Abstract
Prevention of tumors induced by environmental carcinogens has not been achieved. Skin tumors produced by polyaromatic hydrocarbons, such as 7,12-dimethylbenz(a)anthracene (DMBA), often harbor an H-ras point mutation, suggesting that it is a poor target for early immunosurveillance. The application of pyrosequencing and allele-specific PCR techniques established that mutations in the genome and expression of the Mut H-ras gene could be detected as early as 1 d after DMBA application. Further, DMBA sensitization raised Mut H-ras epitope–specific CTLs capable of eliminating Mut H-ras+ preneoplastic skin cells, demonstrating that immunosurveillance is normally induced but may be ineffective owing to insufficient effector pool size and/or immunosuppression. To test whether selective pre-expansion of CD8 T cells with specificity for the single Mut H-ras epitope was sufficient for tumor prevention, MHC class I epitope–focused lentivector-infected dendritic cell– and DNA-based vaccines were designed to bias toward CTL rather than regulatory T cell induction. Mut H-ras, but not wild-type H-ras, epitope-focused vaccination generated specific CTLs and inhibited DMBA-induced tumor initiation, growth, and progression in preventative and therapeutic settings. Transferred Mut H-ras–specific effectors induced rapid tumor regression, overcoming established tumor suppression in tumor-bearing mice. These studies support further evaluation of oncogenic mutations for their potential to act as early tumor-specific, immunogenic epitopes in expanding relevant immunosurveillance effectors to block tumor formation, rather than treating established tumors.
Introduction
Tumors in cancer patients develop because they have successfully evaded immune detection, making them challenging targets for immunotherapy. Indeed, such therapies using combinations of approaches have met with limited success (1). The failure of endogenous immunosurveillance to block tumor development is likely due to many variables (2). Tumor-specific mutations may not be effectively processed and presented as a neoepitope. If the T cell repertoire can recognize the neoepitope, the initial pool of tumor-specific immune effectors is small and limits early and complete eradication of newly emergent neoplasia. Thus, the time it takes to prime and expand a sufficient number of effector cells provides the window of time necessary to drive selection of nonimmunogenic tumor cells. We propose that by bolstering the initial numbers of tumor epitope–focused effector T cells, rapid recognition and aggressive elimination of preneoplastic cells may be possible before immune-resistant tumors can develop. The ability to block tumor development in the early phases would likely meet with greater success than treating established disease. Tumors that depend on the expression of a somatically mutated protein make ideal candidates for immunoprevention to protect individuals who are at increased risk for developing such cancers.
Polyaromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants derived from the incomplete combustion of fossil fuels, cigarette smoke, and other sources. In humans, PAH exposure is associated with epithelial tumors of skin, lung, pharynx, mouth, breast, gastrointestinal tract, and others (3–7). Cutaneous exposure to the carcinogenic PAH 7,12-dimethylbenz(a)anthracene (DMBA) produces tumors that are associated with a characteristic mutation in the 61st codon of the H-ras oncogene (8, 9), which results in constitutive activation of the Ras protein and sustained signal transduction for cell growth (10). Mouse models of multistage skin carcinogenesis, in which DMBA is the initiating agent, develop benign papillomas, some of which progress to squamous cell carcinomas (SCCs), which harbor the H-ras Q61L oncogene mutation, suggesting that it is a poor epitope for immune recognition (11, 12). A large body of work supports the concept that such a point mutation may represent an initiating event generating preneoplastic cells that are intrinsically receptive to promotion mechanisms and drive the pathogenesis of invasive SCC. However, it is not clear to what extent DMBA-induced carcinogenesis depends on the H-ras Q61L mutation, as this strong mutagen may activate alternative transformation pathways. This process of tumor development is remarkably similar to the phenotypic and genotypic characteristics of human SCC development (13).
Contact hypersensitivity (CHS) occurs owing to the elimination of hapten-modified skin cells by cytotoxic CTLs (14). CHS responses to DMBA can be generated efficiently in mice. We have observed that during chemical carcinogenesis, T cell–mediated immunity to DMBA can be elicited, which can influence tumor development. IFN-γ–producing CD8+ T cells are responsible for antitumor immunity, whereas CD4+ T cells that produce IL-10 and IL-17 promote tumor development (15, 16). These observations raise the possibility that DMBA-induced tumors are immunogenic but require an immunologic boost to expand CD8+, rather than CD4+, T cells to overcome the immune evasion and immunosuppression mechanisms. By employing vaccination techniques that promote an appropriate type of T cell response to tumor-specific Ags expressed at the earliest stages of development, tumor immune evasion and outgrowth may be prevented. We investigate whether the peptide epitope of the H-ras Q61L point mutation (Mut H-ras) is presented by MHC class I molecules and represents an early tumor-specific Ag that can be targeted for effective cancer immunoprevention.
In this study, we demonstrate that mutant H-ras Q61L gene expression is detectable in preneoplastic skin cells 24 h after carcinogen application, and the CHS response to DMBA is due, in part, to T cells that recognize the Mut H-ras epitope presented on preneoplastic skin cells. By developing epitope-focused vaccine strategies, we show that effective early immunosurveillance depends on this subset of Mut H-ras–specific T cells. Importantly, immunity to this single epitope provided substantial protection against the formation of chemical carcinogen–induced tumors. The few vaccine-resistant tumors that did develop were largely devoid of Mut H-ras gene expression and possessed low tumorigenicity, demonstrating effective immunoediting without advancing tumor immune evasion. Moreover, adoptive transfer of cells from carcinogenesis-resistant, vaccinated mice into recipient mice with established tumors led to rapid tumor regression, demonstrating that suppressive mechanisms put in place by established tumors can be overcome.
Materials and Methods
Animals and reagents
All animal procedures were performed according to National Institutes of Health guidelines under protocols approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. Female mice on C3H/HeN and A/J backgrounds, 8 to 10 wk of age, were used in all experiments. DMBA and acetone were purchased from Sigma-Aldrich, St. Louis, MO. Phorbol 12-myristate 13-acetate [12-O-tetradecanoylphorbol-13-acetate (TPA)] with >99.5% purity was purchased from LC Laboratories, Woburn, MA.
The XS106 Langerhans cell line (obtained as a gift from Dr. A. Takashima, University of Toledo College of Medicine, Toledo, OH) has been established from the epidermis of newborn A/J mice and maintained in vitro, as described previously (17), and demonstrates potent APC function in vitro and in vivo (18). The plasmid pUb-vv-GFP backbone was a kind gift from Dr. Michael A. Barry (University of Minnesota) (19).
Detection of Mut H-ras–specific cytokine production
The lymph nodes (LNs) of DMBA-sensitized mice were harvested after 5 d, and T cells were purified by CD90.2 MACS beads (Miltenyi Biotec). Naive T cells were also obtained in parallel. Bone marrow dendritic cells (BMDCs) from naive mice were cultured with GM-CSF and IL-4 for 7 d, then pulsed with or without 100 μg DMBA or the indicated peptides at 300 μg/ml for 30 min at 37°C in PBS. Ag-pulsed BMDCs were washed, then added to DMBA T cells at a 1:10 ratio (100 μl each: mixing 2 × 104 BMDCs with 2 × 105 T cells) for 48 h. The mean and range of concentrations determined from duplicate wells of IL-17– and IFN-γ–specific ELISAs are shown.
Ag-specific delayed-type hypersensitivity response
Elicitation of specific responses was assessed 5 d following the last immunization with DNA-based or DC-based vaccines, or DMBA sensitization. Elicitation was done by intradermal (i.d.)injection peptide (50 μg in 20 μl PBS) or by DMBA ear painting. Peptide refers to wild-type (WT) or Mut H-ras 9-mer amino acids 59–67, WT with Q at codon 61, and Mut with L at codon position 61. Baseline ear thickness measurements were taken before and after elicitation over 3–5 d using a dial thickness gauge spring-loaded micrometer (Mitutoyo 7301). The maximum increment in ear thickness compared with the baseline pre-elicitation level was used to quantify the magnitude of the response. Naive mice, which were not vaccinated but were challenged with peptide, served as negative controls. In some experiments, positive control mice were immunized with Mut H-ras peptide by i.d. injection into one ear; then elicitation responses were measured after injection into the other ear. When DMBA was used for sensitization and/or elicitation phases of the hypersensitivity response, DMBA was applied on the abdomen for sensitization (100 μl 0.1% w/v DMBA in acetone) and ear for elicitation (20 μl 0.1% w/v DMBA in acetone). Ear measurements were taken as described above.
Generation of genetic immunization plasmid DNA
To ensure that the mutant H-ras epitope efficiently traffics to the MHC-I peptide loading pathway, we have generated the mini-gene sequences encoding the mutant and WT H-ras epitope. The nucleotide sequences encoding codons 59–67 contained a single change in codon 61 (CAA → CTA), resulting in a change from glutamine (Q) to leucine (L) for mutant H-ras. This was achieved by designing complementary pairs of DNA primers, 27 bp in length (mutant AGLEEYSAM: 5′-GCCGGCCTGGAGGAGTACTCCGAAATG-3′; WT AGQEEYSAM: 5′-GCCGGCCAGGAGGAGTACTCCGAAATG-3′). These primers were flanked with sequences containing BamHI restriction enzyme sites compatible for inframe cloning into the genetic-immunization vector pUb-vv-GFP (plasmid backbone was a gift from Dr. Michael A. Barry) (19). The pUb-vv-GFP vector enhances the generation of CD8+ T lymphocytes by virtue of its superior proteosome targeting of the encoded ubiquitin (Ub)/antigen/enhanced GFP fusion protein, which promotes MHC class I Ag processing (Supplemental Fig. 1). The Ub fusion to the H-ras–GFP protein enhances MHC class I peptide processing because the process of Ub-dependent proteolysis and peptide trimming is linked with efficient epitope (peptide) loading (20, 21). The following plasmids were generated, sequence verified, and used as genetic immunization DNA: empty vector (pUb/GFP); WT H-ras vector (pUb/H-ras (Q61)/GFP); and mutant H-ras vector (pUb/H-ras(L61)/GFP). Simplified names for these genetic immunization plasmids used throughout this article are the following: GFP-DNA, WT H-ras-DNA, and Mut H-ras-DNA.
Generation of stable DC transfectant cell lines
The cloned fusion protein DNA segments from these plasmids were subcloned into lentivirus vectors for generating stable XS106 cell lines. A double digest was performed on the plasmids for directional cloning (BamHI and Mfe I for lentivirus; BglII and EcoR1 for Ub-vv-GFP). The inserts containing the Ub-H-ras (Q61L)-GFP gene sequences were purified and ligated with the digested, gel-purified lentiviral vector. The resultant transformant clones were screened and sequence verified. The vectors were transfected, along with helper plasmids, into 293T cells, and supernatants were harvested over 72 h. The supernatants were used to infect the Langerhans-like dendritic cell (DC) line XS106, which was analyzed by flow cytometry 3 d later. Although the Ub-GFP turnover is quite high in lentivirus-infected cells, a dull fluorescent signal allowed us to selectively sort the upper third fluorescence intensity to generate pure GFP-positive XS106 cells to generate stable cell lines (Supplemental Fig. 2). The GFP levels remained stable over many months but, when necessary, were resorted prior to their use in experiments.
Genetic immunization
Genetic immunization using previously published techniques (22–24) was performed with DNA vectors and purified using endotoxin-free QIAGEN kit columns. DNA (100 μg) in distilled water was transferred to a spot adhesive bandage, which was immediately applied to flank skin, previously prepared by shaving and depilatory treatment, and then removed after 18–24 h. Mice received two or three immunizations in 7- to 14-d intervals. A separate cohort of mice from each experimental group (n = 3 per group) were tested for positive delayed-type hypersensitivity (DTH) response to mutant H-ras peptide prior to performing the two-step carcinogenesis protocol.
DC-based vaccination
The following DC lines (XS106-parent) were used to vaccinate A/J mice: DC-Ub/GFP, DC-Ub/WT H-ras/GFP, and DC-Ub/Mut-H-ras/GFP. For simplicity, they are designated in the article as follows: DC-GFP, DC–WT H-ras, and DC–Mut H-ras. For each immunization or boost, cells from each DC line were prepared and checked for >85% viability and then injected into the nape of the neck (1–5 × 106 cells in 1 ml PBS) per Timares et al. (18). As controls for DC-based immunization, either the parental XS106 (DC) or the transduced DC-GFP line was used alone as a negative control, or pulsed with mutant H-ras peptide as a positive control. DCs (107 cells/ml) were pulsed with mutant H-ras peptide (100 μg/ml) in serum-free RPMI 1640 for 30 min, then washed three times with PBS and viability checked prior to injection.
Two-step carcinogenesis
Application of the two-step carcinogenesis protocol in mice has been used to define three distinct and sequential steps of tumorigenesis: 1) Initiation occurs when a carcinogen, such as DMBA, induces mutations in proto-oncogenic/regulatory genes, resulting in preneoplastic “initiated” cells. 2) Promotion refers to the outgrowth of initiated cells, which requires a strong stimulus provided by a tumor promoter. The phorbol ester TPA is an analog of diacyl glycerol and a potent activator of protein kinase C signaling. Proliferation is reversible upon TPA withdrawal, and regression of benign tumors is observed, whereas stably transformed cells will continue to grow. 3) Progression occurs when secondary mutations accumulate during promotion, leading to stable transformation and the acquisition of invasive/metastatic properties (16, 25).
A single application of 400 nmol DMBA (100 μl 0.1% w/v in acetone) was painted on the shaved dorsal skin of immunized C3H/HeN or A/J mice 1 wk after their last immunization. Promotion was started a week later with TPA (40 nmol in acetone) applied to the dorsal skin twice a week. Mice were shaved weekly for monitoring tumor development. Tumor size was determined by measuring tumor length, width, and height using a digital vernier caliper–micrometer. Tumor volume was calculated using the following formula for an ellipsoid dome: Vo = (4/3)π(l/2)(w/2)(h/2) = 0.52(l)(w)(h), where l is length, w is width, and h is height. Tumors >3 mm3 and present for ≥2 wk were tallied.
Immunofluorescence staining of tumor sections
Tumors from various groups were randomly picked and fixed in 10% formalin overnight. The tissues were paraffin embedded, and 5-μm sections were cut and stained with H&E. For immune cell staining in tumors, tissues were embedded in Tissue-Tek solution and frozen in liquid nitrogen. Sections were cut as 7-μm slices on a cryotome, fixed, and rehydrated as described above. They were then stained with conjugated Abs specific for mouse CD4 (FITC) or Foxp3 (PE) or CD8 (PE). Image capture was performed using an Olympus DP70 camera and DP Manager software. All Abs used are listed in Supplemental Table I.
In vivo CTL assay
In vivo CTL assays followed established protocols (26). Briefly, spleen cells from naive A/J or C3H/HeN mice (depending on the immunized recipient strain) were prepared as single-cell suspensions to 107/ml in 2% FBS–RPMI 1640. The specific target population was pulsed with 100 μg/ml Mut H-ras peptide. The negative control target population was either pulsed with the WT H-ras nonamer peptide or not pulsed with peptide. Cells were incubated for 60 min at 37°C, then were washed twice in PBS and brought up in Ca/Mg-free HBSS or PBS for labeling with CFSE (Invitrogen/Molecular Probes). Cells at 50 million per milliliter PBS were incubated with either 5 μM (CFSEhi) or 0.5 μM (CFSElo) concentrations for 8 min at room temperature, and then quenched by addition of FBS (20% final). Cells were washed more three times, then mixed 1:1 prior to injection into recipient mice. A total of 2 million cells per 200 μl Ca/Mg-free PBS (at room temperature) were transferred into mice by i.v. injection into the eye sinus. Recipient mice were euthanized 18–24 h later, and the harvested splenocytes were analyzed by flow cytometry to determine the percentage of CFSEhi and CFSElo cells. Flow cytometric acquisition of all events was collected until 2000 CFSElo cells were detected per sample. The percentage of Mut-H-ras–specific cytotoxicity is calculated as follows: 100 * [1 − ([CFSEhi/Total CFSE+] per sample/Average [CFSEhi/Total CFSE+] of naive mouse group)]. For most experiments there were three mice per group. The mean ± SEM value is shown.
Analysis of IFN-γ–producing CD8 T cells
Immunized mice subjected to the carcinogenesis protocol, or not (control), were sacrificed at the end of 25 wk. Skin-draining LNs (cervical and inguinal) were pooled for each individual mouse for analysis. Single-cell suspensions, prepared by collagenase D (2 mg/ml; Roche) and DNase I (20 μg/ml) digestion, were cultured in the absence or presence of peptides (WT or Mut H-ras 100 μg/ml) for 2 d. A set of cells cultured without peptides was activated with PMA (50 ng/ml; Sigma-Aldrich) plus calcium ionophore A23187 (500 ng/ml; Sigma-Aldrich) for the final 16 h. All cultured cells were treated with GolgiStop (BD Biosciences) for a further 6 h. Cells were first stained with Aqua (405 nm) LIVE/DEAD dye (Invitrogen), then pretreated with Fc-R blocking Ab CD16 (2.4G2) and stained for IFN-γ and CD8, following BD Biosciences intracellular staining kit directions. Supplemental Table I lists details pertaining to the Ab clones used in this study. Flow cytometric analysis was performed on an LSR II, using FlowJo v9.5.2 software.
Detection of H-ras (Q61L) mutation in tumors
Tumors of similar size were carefully harvested and trimmed of uninvolved tissue to reduce contamination of normal cells that can dilute the H-ras (Q61L) signal of the mRNA source. Trimmed tumors were homogenized in TRIzol and processed for mRNA isolation per the manufacturer’s instructions. Then, 1 μg RNA was used to synthesize cDNA, according to the iScript cDNA synthesis kit (Biorad) instructions. We used the method of allele-specific competitive blocker–PCR (ACB-PCR), which suppresses amplification of the WT H-ras allele using a nonextending primer in conjunction with a primer that specifically amplifies the mutant sequence allele, as described originally by Parsons et al. (27) and recently improved upon by Morlan et al. (28). SCCs that arise by DMBA exposure are associated with a mutation in H-ras that changes the 61st codon from CAA to CTA. The melting temperature of the allele-specific primer was designed to be ∼10° below the annealing melting temperature of the WT allele blocking primer and during PCR extension (∼60°C). The WT allele blocking primer was phosphorylated at the 3′ end to block primer extension. The primers used were as follows: WT forward, 5′-CAGCAGGTCAAGAAGAGTATAGTGCCA-PO4-3; mutant forward, 5′-CATCTTAGACACAGCAGGTCT-3′; and common reverse, 5′-GCGAGCAGCCAGGTCACAC-3′. The blocker allele and mutant allele–specific primers were used at a 4:1 ratio (WT:Mutant), using RT-PCR protocols optimized for increased specificity and sensitivity in detecting the H-ras Q61L point mutation. Thermocycling conditions were 10 min at 95°C and 35 cycles of 15 s at 95°C, 20 s at 60°C, and 15 s at 72°C. To determine the sensitivity of the assay, CH72 cells (Mut H-ras+/−) were added to murine cell line Pam212 to create a standard curve of Mutant:WT H-ras genomes ranging from 101 to 107 (<1 mutant cell in 105 normal cells can be detected; data not shown). Analysis was performed using an Eppendorf thermocycler. To quantify the amplicon products in gel imaging, National Institutes of Health ImageJ densitometry reading of equivalent area per lane was performed, subtracting gel background noise. Relative Mut H-ras gene expression per sample was normalized as the ratio of Mut H-ras densitometry value to the housekeeping gene GAPDH or β-actin.
Therapeutic cell transfer experiments
Mice with similar tumor burdens in each group were chosen as cell donors. Spleen and LN cells were pooled from three mice per group, and 25 × 106 cells were transferred by i.v. eye sinus injection into recipient GFP-DNA–vaccinated mice in duplicate. Cells were reserved for phenotyping to determine the cellular makeup of donor populations. Recipients displayed similar tumor burdens at the time of injection. Tumor measurements taken just prior to cell injections at 28 wk were considered baseline, time 0. The difference in size for each individual tumor was monitored weekly over the course of 3 wk.
Statistical analysis
All experiments were carried out at least two times to assess reproducibility. All data were processed by the GraphPad Prism 4.0 program (GraphPad Software for Macintosh). Unless otherwise stated, quantitative data are presented as the mean ± SD. Statistical significance for the majority of in vitro experiments was determined by the two-tailed Student t test. Dr. Naomi S. Fineberg (Division of Biostatistics, University of Alabama at Birmingham School of Public Health) performed statistical analysis of tumor development. Two-way ANOVA and Tukey multiple comparison tests were applied. For DC-vaccine studies of tumor development, data were first normalized using logarithmic transformation values for statistical analysis. Any p value ≤ 0.05 was considered significant. All other statistics were determined using Prism 4.0c (GraphPad Software) algorithms for two-way ANOVA and Tukey comparison tests, or two-tailed Student t tests, as indicated in the figure legends.
Results
Early detection and expression of mutant H-ras genes in DMBA-treated skin
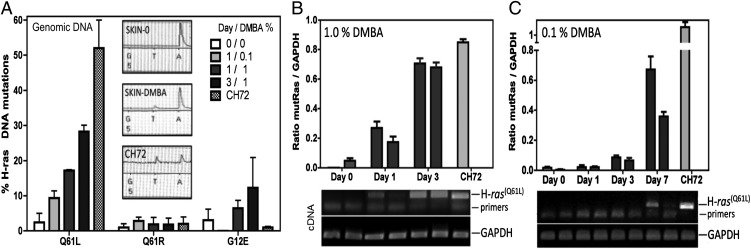
DMBA undergoes enzymatic conversion to a reactive diol epoxide that forms DNA adducts causing the characteristic A → T transversion in the 61st codon of the H-ras oncogene (mutant H-ras Q61L) found in skin tumors (29). Initial studies were conducted to establish that the same H-ras mutations occurred in the skin of C3H/HeN mice. DMBA was applied at doses known to be tumorigenic when followed by chronic TPA treatment, and then genomic DNA and mRNA were isolated from skin to assess the gene mutation load and to determine if mutant genes were expressed. Pyrosequencing-based quantification of the genetic mutation load revealed a dose- and time-dependent increase in the number of mutant H-ras genes (Fig. 1A). After 1 d, the relative frequency of codon 61 mutations was determined to be 10–17% for 0.1–1% DMBA, respectively. Of interest, the frequency of mutant H-ras alleles in 1% DMBA–treated skin increased almost 2-fold to 28% by day 3. This result indicates that early autonomous expansion of mutated cells can occur without TPA-induced signals, which are required for promotion. Within the short time frame studied in this work, the majority of expanded mutant cells most likely reflect transient amplifying cells, which are destined to differentiate and slough off. It is proposed that mutated keratinocyte stem cells remain latent because they cycle infrequently, and that TPA is required to promote their outgrowth (30); however, it is not clear to what extent they might proliferate as part of a wound-healing response to repair skin damaged by DMBA treatment or as a consequence of direct activation resulting from the induced oncogenic H-ras point mutation (31).
FIGURE 1.
Early detection of H-ras mutations and oncogene expression in DMBA-treated skin. (A) Identification of DMBA-induced H-ras codon 61 mutations by pyrosequencing. Quantification of Q61L, Q61R, and G12E mutant H-ras alleles in genomic DNA from samples of skin treated with 0, 0.1 (400 nmol), or 1% (4 μmol) DMBA after 1 or 3 d (n = 2 mice per treatment; mean ± range is shown). Insets: pyrosequence tracing peaks for missense codon 61 mutation A → T. Pyrosequence quantification validation: the CH72 cell line, heterozygous for the H-ras Q61L allele, indicates 50% of its H-ras alleles are mutated. (B and C) Relative expression of Mut H-ras mRNA in (B) 1% or (C) 0.1% DMBA-treated skin samples. Skin samples from two mice per condition were examined. Densitometry of amplicons generated by H-ras Q61L-specific ACB-PCR and GAPDH-specific PCR of cDNA made from individual skin tissues. CH72 mRNA serves as a positive control. Bar graphs display Mut-H-ras gene expression levels when normalized to expression levels of a housekeeping gene GAPDH, as shown. The corresponding gel used to provide densitometry readings is shown below each bar graph (mean ± range of duplicate densitometry readings for each sample is shown). The data are from one of two separate experiments.
Mutant H-ras oncogenes were expressed in an early time frame, with kinetics similar to detection of genomic mutations. We used an established ACB-PCR (27, 28, 32) and observed abundant levels of Mut H-ras mRNA expression in skin cells 1 d following 1% DMBA application (Fig. 1B). In contrast, after 0.1% DMBA application, the undetectable frequency of Mut H-ras cells reaches detectable levels, in the absence of promotion with TPA, after 7 d, suggesting that clonal expansion of mutant cells occurred (Fig. 1C). Thus, early expression of the activated H-ras Q61L oncogene indicates that it may serve as an early transformation target for recognition by immunosurveillance.
Elicitation of cell-mediated immunity to DMBA with mutant H-ras peptides
We, and others, have shown that topical application of DMBA results in the development of a cell-mediated immune response that is dependent on DMBA metabolism (15, 33). This finding raises the question as to the actual antigenic moiety responsible for DMBA CHS, which is not known. As a classic hapten, the DMBA metabolite decorates cellular proteins, which are processed and presented as hapten-conjugated peptides to DMBA-specific T cells (34, 35). Alternatively, gene mutations introduced by repair of carcinogenic DMBA-DNA adducts (9) may be expressed as neoantigens. These mutant neoepitopes may contribute to DMBA-specific CHS responses and may serve an important role in marking damaged, preneoplastic cells for elimination by immunosurveillance.
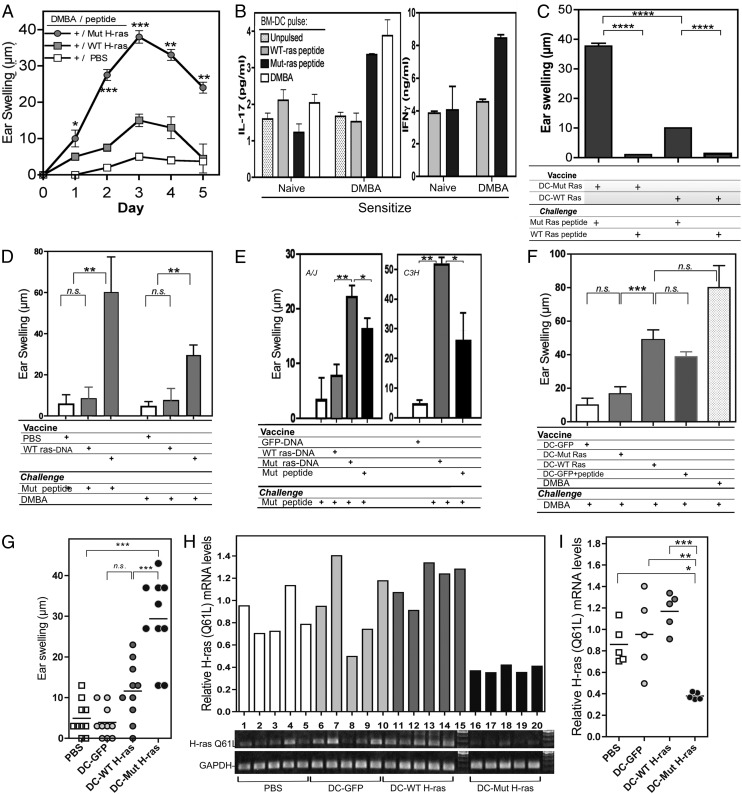
To investigate whether part of the normal CHS response to DMBA is directed to the DMBA-associated neoantigen encoded by the H-ras Q61L mutation, we challenged DMBA-sensitized C3H/HeN mice with mutant H-ras (Mut H-ras) peptide in the ear, then measured DTH ear swelling responses. Mice sensitized with DMBA developed significant responses, whereas mice injected with WT H-ras peptide or vehicle (PBS) exhibited diminished responses (Fig. 2A). The level of swelling observed in response to WT H-ras peptide is comparable to swelling elicited by any nonspecific Ag (e.g., OVA peptide; data not shown). In addition, purified T cells from DMBA-sensitized mice could be stimulated to produce IL-17 and IFN-γ in vitro by BMDCs pulsed with Mut H-ras peptides (but not by unpulsed or WT H-ras peptide–pulsed BMDCs) (Fig. 2B). The results reveal that DMBA-specific cellular immunity is directed, in part, to neoepitopes that are generated by expression of DMBA-induced mutant oncoproteins, rather than (or in addition to) the DMBA moiety itself.
FIGURE 2.
Specificity of DMBA and Mut H-ras T cell responses. (A) DMBA-sensitized T cells respond to Mut H-ras peptide elicitation. (A) DTH responses. Ear-swelling responses elicited by the indicated peptide after DMBA sensitization are shown (n = 5 per group). (B) Cytokine production by DMBA-sensitized T cells stimulated by Mut H-ras peptide–pulsed BMDCs. T cells from the LNs of DMBA-sensitized mice were cocultured with DMBA-pulsed or unpulsed BMDCs from naive mice at a 1:10 ratio for 48 h. The mean and range concentration values of duplicate wells for IL-17 and IFN-γ ELISAs are shown. (C–F) Mut H-ras vaccines induce responses that can be elicited by DMBA. (C) Specificity of DC-based vaccine. Mice were vaccinated, then challenged by i.d. injection of the ear with WT or Mut H-ras peptide. Specific ear-swelling responses on day 3 are shown (n = 3 per group, repeated twice). (D) Engineered DC vaccination induces responses that are elicited by DMBA challenge. DC-GFP serves as a specificity control. Note that day 3 responses induced by Mut H-ras–engineered DC and peptide-pulsed DC-GFP to DMBA elicitation are equivalent. DC-GFP serves as a specificity control. (E) Specificity of DNA-based vaccine in A/J and C3H/HeN mice. A/J (left) or C3H/HeN (right) mice receiving DNA-based vaccines generate comparable responses to Mut H-ras peptide. Day 3 response is shown. (F) Mut H-ras DNA vaccination responses can be elicited by Mut H-ras peptide or DMBA. Day 3 response is shown (n = 5 per group, more than two experiments each). (G–I) DC–Mut H-ras vaccines eradicate Mut-H-ras mRNA expression in DMBA-treated skin. DC-vaccinated mice were challenged by either (G) i.d. injection of Mut H-ras peptide (100 μg) or (H and I) painting the dorsal skin with 100 μL 0.1% DMBA (400 nmol) (n = 5 mice per group). (G) Specific ear-swelling responses on day 3 and (H and I) Mut H-ras mRNA expression levels in skin, assessed by ACB-PCR on day 7. (H) gel (bottom). GAPDH expression is a loading control (bottom row). (H, top) Relative H-ras (Q61L) mRNA expression: the ratio of Mut H-ras over the GAPDH densitometry values is shown for tumors per mouse (3 tumors pooled per mouse, n = 5 mice per group). (I) Scatter plot of data in (H). Significant differences (by Student t test) between Mut H-ras–immunized samples from controls are shown. *p < 0.05, **p < 0.01, ***p < 0.001. n.s., not significant.
Development of epitope-focused vaccines
On the basis of these findings, we set out to test whether immunization with the mutant H-ras epitope can raise an effective immunoprevention response against development of carcinogen-induced skin tumors. We anticipated that to generate an effective response, we would need to develop a strategy that would focus TCR recognition exclusively toward the neoepitope, consisting only of the point mutation, without also inducing an unwanted side effect of generating autoimmune responses to normal self-epitopes of H-ras. Raising robust cellular immune responses to a point mutation epitope is a challenge that may be overcome by harnessing the effective Ag-presenting capacity of DCs. Furthermore, epitope expression and Ag presentation were engineered to selectively present to CD8+ T cells rather than regulatory CD4+ T cells, which are normally favored by DMBA (16). We first tested whether the XS106 DC line (A/J strain origin) could be effective at inducing a response to the mutant, rather than the WT peptide, and found that peptide-pulsed cells could activate a DTH response specific for the mutant, rather than WT peptide epitope challenge (data not shown). As proof-of-principle, we developed a panel of genetically modified DC lines to selectively expand CD8 T cells with focused recognition of the single Q(61)L point mutation of the H-ras oncogene. We constructed a chimeric gene encoding the DNA sequence for the H-ras Q61L point mutation (and nonmutant sequence as a control [WT H-ras]) 9-mer epitope flanked inframe to an N-terminal Ub sequence, optimized for targeting to the proteasome (19), and the GFP reporter gene at the C terminus, to identify cells that express the transgene. Because Ub-dependent proteolysis is linked with efficient peptide loading onto MHC class I molecules (21), the embedded H-ras epitope in the chimeric protein should undergo enhanced processing and presentation to CD8+ T cells (Supplemental Fig. 1A). Details regarding the cloning strategy are shown in Supplemental Fig. 1B. The chimeric gene construct was subsequently cloned into a lentivector system and used to infect XS106 cells, a well-characterized skin DC line derived from A/J mice (17, 18). GFP+ XS106 cells (hereafter referred to as engineered DCs) were sorted such that all lines expressed comparable GFP levels (Supplemental Fig. 2A). Confirmation of equivalent GFP expression levels was done prior to immunization to eliminate that variable as a confounding issue. Peptide-pulsed DCs and engineered DCs were equally potent in generating DTH responses to Mut H-ras peptide (Supplemental Fig. 2B). In contrast, DCs transduced with WT H-ras lentivector or with the empty vector failed to induce significant DTH responses. The results provide evidence that the epitope embedded into the chimeric protein is efficiently processed and presented as an immunogenic Mut H-ras peptide–MHC complex. For simplicity, the engineered lines are hereafter referred to as DC–WT H-ras, DC–Mut H-ras, and DC-GFP (empty vector).
Specificity of the response induced by the H-ras point mutation epitope
To determine if the epitope-focused vaccine posed a threat for generating autoreactive T cell responses to endogenous WT H-ras, A/J mice were vaccinated and boosted with either DC–Mut H-ras or DC–WT H-ras, then, after 5 d, challenged with mutant or WT peptides (Fig. 2C). Only when the specificity of the immunizing DC and peptide challenge were matched for Mut H-ras did a significant DTH response develop. A similar pattern of swelling responses was detected in DC-vaccinated A/J mice when DMBA was used to elicit the response (Fig. 2D).
Although DC-based vaccination represents the most potent way to immunize animals, DC-based vaccination has practical limitations for use as a general approach to human immunization. Therefore, we tested whether a genetic immunization protocol, using plasmid DNA vectors (diagramed in Supplemental Fig. 1B), could be optimized to generate comparable levels of Mut H-ras–specific immunity. We compared standard s.c. injection of peptide with epicutaneous immunization using DNA patches loaded with plasmids encoding fusion genes: Ub-Mut H-ras-GFP, Ub-WT H-ras-GFP, and Ub-GFP. Genetic immunization with the Mut H-ras chimeric gene generated significant ear-swelling responses to Ag challenge with Mut H-ras peptide injection into the ear pinnae of A/J and C3H/HeN strain mice (Fig. 2E, 2F), or elicitation with DMBA, but to a lesser magnitude (Fig. 2F). Thus, DNA-based and DC-based vaccines were both effective at generating T cells that could be recalled by elicitation with DMBA 5 d after finishing the immunization procedure.
To see if DC-vaccinated mice affected the frequency of DMBA-induced Mut H-ras–expressing skin cells, we assessed expression levels of the mutant allele by ACB-PCR. Following immunization, a response was confirmed 3 d after DMBA elicitation by measuring specific ear-swelling responses in a separate cohort of mice (Fig. 2G). Mutant H-ras mRNA expression levels were assessed in DMBA painted skin after 7 d. DMBA-treated skin samples taken from unvaccinated (PBS) and control vaccinated cohorts of mice expressed high levels of mutant H-ras mRNA (Fig. 2H, 2I). In contrast, DMBA-treated skin from DC–Mut H-ras–vaccinated mice expressed only low levels of mutant H-ras mRNA, consistent with rapid elimination of H-ras oncogene–positive skin cells. The results indicate that 1) Mut H-ras vaccines can induce an H-ras point mutation–specific immune response; 2) the mutant oncoprotein epitope is naturally processed and presented by preneoplastic cells generated by DMBA; and 3) presentation of the neoepitope is rapid, occurring at the earliest stages of cell transformation.
Mut H-ras vaccination generates specific CD8 T cells with potent CTL activity
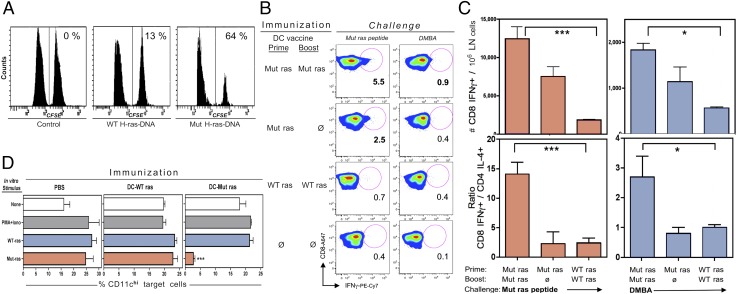
To determine if reduced mutant mRNA expression in DMBA-treated skin was due to vaccine-induced cell-mediated cytotoxicity, we measured mutant peptide–specific CTL activity in vaccinated A/J and C3H/HeN mice by in vivo and in vitro approaches. Specific CTL activity was observed in an in vivo CTL assay. A selective loss of mutant peptide–pulsed CFSE high (CFSEhi)–labeled spleen cells was observed 18 h after transfer of these cells, mixed 1:1 with WT peptide–pulsed CFSE low (CFSElo)–labeled spleen cells, only in Mut H-ras DNA–vaccinated mice (64% cytotoxicity), but not in control WT H-ras DNA–vaccinated (13%) or unvaccinated (0%) mice (Fig. 3A).
FIGURE 3.
Vaccine-induced CTL activity and CD8 T cell expansion. (A) In vivo CTL assay. C3H/HeN mice were immunized and boosted with DNA-based vaccines or PBS (Control), then injected with CFSE-labeled splenocytes pulsed with WT H-ras (CFSElo) or Mut H-ras (CFSEhi) peptides. The CFSE-stained cells remaining in spleen were analyzed the following day. The % specific cytotoxicity is indicated. (B and C) DC-based immunization raises DMBA-responsive Mut H-ras–specific effector T cells. A/J mice were immunized with DC vaccines once (Prime only) or twice (Prime/Boost), as indicated. At 10 d later, mice were challenged with either Mut H-ras peptide or DMBA. At 3 d later, LN cells were prepared and cultured with PMA + ionophore (B and C) or the indicated stimulus (D) and then stained for intracellular cytokines and phenotypic markers. (B and C) IFN-γ+ CD8 T cells expand in a vaccine dose–dependent manner. (B) IFN-γ+ CD8+ T cells. Mut ras peptide or DMBA elicits IFN-γ+ CD8 cells. The % of IFN-γ+ gated CD8 cells is shown. (C) Selective expansion of CD8 over CD4 T cells. The absolute number of IFN-γ+ CD8 T cells per million LN cells expanded in vivo by either Mut ras peptide or DMBA challenge (top row). CD8/CD4 effector cell ratio = the number of IFN-γ+ CD8 T cells over the number of IL-4+ CD4 T cells per million LN cells (mean ± SEM of triplicate mice). (D) Ag specificity of CTL. Bar graphs display the % of CD11chi target cells within the CD11c+ gate for each set of stimuli per immunized group. LN cells from immunized mice were harvested 3 d after DMBA Ag elicitation and then cultured for 6 h with PMA + ionophore or overnight with peptides, as indicated. The percentage of CD11chi target cells in the CD11c DC gate was determined by flow cytometric analysis. The presence of WT H-ras peptide or PMA + ionophore did not affect DC profiles; however, Mut H-ras peptide cultures lost 87% of CD11chi cells. (n = 3 per group). Student t test: *p < 0.05, ***p < 0.001 (one of more than three similar experiments).
The detection of IFN-γ+ CD8 T cells is a correlate of mature CTL activity (36). DC–Mut H-ras–immunized mice generated increased numbers of IFN-γ–producing CD8 T cells in response to Ag challenge by either mutant peptide or DMBA elicitation, in a vaccine dose–responsive manner (Fig. 3B, 3C). The ratio of expanded IFN-γ+ CD8+ T cells to IL-4+ CD4+ T cells increased 14-fold in response to mutant peptide and 2.7-fold in response to DMBA (Fig. 3C). These data provide evidence that engineered Ub-tagged chimeric protein expressed by genetically modified DCs favors presentation to CD8 rather than CD4 T cells, with exquisite specificity for the Mut H-ras epitope.
Evidence for the Ag specificity of CTL activity was observed in vitro when peptide-presenting DCs, which serve as target cells, were enumerated in cultures of splenocytes from vaccinated mice following overnight stimulation with nonspecific PMA + ionophore, or peptides of WT H-ras or Mut H-ras epitopes (Fig. 3D). The presence of Mut H-ras peptide resulted in a profound loss (87%) of CD11chi DCs only in cultures from DC–Mut H-ras–vaccinated mice (in which only 2% of the initial 22% of CD11chi cells within the CD11c gate remained). Selective loss of CD11chi cells was not seen in the presence of any other stimuli or in any of the other cell cultures. These results suggest that CD11chi peptide–pulsed APCs serve as target cells for vaccine-induced Mut H-ras–specific CTLs.
Immunoprevention against chemical carcinogenesis by DNA- and DC-based epitope-focused vaccines
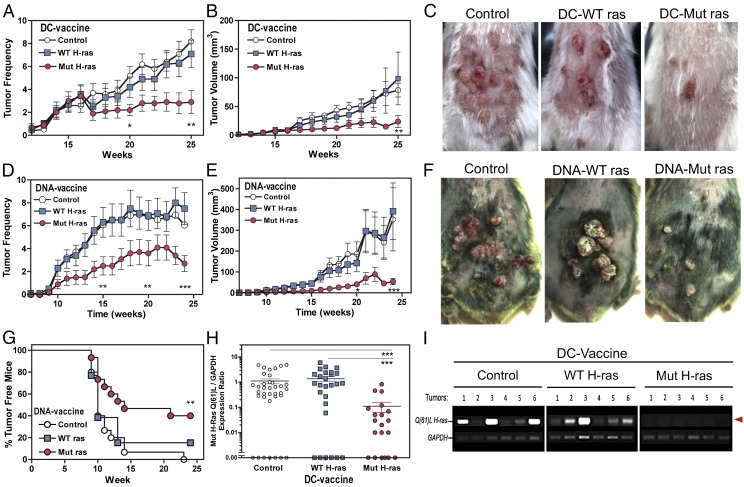
To investigate whether immunization to a single mutant epitope can protect against the development of carcinogen-induced tumors, we subjected optimally immunized cohorts of mice to the standard two-step DMBA/TPA carcinogenesis protocol for 25 wk. We first tested our panel of engineered DC vaccines in A/J mice. The average number and size of tumors that developed per mouse in each group was monitored over time (Fig. 4A–C). Both tumor numbers and tumor growth were effectively restricted only in mice immunized with the DC–Mut H-ras vaccine.
FIGURE 4.
Immunoprevention against chemical carcinogenesis in two mouse strains and evidence for immunoediting. (A–C) Engineered DC-based vaccination of A/J mice. Mice were immunized twice with the indicated DC vaccine, then subjected to DMBA/TPA carcinogenesis over 25 wk (n = 20 per group). The individual number of tumors per mouse (A) and the average tumor volume per mouse (B) per group are shown (mean ± SEM). Tumor incidence and growth rates were inhibited by 70% and 90%, respectively (the Tukey significance test, p < 0.01). (A and B) Two-way ANOVA and the Tukey posttest calculated significance are indicated: *p < 0.05, **p < 0.01. (C) Photodocumentation of representative mice per vaccination group is shown. (D–G) DNA-based epicutaneous vaccination of C3H/HeN mice. Mice received three vaccinations over 15 d, then were subjected to DMBA/TPA carcinogenesis over 24 wk (n = 20 per group). (D) The average tumor numbers per mouse in each group. (E) The average tumor volume per mouse in each group. The Mut H-ras DNA–vaccinated group developed 50% fewer tumors, and tumor growth was inhibited by >85% (p < 0.01, p < 0.001, respectively). (F) Representative photographs of tumors that develop in DNA-vaccinated mice. (G) Tumor-free plot. Tumor penetrance was inhibited in C3H/HeN mice immunized with Mut H-ras DNA–based vaccine. Tumor-free mice are defined as having no tumors or only small tumors < 3 mm3. (H and I) Immunoediting in tumors from Mut H-ras–vaccinated mice. (H) Relative quantification of Mut H-ras mRNA expression levels in individual tumors. The densitometry ratio for Mut H-ras ACB-PCR and matched GAPDH amplicon products of each tumor sample, as shown for six tumors in (I), as individual data points. The number of tumors screened was as follows: PBS, n = 30; DC–WT H-ras, n = 25; DC–Mut H-ras, n = 20. Expression was negligible in 45% of tumors from Mut H-ras–vaccinated mice and in 15–22% of tumors from the control groups. (G and H) Two-way ANOVA statistical significance is indicated: **p < 0.01, ***p < 0.001. (I) Loss of H-ras Q61L mutation expression in tumors from DC–Mut H-ras–vaccinated mice. The Q61L H-ras allele (upper band, red arrow) detected by ACB-PCR in mRNA from individual tumors harvested from A/J mice after 25 wk of carcinogenesis. GAPDH serves as an internal gene expression control.
Then we tested whether DNA-based vaccines could similarly inhibit chemical carcinogenesis in C3H/HeN mice, a strain that is highly susceptible to DMBA-induced tumor formation. The average tumor number and tumor volume per mouse over time are shown in Fig. 4D–F. The appearance of initiated tumors was slightly delayed, and their number and growth were significantly reduced only in mice that received the Mut H-ras DNA vaccine. Further, ≤40% of Mut H-ras–vaccinated mice remained free of tumors >3 mm3 (Fig. 4G).
Evidence for immunoediting
The tumors were screened for expression of mutant H-ras mRNA, using the previously established ACB-PCR method. Mutant H-ras mRNA was expressed in 80% of the tumors from control and WT-ras–vaccinated mice, but was low or undetectable in the majority of tumors from mice receiving DC–Mut H-ras vaccines (Fig. 4H, 4I), suggesting that vaccine boosted immune-mediated eradication of mutant H ras oncogene–transformed skin cells or that tumor cells expressing only the lowest levels of the oncogene were immune selected.
The histologic pathology of remaining tumors (14–57 tumor specimens per group), read in a blinded fashion by experienced dermatopathologists, revealed no significant differences in the distribution of tumor types or stages (data not shown). However, in contrast to the lines established from the tumors of all control cohorts, tumor-derived cultures from Mut H-ras–immunized mice were difficult to establish and did not grow well as orthotopic transplants in nude mice (data not shown). Thus, immunoediting did not result in tumors with increased metastatic properties.
Collectively, these experiments show that engineered DC-based and epicutaneous DNA-based vaccines provide prophylactic protection against development of DMBA-induced skin tumors in both A/J and C3H/HeN mice. Evidence for the development of autoimmune disorders was not observed in mice immunized with control, WT H-ras (or Mut H-ras)–encoding DCs, or plasmid vaccines (data not shown).
DMBA presensitization as a protective vaccine
Because a portion of the DMBA-specific CHS response recognizes the Mut H-ras epitope, we tested whether sensitizing mice with subcarcinogenic doses of DMBA (25 μg or 100 nmol) could induce immunity against Mut H-ras mRNA–expressing skin cells. DMBA-treated skin of DMBA-sensitized mice, as compared with nonsensitized mice, expressed half the levels of Mut H-ras mRNA (Supplemental Fig. 3A). This result prompted us to directly compare the efficacy of low-dose DMBA presensitization with that of Mut H-ras DNA–based immunization in C3H/HeN mice. Low-dose DMBA presensitization in these mice led to low tumor numbers; however, tumor volumes were significantly higher than in Mut H-ras–immunized mice (Supplemental Fig. 3B, 3C).
Immune status of vaccinated tumor-bearing mice
Mice immunized with DNA-based vaccines were subjected to DMBA/TPA carcinogenesis for 20 wk, and then rested for 8 wk to allow transformed tumors to stabilize and benign tumors to regress. A cohort of vaccinated mice was treated in parallel with DMBA at initiation but not TPA promotion; therefore, no tumors were formed (Control). Naive mice were untreated. After 28 wk, LN and spleen cells were processed from mice with similar tumor burdens. Samples from individual mice were stained for phenotypic analysis, using flow cytometry.
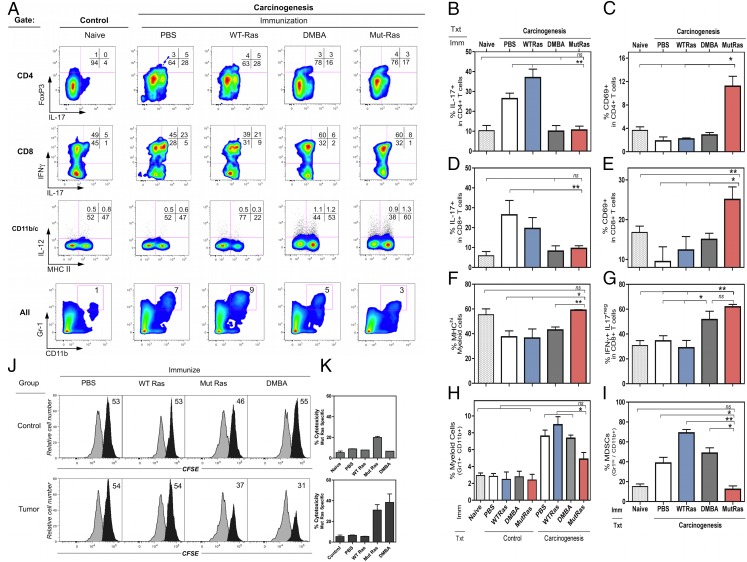
Flow cytometric profiles (Fig. 5A) and quantification of gated subsets (Fig. 5B–I) reveal that two distinct phenotypic patterns emerge, depending on the vaccine. Mut H-ras–vaccinated and DMBA-presensitized mice contained fewer IL-17+ cells in both CD4 and CD8 subsets (Fig. 5A, upper two rows, 5B, 5D). Mice that received PBS or WT H-ras contained 2.6- to 4.7-fold greater IL-17–producing T cells in CD4 and CD8 subsets compared with either naive mice or tumor-bearing Mut H-ras–vaccinated or DMBA-sensitized mice. Mut H-ras–vaccinated mice developed 100% more IFN-γ–producing CD8 T cells (Fig. 5A, row 2, 5G), and a larger proportion of CD4 (Fig. 5C) and CD8 T cells (Fig. 5E) were functionally activated, as indicated by CD69 expression. In contrast, T cell CD69 levels were reduced in all other tumor-bearing mice. No differences in Foxp3+ CD4 T cells were detected (Fig. 5A, upper row). The frequency of MHC-II+ myeloid cells increased by 40% in Mut H-ras DNA–vaccinated mice (26% DMBA-sensitized mice) compared with naive mice, and decreased by almost 50% in mice immunized with WT H-ras vaccine (Fig 5A, row 3, lower right quadrant). Myeloid-derived suppressor cells (Gr-1hi, CD11b+) in tumor-bearing mice immunized with the WT H-ras vaccine contained 3.9-fold more than naive mice and 2-fold more than mock-vaccinated (PBS) or DMBA-sensitized mice (Fig. 5A, bottom row, 5H, 5I). This result suggests that vaccination with the canonical self-epitope of H-ras induces a peripheral suppression network that may be important for maintaining tolerance to this self-epitope.
FIGURE 5.
Phenotypic and functional characterization of tumor-bearing vaccinated mice: (A) Flow cytometry multivariate plots. (B–I) Subset quantification in LNs (duplicate mice; mean ± range): (B) IL-17+ CD4 T cells; (C) CD69+ CD4 T cells; (D) IL-17+ CD8 T cells; (E) CD69+ CD8 T cells; (F) IL-12+ MHChi cells; (G) IFN-γ+ IL-17− CD8 T cells; (H) Gr1+ CD11c+ cells. Control: mice are vaccinated, DMBA treated, but not TPA treated. Carcinogenesis: mice are vaccinated, DMBA and TPA treated for 20 wk. (I) Gr-1hi, CD11b+ cells. Fold change from control (dotted line). Student t test: *p < 0.05, **p < 0.01. (J and K) In vivo CTL activity detected in immunized control and tumor-bearing mice. (J). Histogram display of CFSElo and CFSEhi target cells in the spleen, 1 d after target cell transfer. Gated CFSE-labeled target cells: CFSElo control (white) and the % of CFSEhi peptide–pulsed population (black) per sample (n = 3). (K) The % of Mut-H-ras–specific cytotoxicity for each group (mean ± SEM).
CTL activity in immunized normal and tumor-bearing mice.
CTL memory responses to Mut-H-ras peptide were determined 28 wk after DMBA initiation, using an in vivo CTL assay. For control mice [vaccinated and treated with DMBA, but not TPA (Fig. 5J, 5K, top panels)], Ag-specific CTL activity was observed only by Mut H-ras–immunized mice. Tumor-bearing mice (Fig. 5J, 5K, bottom panels) produced a robust Ag-specific elimination of target cells, but only by those mice immunized with Mut H-ras DNA and, to a lesser extent, by DMBA-presensitized mice. These results were consistent with the levels of tumor regression observed after TPA treatment ceased at 20 wk (data not shown).
Therapeutic regression of established tumors.
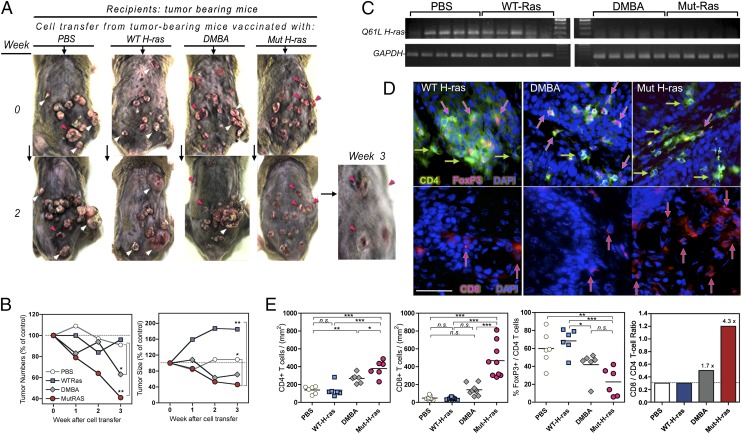
We examined what effect the transfer of cells from vaccinated, carcinogenesis-treated mice might have on recipient mice with established tumors (control GFP-DNA–vaccinated, carcinogenesis-rested mice). On the basis of phenotypic analysis, the transferred donor cells from each group contained similar numbers of CD4 or CD8 T cells. Tumor number and size (growth or regression) were monitored for recipients of each cell transfer group. Representative photos in Fig. 6A show similar tumor burdens at time 0. The differences from baseline tumor burden after 2 and 3 wk is shown (Fig. 6A, 6B). Transfer of cells from tumor-bearing PBS-treated mice had no effect, whereas cells from Mut H-ras mice resulted in dramatic tumor regression. Cells from DMBA-sensitized mice also induced regression, but to a lesser extent. In contrast, transfer of cells from WT H-ras–vaccinated, tumor-bearing mice accelerated existing tumor growth but did not affect tumor numbers. Mut H-ras mRNA was present in all tumors from recipient GFP mice except those that received cells from DMBA and Mut H-ras–vaccinated tumor-bearing mice (Fig. 6C). An abundance of infiltrating CD4+ Foxp3+ T cells was detected in tumor sections from recipients of cells from WT H-ras–immunized, and DMBA-sensitized mice (Fig. 6D, enumerated in Fig. 6E). In contrast, a greater number of infiltrating CD8 T cells were observed in tumors from recipients of cells from Mut H-ras–immunized mice, consistent with tumor regression and CTL activity. The frequency of Foxp3+ cells in tumor sections was not significantly different (data not shown), suggesting their presence may reflect pre-established tumor suppression in the host. The 4.3-fold increase in the CD8/CD4 ratio of tumor-infiltrating cells from Mut H-ras–vaccinated mice (Fig. 6E, right panel) provides supportive evidence that our vaccine effectively biases presentation of the epitope to CD8 T cells. Moreover, the rapid regression of established tumors, seen just weeks after cell transfer, demonstrates the capacity of Mut H-ras–specific effectors to overcome or overwhelm the pre-existing tumor suppressor cells.
FIGURE 6.
Therapeutic regression of established tumors by transfer of cells from Mut H-ras–immunized, tumor-bearing mice. LN and splenocytes (1:4 cells) from carcinogenesis-treated vaccinated mice were harvested at 28 wk and pooled for infusion into control GFP-vaccinated tumor-bearing mice (n = 2 per group). Phenotypic analysis indicated there were no significant differences in T cells contained in each pool: 3 million CD4 (range: 2.8–3.3) and 1 million CD8 (range: 1.4–1.6) T cells were transferred. (A) Effect of cell transfer on established tumors. Photos of tumors in recipient mice over 3 wk; arrows highlight tumor progression (white) and regression (red). Magnified view (original magnification ×3.6): tumor regression in Mut H-ras recipient mice at 3 wk. (B) Kinetics of tumor growth and regression. Tumor change from week 0: average tumor number per mouse (left), average tumor size per tumor (right). (C) Immunoediting of tumor H-ras expression. Tumor biopsy specimens obtained at 30 wk were analyzed for Mut H-ras mRNA expression by ACB-PCR (n = 5 per group). GAPDH gene expression of each sample is a loading control (upper panels). (D) Skin tumor infiltrating CD4 T cells. Cryosections of tumors stained with conjugated Abs: mouse CD4 (FITC+, green), Foxp3+ (PE), and DAPI (blue nuclei). Foxp3+ cells have pink nuclear staining (pink arrows). Foxp3− CD4+ T cells are indicated by green arrows (upper panels). CD8+ T cell infiltration in and around cutaneous tumors. Mouse CD8+ T cells [PE, red arrows (lower panels)]. Scale bar, 50 μm. (E) Enumeration of T cell subsets in tumor sections. Tumors infiltrating CD8, CD4, and Foxp3+ T cells were counted in at least five non-overlapping views per section. (n = 6–8 tumors per group). The % of CD4 (green) T cells stained with Foxp3 (pink)+ nuclei was determined in the same view, as described. Bar graph depicts the ratio of CD8:CD4 average values per group. Text indicates the fold increase in the CD8:CD4 ratio compared with mock (PBS)–vaccinated mice. *p < 0.05, **p < 0.01, ***p < 0.001. n.s., not significant.
Discussion
These studies address fundamental questions regarding the nature of the immunogenic epitopes that are recognized in primary immune responses to DMBA and, more specifically, the immunogenicity of the tumor-associated activating mutation Q61L of the H-ras oncogene and its role as an immunosurveillance target during initiation stages of DMBA-induced chemical carcinogenesis. Although it is well established that the majority of papillomas induced by DMBA/TPA treatment harbor the H-ras Q61L-activating mutation (8), detection and quantification of genomic DNA mutations in the H-ras gene at preneoplastic stages have been difficult. Previous studies required between 1 and 12 wk of TPA promotion to detect a signal (37, 38). More recently, using sensitive PCR-amplification methods, other investigators have successfully detected a low frequency of H-ras Q61L mutations in the genome 1 d after application of another PAH, dibenzo[a]pyrene, in SENCAR mice (39), or in purified CD34+ hair bulge stem cells in FVB mice following DMBA (40). In contrast to these previous studies, we report that DMBA quickly mutates a significant number of H-ras genes upon contact. Pyrosequencing data of genomic DNA from DMBA-treated skin revealed that by 24 h there is a dose-dependent increase in the frequency of H-ras alleles carrying the Q61L mutation. The DMBA mutation frequency of 10%, induced by 0.1% DMBA, is much higher than previously reported, and this finding may reflect the increased sensitivity of direct pyrosequencing technology over indirect quantification approaches. Surprisingly, we observed that without tumor promoter TPA application, the mutation load doubled over 2 d, consistent with autonomous clonal expansion of initiated cells. This finding is in contrast to previous studies that showed TPA was required for such expansion (37, 38), but this may depend on the mouse strain (41, 42). Thus, we show that the mutated genes are immediately transcribed and translated for expression of H-ras oncoproteins, which may be at levels sufficient to stimulate autonomous clonal expansion in a subset of cells. In addition, the ability to elicit swelling responses to Mut H-ras peptide in DMBA-sensitized mice and to eliminate Mut H-ras mRNA–expressing cells in DMBA-treated skin demonstrates that the oncoprotein epitope is appropriately processed and presented on preneoplastic skin cells at the earliest stages of transformation.
It is known that chemical carcinogenesis generates tumors that are initially immunogenic because the carcinogen-induced tumors from immunodeficient mice are rejected upon orthotopic transfer into immunocompetent syngeneic mice (43). It is proposed that the most immunogenic tumor epitopes undergo a slow process of immunoediting during the initiation and promotion stages, which provides the selective pressure necessary to select for nonimmunogenic tumors within immunocompetent mice. By comparing exome sequence data from two methylcolanthrene (MCA)–induced sarcomas derived from immunodeficient mice versus immunocompetent mice, Matsushita et al. (44) identified >2000 somatic missense mutations in each tumor line, but only 5% of the mutations were shared. However, following transfer of the unedited line into immunocompetent mice, the resultant “progressor” tumors lost expression of only 1% of their initial mutations, demonstrating that only a fraction of the large number of induced missense mutations have the potential to act as immunosurveillance targets. However, the identification of mutations in “unedited” established tumors does not delineate which mutations are critical for initiation at the earliest stages in preneoplastic cells. Furthermore, no one has demonstrated that immunization to any tumor-specific Ag can provide protection against tumor development caused by exposure to mutagenic chemical carcinogens.
Our study shows that a driver mutation associated with fully developed chemical carcinogen–induced tumors (and therefore thought to be nonimmunogenic) is indeed immunogenic at the earliest phases of cell transformation. We show that chemical carcinogen–induced mutations are generated in a significant proportion of H-ras genes (10% by 0.1% DMBA) and within 24 h are expressed by at least a subset of preneoplastic cells, leading to the efficient induction of neoepitope-specific effector T cells. It will be of interest to determine whether directly mutated cutaneous DCs play a role in this induction. Specifically, our findings reveal that recognition of the Mut H-ras neoepitope is a normal part of the DMBA-specific immune response because DMBA sensitization induces expansion of effectors that eliminate Mut H-ras–positive skin cells at the site of DMBA-induced elicitation of the CHS response.
The nature of the immunosurveillance-targeted mutant gene is critical in determining whether immunoediting will drive tumor eradication or immune evasion. For example, the MCA-induced mutation in spectrin-β2, a tumor-specific but nonessential gene, was shown to undergo immunoediting that drove tumor immune evasion (44). In contrast to spectrin-β2, the Q61L mutation activates the H-ras oncoprotein, which is thought to be an important driver of DMBA-induced transformation during initiation of benign papillomas and progression into malignant SCCs. We show that immunoediting was evident only in tumors that eventually developed in Mut H-ras–immunized mice, leading to a profound reduction in tumor burden. Thus, expanding CD8 T cells that recognize tumor-specific driver mutations expressed during the earliest phases of tumor initiation results in immunoediting that leads to tumor regression rather than immune evasion.
We demonstrate that DMBA sensitization induces T cells with specificity for the Mut H-ras epitope that are capable of eliminating neoplastic skin cells at the site of DMBA elicitation. However, low-dose DMBA presensitization provided only limited protection against tumor formation. To increase the efficacy of tumor prevention, we used two vaccination approaches (DC based and DNA based) that used a chimeric gene engineered to favor MHC class I presentation of the Mut H-ras epitope (19). The augmented protection induced by this single epitope may be due to the engineered vaccine’s ability to bias selective expansion of CD8 T cells over regulatory CD4 T cells [which typically predominate in DMBA-induced responses (15, 16)]. About 20% of DMBA-induced tumors are negative for the Mut H-ras mutation, suggesting that alternative pathways can drive tumorigenesis; however, if other DMBA-induced mutant genes are presented as neoantigens, their role in broadening the T cell repertoire against initiated cells is not clear. Exome sequencing of unedited regressor versus progressor DMBA-induced tumor cells has yet to be performed, but it is likely that this carcinogen is similar to MCA in causing mutations in many gene loci. Thus, Mut H-ras–vaccinated mice could have favored the outgrowth of DMBA-induced tumors activated by alternative tumorigenic pathways. However, the skin tumors that developed were smaller and slow growing (Figure 5); further, they were less, not more, tumorigenic than the tumors that formed in the other cohorts (data not shown).
The mutations present in Mut H-ras–negative tumors require further investigation. The other mutations may not contribute to immune surveillance because they are: 1) not expressed, 2) nonimmunogenic, 3) inducers of suppression, or 4) nonessential for tumor fitness (growth or survival). Raising immune responses to driver mutations that are expressed during tumor initiation may be critical for the success of preventative vaccines. The ability to provide substantial protection against carcinogen-induced tumor development with a single epitope confirms the importance of oncogenic ras in tumor initiation and maintenance (45); however, further optimization must be pursued to increase the breadth of tumor epitopes and the accumulation of neoepitope-specific skin resident memory cells to readily provide complete eradication of newly emergent transformed cells.
A systemic immunosuppressive environment was established in all carcinogen-induced tumor-bearing mice in control cohorts, indicated by increased numbers of MDSCs and regulatory T cells detected in tumor tissue, draining LNs, and spleen; however, these cells were largely absent in Mut H-ras–vaccinated mice. More importantly, we found that when cells from carcinogenesis-resistant Mut H-ras–vaccinated mice were transferred into tumor-bearing recipients, they overcame established immunosuppressive environments and induced rapid tumor regression. Transfer of DMBA-sensitized cells resulted in a mixed response, causing regression in some, but augmenting growth in other tumors of the same recipient. Tumor biopsy specimens from mice that received cells from Mut H-ras–vaccinated mice contained a high number of infiltrating CD8 T cells. In contrast, tumors from other groups contained high numbers of Foxp3+ CD4 regulatory T cells. These data, together with flow cytometric results, suggest that our immunization procedure was successful in biasing presentation of the Mut H-ras epitope to CD8 rather than CD4 T cells, generating effectors that can overcome the mechanisms of tumor-induced suppression.
In contrast, the transfer of cells from WT H-ras–vaccinated mice manifested accelerated tumor growth in recipient mice, suggesting that networks involved in maintaining tolerance to self were expanded. This observation is similar to those reported by Siegel et al. (46), demonstrating that active immunization with an alternate peptide of H-ras (containing both class I and II epitopes of codon 12 Val → Arg mutation) did not protect but instead promoted tumor development in an inducible transgenic mouse model. Similar epitope recognition by human T cells has been reported (47, 48). It is clear that in our model, immunization with the endogenous self-epitope (WT H-ras) does not evoke autoimmunity but instead may expand tolerogenic immune networks. These results highlight the importance of setting appropriate criteria in choosing tumor-specific epitopes that favor MHC-I presentation for effective preventative vaccines.
In this era of cancer cell genomics and personalized medicine, our knowledge of critical driver mutations and initiating tumor-specific epitopes will increase dramatically, concomitantly increasing the potential to boost immunosurveillance against a broader spectrum of neoplasias. The results from our studies support further research in the development of epitope-focused and multiepitope-focused vaccination strategies that may be used to protect individuals at risk for ras-driven, or other potential mutant oncogene–driven cancers, caused by exposure to environmental carcinogens.
Supplementary Material
Acknowledgments
We thank the following colleagues: Michael A. Barry (University of Minnesota) for the Ub/vv/GFP genetic immunization vector, Justin Roth (University of Alabama at Birmingham) for HIV-based lentivector construct and molecular biology guidance (also provided by Angel Rivera [Emory University]), Nabiha Yusuf (University of Alabama at Birmingham) and Aton Holzer for initial pilot studies (N.Y., A.H.), George Twitty for assistance with setting up immunizations and CTL assays, Dr. Aleodor Andea for dermatopathology, Michael R. Crowley (University of Alabama at Birmingham Heflin Center Genomic Core and the Cancer Center Core) for help with pyrosequencing, and the University of Alabama at Birmingham Comprehensive Flow Cytometry Core of the Rheumatic Diseases Core Center.
This work was supported by a VA Merit Award (to C.A.E. and L.T.); the National Institutes of Health Skin Diseases Research Center Core Grant P30AR050948 (to C.A.E. and L.T.); National Institutes of Health National Cancer Institute Award R01CA138988 (to M.A. and L.T.); National Institutes of Health Grant P30CA13148-40 to the University of Alabama at Birmingham Heflin Center Genomic Core and Cancer Center Core; and National Institutes of Health Grants P30AR048311 and P30AI2766 to the University of Alabama at Birmingham Comprehensive Flow Cytometry Core of the Rheumatic Diseases Core Center.
The online version of this article contains supplemental material.
- ACB-PCR
- allele-specific competitive blocker–PCR
- BMDC
- bone marrow dendritic cell
- CHS
- contact hypersensitivity
- DC
- dendritic cell
- DMBA
- 7,12-dimethylbenz(a)anthracene
- DTH
- delayed-type hypersensitivity
- i.d.
- intradermal
- LN
- lymph node
- MCA
- methylcolanthrene
- PAH
- polyaromatic hydrocarbon
- SCC
- squamous cell carcinoma
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Rosenberg S. A., Yang J. C., Restifo N. P. 2004. Cancer immunotherapy: moving beyond current vaccines. Nat. Med. 10: 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnet F. M. 1970. The concept of immunological surveillance. Prog. Exp. Tumor Res. 13: 1–27. [DOI] [PubMed] [Google Scholar]
- 3.Everall J. D., Dowd P. M. 1978. Influence of environmental factors excluding ultra violet radiation on the incidence of skin cancer. Bull. Cancer 65: 241–247. [PubMed] [Google Scholar]
- 4.Hoffmann D., Rivenson A., Hecht S. S. 1996. The biological significance of tobacco-specific N-nitrosamines: smoking and adenocarcinoma of the lung. Crit. Rev. Toxicol. 26: 199–211. [DOI] [PubMed] [Google Scholar]
- 5.Hecht S. S. 2002. Cigarette smoking and lung cancer: chemical mechanisms and approaches to prevention. Lancet Oncol. 3: 461–469. [DOI] [PubMed] [Google Scholar]
- 6.Vineis P., Alavanja M., Buffler P., Fontham E., Franceschi S., Gao Y. T., Gupta P. C., Hackshaw A., Matos E., Samet J., et al. 2004. Tobacco and cancer: recent epidemiological evidence. J. Natl. Cancer Inst. 96: 99–106. [DOI] [PubMed] [Google Scholar]
- 7.Shields P. G. 2000. Epidemiology of tobacco carcinogenesis. Curr. Oncol. Rep. 2: 257–262. [DOI] [PubMed] [Google Scholar]
- 8.Balmain A., Pragnell I. B. 1983. Mouse skin carcinomas induced in vivo by chemical carcinogens have a transforming Harvey-ras oncogene. Nature 303: 72–74. [DOI] [PubMed] [Google Scholar]
- 9.Tang M. S., Vulimiri S. V., Viaje A., Chen J. X., Bilolikar D. S., Morris R. J., Harvey R. G., Slaga T. J., DiGiovanni J. 2000. Both (+/-)syn- and (+/-)anti-7,12-dimethylbenz[a]anthracene-3,4-diol-1,2-epoxides initiate tumors in mouse skin that possess -CAA- to -CTA- mutations at Codon 61 of c-H-ras. Cancer Res. 60: 5688–5695. [PubMed] [Google Scholar]
- 10.Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. 1997. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277: 333–338. [DOI] [PubMed] [Google Scholar]
- 11.Clark C. E., Hingorani S. R., Mick R., Combs C., Tuveson D. A., Vonderheide R. H. 2007. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 67: 9518–9527. [DOI] [PubMed] [Google Scholar]
- 12.Tran Thang N. N., Derouazi M., Philippin G., Arcidiaco S., Di Berardino-Besson W., Masson F., Hoepner S., Riccadonna C., Burkhardt K., Guha A., et al. 2010. Immune infiltration of spontaneous mouse astrocytomas is dominated by immunosuppressive cells from early stages of tumor development. Cancer Res. 70: 4829–4839. [DOI] [PubMed] [Google Scholar]
- 13.Schneider B. L., Bowden G. T., Sutter C., Schweizer J., Han K. A., Kulesz-Martin M. F. 1993. 7,12-Dimethylbenz[a]anthracene-induced mouse keratinocyte malignant transformation independent of Harvey ras activation. J. Invest. Dermatol. 101: 595–599. [DOI] [PubMed] [Google Scholar]
- 14.Kehren J., Desvignes C., Krasteva M., Ducluzeau M. T., Assossou O., Horand F., Hahne M., Kägi D., Kaiserlian D., Nicolas J. F. 1999. Cytotoxicity is mandatory for CD8(+) T cell-mediated contact hypersensitivity. J. Exp. Med. 189: 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yusuf N., Timares L., Seibert M. D., Xu H., Elmets C. A. 2007. Acquired and innate immunity to polyaromatic hydrocarbons. Toxicol. Appl. Pharmacol. 224: 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yusuf N., Nasti T. H., Katiyar S. K., Jacobs M. K., Seibert M. D., Ginsburg A. C., Timares L., Xu H., Elmets C. A. 2008. Antagonistic roles of CD4+ and CD8+ T-cells in 7,12-dimethylbenz(a)anthracene cutaneous carcinogenesis. Cancer Res. 68: 3924–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu S., Bergstresser P. R., Takashima A. 1995. Phenotypic and functional heterogeneity among murine epidermal-derived dendritic cell clones. J. Invest. Dermatol. 105: 831–836. [DOI] [PubMed] [Google Scholar]
- 18.Timares L., Takashima A., Johnston S. A. 1998. Quantitative analysis of the immunopotency of genetically transfected dendritic cells. Proc. Natl. Acad. Sci. USA 95: 13147–13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson H. A., Barry M. A. 2004. Maximizing antigen targeting to the proteasome for gene-based vaccines. Mol. Ther. 10: 432–446. [DOI] [PubMed] [Google Scholar]
- 20.Brouwenstijn N., Serwold T., Shastri N. 2001. MHC class I molecules can direct proteolytic cleavage of antigenic precursors in the endoplasmic reticulum. Immunity 15: 95–104. [DOI] [PubMed] [Google Scholar]
- 21.Serwold T., Gonzalez F., Kim J., Jacob R., Shastri N. 2002. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. [see comment] Nature 419: 480–483. [DOI] [PubMed] [Google Scholar]
- 22.Fan H., Lin Q., Morrissey G. R., Khavari P. A. 1999. Immunization via hair follicles by topical application of naked DNA to normal skin. Nat. Biotechnol. 17: 870–872. [DOI] [PubMed] [Google Scholar]
- 23.Peachman K. K., Rao M., Alving C. R. 2003. Immunization with DNA through the skin. Methods 31: 232–242. [DOI] [PubMed] [Google Scholar]
- 24.Belyakov I. M., Hammond S. A., Ahlers J. D., Glenn G. M., Berzofsky J. A. 2004. Transcutaneous immunization induces mucosal CTLs and protective immunity by migration of primed skin dendritic cells. J. Clin. Invest. 113: 998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abel E. L., Angel J. M., Kiguchi K., DiGiovanni J. 2009. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat. Protoc. 4: 1350–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson D., Bundell C., Robinson B. 2000. In vivo cross-presentation of a soluble protein antigen: kinetics, distribution, and generation of effector CTL recognizing dominant and subdominant epitopes. J. Immunol. 165: 6123–6132. [DOI] [PubMed] [Google Scholar]
- 27.Parsons B. L., McKinzie P. B., Heflich R. H. 2005. Allele-specific competitive blocker-PCR detection of rare base substitution. Methods Mol. Biol. 291: 235–245. [DOI] [PubMed] [Google Scholar]
- 28.Morlan J., Baker J., Sinicropi D. 2009. Mutation detection by real-time PCR: a simple, robust and highly selective method. PLoS ONE 4: e4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakravarti D., Pelling J. C., Cavalieri E. L., Rogan E. G. 1995. Relating aromatic hydrocarbon-induced DNA adducts and c-H-ras mutations in mouse skin papillomas: the role of apurinic sites. Proc. Natl. Acad. Sci. USA 92: 10422–10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris R. J. 2000. Keratinocyte stem cells: targets for cutaneous carcinogens. J. Clin. Invest. 106: 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapouge G., Youssef K. K., Vokaer B., Achouri Y., Michaux C., Sotiropoulou P. A., Blanpain C. 2011. Identifying the cellular origin of squamous skin tumors. Proc. Natl. Acad. Sci. USA 108: 7431–7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons B. L., Heflich R. H. 1998. Detection of a mouse H-ras codon 61 mutation using a modified allele-specific competitive blocker PCR genotypic selection method. Mutagenesis 13: 581–588. [DOI] [PubMed] [Google Scholar]
- 33.Esser C., Bargen I., Weighardt H., Haarmann-Stemmann T., Krutmann J. 2013. Functions of the aryl hydrocarbon receptor in the skin. Semin. Immunopathol. 35: 677–691. [DOI] [PubMed] [Google Scholar]
- 34.Palm N. W., Medzhitov R. 2009. Immunostimulatory activity of haptenated proteins. Proc. Natl. Acad. Sci. USA 106: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weltzien H. U., Martin S. F., Nicolas J. F. 2014. T cell responses to contact allergens. EXS 104: 41–49. [DOI] [PubMed] [Google Scholar]
- 36.Denton A. E., Russ B. E., Doherty P. C., Rao S., Turner S. J. 2011. Differentiation-dependent functional and epigenetic landscapes for cytokine genes in virus-specific CD8+ T cells. Proc. Natl. Acad. Sci. USA 108: 15306–15311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson M. A., Futscher B. W., Kinsella T., Wymer J., Bowden G. T. 1992. Detection of mutant Ha-ras genes in chemically initiated mouse skin epidermis before the development of benign tumors. Proc. Natl. Acad. Sci. USA 89: 6398–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finch J. S., Albino H. E., Bowden G. T. 1996. Quantitation of early clonal expansion of two mutant 61st codon c-Ha-ras alleles in DMBA/TPA treated mouse skin by nested PCR/RFLP. Carcinogenesis 17: 2551–2557. [DOI] [PubMed] [Google Scholar]
- 39.Khan G. A., Bhattacharya G., Mailander P. C., Meza J. L., Hansen L. A., Chakravarti D. 2005. Harvey-ras gene expression and epidermal cell proliferation in dibenzo[a,l]pyrene-treated early preneoplastic SENCAR mouse skin. J. Invest. Dermatol. 125: 567–574. [DOI] [PubMed] [Google Scholar]
- 40.Kim D. J., Kataoka K., Rao D., Kiguchi K., Cotsarelis G., Digiovanni J. 2009. Targeted disruption of stat3 reveals a major role for follicular stem cells in skin tumor initiation. Cancer Res. 69: 7587–7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hennings H., Devor D., Wenk M. L., Slaga T. J., Former B., Colburn N. H., Bowden G. T., Elgjo K., Yuspa S. H. 1981. Comparison of two-stage epidermal carcinogenesis initiated by 7,12-dimethylbenz(a)anthracene or N-methyl-N′-nitro-N-nitrosoguanidine in newborn and adult SENCAR and BALB/c mice. Cancer Res. 41: 773–779. [PubMed] [Google Scholar]
- 42.Wakabayashi Y., Mao J.-H., Brown K., Girardi M., Balmain A. 2007. Promotion of Hras-induced squamous carcinomas by a polymorphic variant of the Patched gene in FVB mice. Nature 445: 761–765. [DOI] [PubMed] [Google Scholar]
- 43.Shankaran V., Ikeda H., Bruce A. T., White J. M., Swanson P. E., Old L. J., Schreiber R. D. 2001. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410: 1107–1111. [DOI] [PubMed] [Google Scholar]
- 44.Matsushita H., Vesely M. D., Koboldt D. C., Rickert C. G., Uppaluri R., Magrini V. J., Arthur C. D., White J. M., Chen Y.-S., Shea L. K., et al. 2012. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482: 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chin L., Tam A., Pomerantz J., Wong M., Holash J., Bardeesy N., Shen Q., O’Hagan R., Pantginis J., Zhou H., et al. 1999. Essential role for oncogenic Ras in tumour maintenance. Nature 400: 468–472. [DOI] [PubMed] [Google Scholar]
- 46.Siegel C. T., Schreiber K., Meredith S. C., Beck-Engeser G. B., Lancki D. W., Lazarski C. A., Fu Y. X., Rowley D. A., Schreiber H. 2000. Enhanced growth of primary tumors in cancer-prone mice after immunization against the mutant region of an inherited oncoprotein. J. Exp. Med. 191: 1945–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gjertsen M. K., Bjorheim J., Saeterdal I., Myklebust J., Gaudernack G. 1997. Cytotoxic CD4+ and CD8+ T lymphocytes, generated by mutant p21-ras (12Val) peptide vaccination of a patient, recognize 12Val-dependent nested epitopes present within the vaccine peptide and kill autologous tumour cells carrying this mutation. Int. J. Cancer 72: 784–790. [DOI] [PubMed] [Google Scholar]
- 48.Gjertsen M. K., Saeterdal I., Saebøe-Larssen S., Gaudernack G. 2003. HLA-A3 restricted mutant ras specific cytotoxic T-lymphocytes induced by vaccination with T-helper epitopes. J. Mol. Med. 81: 43–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.