB subunit with histone deacetylase 1. Mol Cell Biol. 2003;23:4713–4727. doi: 10.1128/MCB.23.13.4713-4727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shanmugam R, Gade P, Wilson-Weekes A, Sayar H, Suvannasankha A, Goswami C, Li L, Gupta S, Cardoso AA, Al Baghdadi T, Sargent KJ, Cripe LD, Kalvakolanu DV, Boswell HS. A noncanonical Flt3ITD/NFκB signaling pathway represses DAPK1 in acute myeloid leukemia. Clin Cancer Res. 2012;18:360–369. doi: 10.1158/1078-0432.CCR-10-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]