Abstract
BRAF(V600E) is the most common oncogenic lesion in melanoma and results in constitutive activation of the mitogen-activated protein kinase (MAPK) pathway and uncontrolled cell growth. Selective BRAF inhibitors such as vemurafenib have been shown to neutralize oncogenic signaling, restrain cellular growth and improve patient outcome. Although several mechanisms of vemurafenib resistance have been described, directed solutions to overcome these resistance lesions are still lacking. Herein, we found that vemurafenib resistance can be (i) mediated by EphA2- a member of the largest receptor tyrosine kinases (RTK) subfamily erythropoietin-producing hepatocellular (Eph) receptors and (ii) associated with a greater phenotypic dependence on EphA2. Furthermore, we developed a series of first-in-class EphA2 inhibitors and show that these new compounds potently induce apoptosis, suppress viability and abrogate tumorigenic growth of melanoma cells, including those that are resistant to vemurafenib. These results provide proof-of-concept that RTK-guided growth, and therapeutic resistance, can be prospectively defined and selectively targeted.
Keywords: EphA2, vemurafenib, resistance, melanoma, tumor growth
INTRODUCTION
Cutaneous melanoma is the most aggressive form of skin cancer. For patients with metastatic disease, only about 10% are expected to survive to 5 years (1). The identification of activating BRAF mutations in melanomas (2) followed by the directed pursuit of selective BRAF(V600E) inhibitors (BRAFi’s) have led to a campaign of highly successful clinical trials in metastatic melanoma (3–5). At the current time, two BRAF inhibitors-vemurafenib (VEM) and dabrafenib have been approved by the U.S. Food and Drug Administration. Despite the rapid and early control achieved with these compounds, response to the BRAFi’s can be transient and eventual relapse is the general rule (3–5). The addition of a selective MEK inhibitor to a BRAF inhibitor improves efficacy (6), but emergence of resistance to all BRAF and MEK inhibitors remains a problem for which there is no current treatment. Moreover, an effective molecularly-targeted therapy has yet to be established for the half of the metastatic population without activating BRAF mutations (BRAF(WT)) (7). In balance, these findings underscore the power of targeted approaches and the unmet need to identify novel “druggable” targets which can be exploited in the setting of therapeutically intractable melanomas (e.g. BRAF(WT) and therapy-resistant BRAF(V600E) tumors).
Activated receptor tyrosine kinases (RTKs) have also come into view as both key drivers of melanoma tumorigenesis and central mediators of resistance of selective BRAF(V600E) inhibitors. Copy number and sequence alterations at the KIT locus have specifically been associated with acral and mucosal subtypes of melanoma (8) while mutations in ERBB4 and multiple EPH family receptors have also been reported (9). Some RTKs, such as EphA2 (10) and MERTK (11), have been functionally shown to be crucial mediators of melanoma cell survival and tumorigenesis. Activation of RTKs, such as PDGFRβ (12), PDGFRα (13), IGF-1R (14), HGF/MET(15), FGFR3 (16) and EGFR (17,18), have also been shown to mediate resistance to BRAF inhibition. Thus, RTK signaling appears to be a central determinant of melanoma progression and drug responsiveness.
We recently showed that one RTK, EphA2, is highly expressed in both BRAF(V600E) and BRAF(WT) melanomas and that depletion of this molecule leads to dramatic loss of cellular viability, heightened apoptosis in vitro and abrogation of tumor growth in vivo (10). EphA2 is a member of the Eph family of RTKs and is known to contribute to mammary gland, brain and limb development and patterning of the visual system (17–25). Furthermore, EphA2 overexpression has been observed in numerous cancer types including melanoma (10, 26–30) and in breast cancer cell lines exhibiting resistance to trastuzumab (31). Signaling by the Eph system is complex, impinges on both Ras-PI3K-AKT and RAS-MAPK pathways and is layered in a network of signaling cross-talk (32). Nevertheless, given its integral role in melanoma survival (10), its role in growth factor signaling (33–36), and its contribution to therapeutic resistance in breast cancer (31), EphA2 is a strong candidate for mediating selective BRAF inhibitor resistance in melanoma. To explore this hypothesis, we generated VEM-resistant melanoma lines in vitro and observed a dramatic upregulation of EphA2 in the resistant lines. Moreover, examination of tumors from patients with acquired resistance to BRAFi’s also revealed evidence of increased EphA2 expression. Depletion of EphA2 preferentially induced apoptosis in VEM-resistant cells and partially restored VEM sensitivity. In addition, we developed first generation ATP-competitive inhibitors against EphA2 and demonstrated significant activity in overcoming VEM-resistance both in vitro and in vivo.
RESULTS
EphA2 upregulation occurs during the course of acquired resistance to BRAF inhibitors
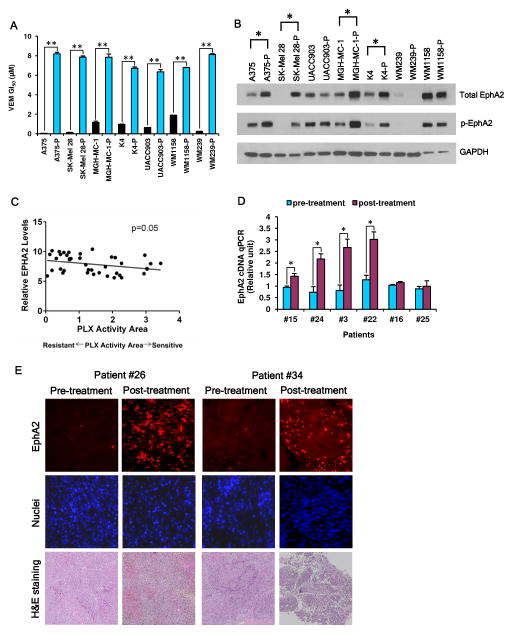
In order to determine the role of EphA2 in VEM response, we performed three lines of investigation. First, we subjected seven melanoma cell lines (A375, SK-Mel 28, UACC903, MGH-MC-1, K4, WM239 and WM1158) to escalating doses of VEM, which resulted in significant increases in VEM GI50’s (Fig. 1A, fig. S1). Since EphA2 has been shown to be a critical melanoma survival factor (10) and since RTKs are known to mediate BRAFi resistance (12, 14, 37), we hypothesized that EphA2 may also contribute to acquired VEM insensitivity. As shown in Fig. 1B, there was a clear upregulation of both total EphA2 and Ser897 phosphorylated EphA2 (p-EphA2Ser897) in 4/7 VEM resistant lines (A375-P, SK-Mel 28-P, MGH-MC-1-P and K4-P) compared to their matched parental controls (A375, SK-Mel 28, MGH-MC-1 and K4) though 3/7 pairs exhibited little difference in EphA2 expression. Second, we used data available for 40 melanoma lines in the Cancer Cell Line Encyclopedia (CCLE) in order to delineate any relationship between EphA2 and intrinsic resistance. As shown in Fig. 1C, there was a significant negative correlation between PLX4720 sensitivity (i.e. higher PLX activity area) and EPHA2 RNA levels (P=0.05). Third, we assessed EphA2 levels in clinical specimens from patients undergoing clinical trials with vemurafenib or dabrafenib/trametinib (patient information in Table S1). In six patients with available matched pre-treatment/post-relapse tumor RNA, four post-relapse samples showed significantly higher EPHA2 RNA levels compared to the pre-treatment specimens (Fig 1D). In two patients with only pathology specimens, both post-relapse specimens demonstrated a notable increase in EphA2 protein levels (Fig 1E). Taken together, these data suggest that EphA2 may contribute to primary and secondary BRAFi ± MEKi resistance both in vitro and in clinical settings.
Fig. 1. EphA2 upregulation occurs during the course of induced resistance to BRAF inhibitors both in vitro and in vivo.
(A) A set of 7 melanoma cell lines were induced into VEM resistance by sequential exposure to escalating doses of VEM. Cell viability assay was performed and the cellular GI50 values for VEM in matched sensitive and resistant lines are shown. Error bars indicate mean viability ± SEM (n=3). **P<0.001 by Students’ t test. (B) Increased total EphA2 and phosphorylated EphA2 (Ser897) in matched VEM sensitive and resistant lines were assessed by western blotting. “*” indicates an apparent upregulation of EphA2 in VEM resistant lines compared to control lines. (C) The CCLE database was used to examine PLX sensitivity (PLX activity area) and relative EPHA2 expression. There is a negative correlation between PLX activity area and EPHA2 levels (P=0.05). (D) EPHA2 mRNA levels are elevated in post-relapse tumor samples compared to patient-matched pre-treatment specimens during the course of treatment with BRAF±MEK inhibitors. Of note, a BRAF splice product has also been detected in the post-relapse specimen for patient #22. The BRAF splice variant may explain resistance to dabrafenib but not necessarily trametinib. RNA levels were measured using q-PCR assay (in triplicate) with GUSB as control. V, Vemurafenib; D+T, Dabrafenib+Trametinib. Error bars indicate mean normalized levels ± SEM (n=3) *P<0.05 by Students’ t test. (E) Immunohistochemistry showed increased EphA2 levels in post-relapse tumor samples compared to pre-treatment tumor specimens from patients #26 and #34.
The resistance phenotype is dependent on EphA2
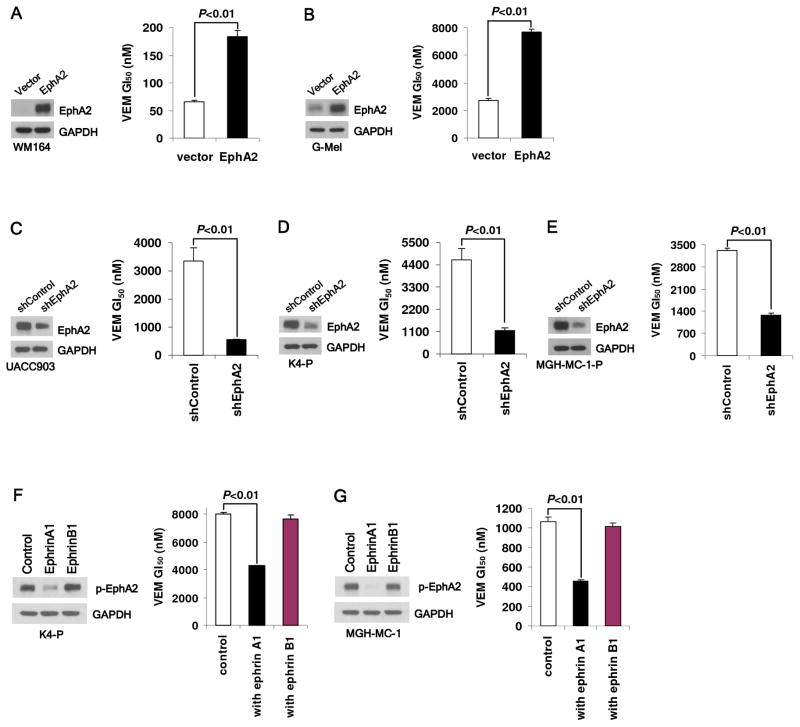
We next set out to determine whether EphA2 is functionally mediating VEM resistance. EphA2 was first overexpressed in WM164 and G-mel melanoma cells (both with low EphA2 levels) and found to confer a moderate but consistent gain in VEM GI50’s from 66.39 nM (WM164vector) to 183.65 nM (WM164EphA2) and 2.73 μM (G-melvector) to 7.69 μM (G-melEphA2) (Fig. 2A,B). We also depleted EphA2 in one naive (UACC903) and two VEM-resistant lines (K4-P and MGH-MC-1-P) using small hairpin (shRNA). In each case, there was a reduction in VEM GI50- 3.35 μM (UACC903shControl) to 0.56 μM (UACC903shEphA2), 4.64 μM (K4-PshControl) to 1.15 μM (K4- PshEphA2) and 3.32 μM (MGH-MC-1-PshControl) to 1.27 μM (MGH-MC-1-PshEphA2) (Fig. 2C-E). As Ser897 phosphorylation was associated with VEM resistance (Fig. 1B) and known to become dephosphorylated upon ephrin-A1 binding (38), we next investigated the effects of ephrin-A1 on drug response. We treated a VEM-sensitive (MGH-MC-1) line and a VEM-resistant (K4-P) line with 100 ng/ml ephrin-A1/Fc and observed a decrease in both EphA2Ser897 phosphorylation and VEM resistance (Fig. 2F, G). In contrast, ephrin-B1, a non-EphA2 ligand, was ineffective in abrogating Ser897 phosphorylation or altering VEM sensitivity. These findings suggest that EphA2 and its concomitant phosphorylation at Ser897 contribute, at least in part, to the development of VEM resistance.
Fig. 2. EphA2 mediates resistance to vemurafenib.
EphA2 overexpression in WM164 (A) and G-mel (B) cells leads to VEM resistance while EphA2 depletion in UACC903 (C), K4-P (D) and MGH-MC-1-P (E) cells by EPHA2 shRNA partially restores VEM sensitivity. Ephrin-A1 (100 ng/ml), but not ephrin-B1 (100 ng/ml), suppresses EphA2Ser897 phosphorylation and partially reverses VEM resistance in K4-P (F) and MGH-MC-1 (G) cells. All error bars indicate mean viability ± SEM (n=3).
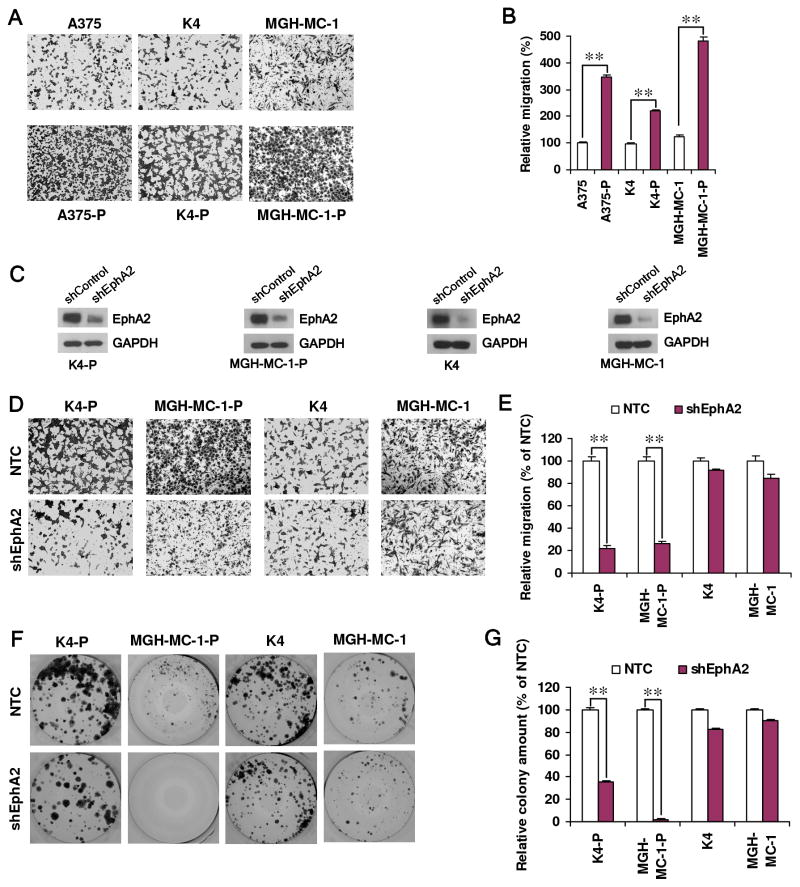
With the heightened levels of EphA2 protein and phosphorylation, one could speculate that VEM resistant cells may adopt a greater phenotypic dependence on EphA2. One known function of EphA2 is to trigger cell migration (38). As shown in Figs. 3A and B, EphA2-enriched resistant cell lines (A375-P, K4-P and MGH-MC-1-P) all demonstrated increased transwell migration compared to their matched parental cells. To evaluate the functional involvement of EphA2 in this phenomenon, we depleted EphA2 using shRNAs in these lines and observed a strong and selective attenuation of cell migration in the VEM-resistant, but not VEM-sensitive cells (Fig. 3C–E). Furthermore, we also found that resistant cells were more dependent on EphA2 compared to sensitive cells in their colony forming capacities (Fig. 3F, G). These data support the idea that the upregulation of EphA2 in VEM-resistant lines renders these cells more dependent on this RTK compared to their sensitive counterparts. It also raises the possibility that pharmacologic inhibition of EphA2 could be an effective approach to overcome resistance to BRAF inhibition and perhaps melanoma growth in general.
Fig. 3. VEM resistant cells exhibit a dependence on EphA2 for migration and colony formation.
VEM resistant cells (A375-P, K4-P, MGH-MC-1-P) show greater motility and invasiveness than their VEM sensitive counterpart (A375, K4, MGH-MC-1). Representative images of the cells that have migrated through to the bottom chamber (A) and quantitation are shown (B). Error bars indicate mean absorbance ± SEM (n=2); see Methods. (C) Depletion of EphA2 in both parental (MGH-MC-1 and K4) and VEM-resistant daughter lines (MGH-MC-1-P and K-4-P) reveal greater effects on migration in the VEM resistant lines relative to the parental lines. Representative images (D) and quantitation are shown (E). Error bars indicate mean absorbance ± SEM (n=2). Similarly, EphA2 suppression leads to a dramatic loss of colony forming capacity in the VEM resistant lines (K4-P and MGH-MC-1-P) compared to the VEM sensitive parents (K4 and MGH-MC-1). Representative images (F) and quantitation (G) are shown. Error bars indicate mean colony count ± SEM (n=2). **P<0.001 by Students’ t test.
First-in-class EphA2 inhibitors effectively suppress vemurafenib-resistant melanoma cells
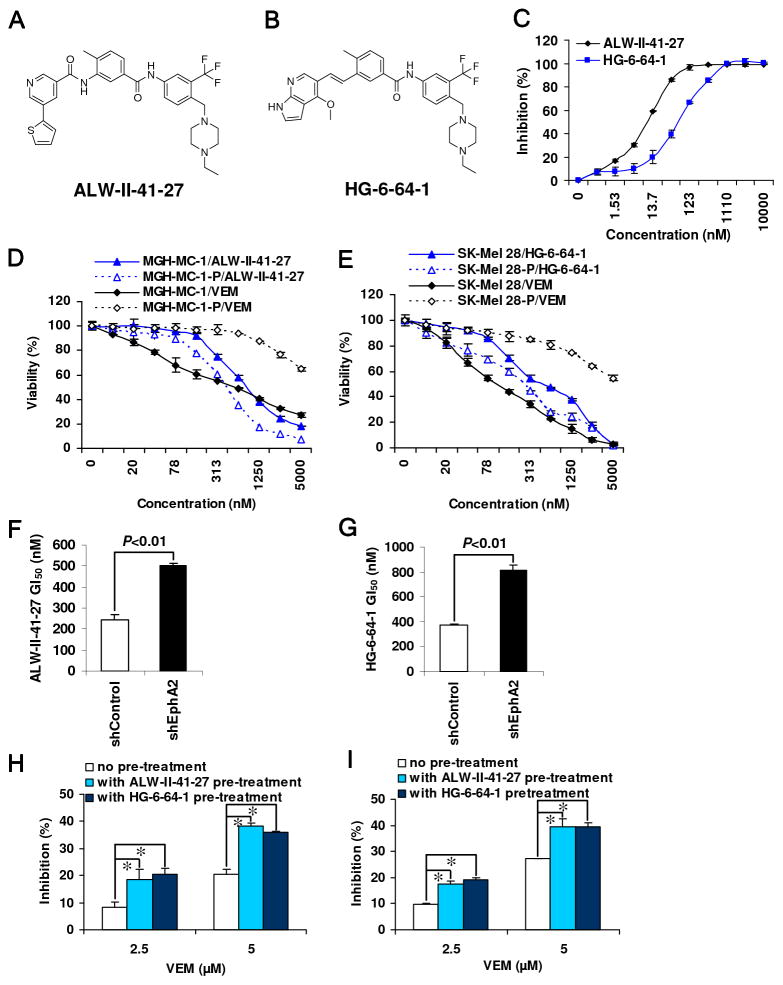
Given EphA2’s potential role in sustaining growth (10) and in mediating VEM resistance, a small molecule inhibitor of EphA2 would serve as both a pharmacological means to interrogate the dependence on EphA2 kinase activity and a starting point for a potential drug discovery campaign. To this end, we developed a library of ATP-competitive inhibitors that was designed to bind to the inactive, so-called ‘DFG-out’ conformation of kinases and broadly screened this library against a panel of several hundred kinases including EphA2. Two compounds which emerged from this screen included ALW-II-41-27 and HG-6-64-1 (Fig. 4A, B). As shown in Fig. 4C, both ALW and HG strongly inhibited EphA2 activity with biochemical IC50’s of 11.0 nM and 77.1 nM for ALW-II-41-27 and HG-6-64-1, respectively. Both compounds were then subjected to kinase selectivity profiling using the KinomeScan. As expected, both compounds inhibited a number of additional kinases with ALW-II-41-27 and HG-6-64-1 returning S(1)-scores of 0.10 and 0.06 (fig. S2A, B), respectively, which are comparable to other clinically-approved kinase inhibitors (39).
Fig. 4. EphA2 inhibitors are highly effective in vemurafenib resistant cell lines.
Structures of ALW-II-41-27 (A) and HG-6-64-1 (B). (C) Both ALW-II-41-27 and HG-6-64-1 strongly inhibited EphA2 activity in a cell-free system using the SelectScreenTM Kinase Profiling Service. VEM- resistant cells (MGH-MC-1-P and SK-Mel-28-P) are more sensitive to ALW-II-41-27 (D) and HG-6-64-1 (E) than their sensitive parental lines (MGH-MC-1 and SK-Mel-28). Depletion of EphA2 by shRNA (Fig 3C western blot) increased resistance to ALW-II-41-27 (F) and HG-6-64-1 (G). Pre-treatment (2 μM for 2 hrs) with ALW-II-41-27 and HG-6-64-1 restored VEM sensitivity in EphA2-upregulated VEM-resistant A375-P (H) and SK-Mel 28-P (I) cells. *P<0.01 by Students’ t test.
In order to establish EphA2 as an intracellular target for these compounds, we first examined whether the inhibitory activities of ALW-II-41-27 and HG-6-64-1 correlated with EphA2 protein levels. Both ALW-II-41-27 and HG-6-64-1 were more effective in CHL-1 cells (high EphA2) compared to SK-mel-119 (low EphA2) (fig. S3A–C). There was also a consistent decrease in the GI50’s for both compounds when EphA2 was overexpressed in SK-mel-119 and WM164 cells (fig. S3D–I). In contrast, depletion of EphA2 in UACC903 and CHL-1 cells by shRNA partially attenuated the inhibitory activity of both EphA2 inhibitors (fig. S3J–O). Moreover, a weaker EphA2 inhibitor NG-25 with a biochemical IC50 of 773 nM also exhibited less potent cellular inhibition compared to more active ALW-II-41-27 (fig. S3P–R). These results show a positive correlation between EphA2 levels and inhibitor sensitivity thereby providing supportive evidence that EphA2 is a functionally relevant target of these compounds in melanoma cells.
In the context of VEM resistance, both EphA2 inhibitors showed similar, if not greater, activity against VEM-resistant lines (i.e. lines with higher EphA2) compared to the sensitive parental lines (i.e. lines with lower EphA2; Fig. 4D,E, fig. S4A, B). Even under conditions of VEM resistance, EphA2 appears to be the target of ALW-II-41-27 and HG-6-64-1 as depletion of EphA2 in MGH-MC-1-P led to increased resistance to both EphA2 compounds (Fig. 4F, G). Morphologically, EphA2 inhibition resulted in cell rounding, loss of adhesion and cell death. In contrast, VEM caused no significant cell death in BRAF(WT) and resistant lines (fig. S4C).
Beyond single agent efficacy, ALW-II-41-27 also demonstrated significant synergism with VEM in both sensitive A375 cells (Table S2; lowest combination index (CI)=0.12, “strong” synergism) and resistant A375-P cells (lowest CI=0.22, “strong” synergism). Overall, 11/16 ALW-II-41-27+VEM dose points exhibited at least “moderate” synergism (i.e. CI<0.85). Furthermore, when the MEK inhibitor AZD6244 was tested in combination with ALW-II-41-27 or HG-6-64-1, 15/16 and 16/16 dose points, respectively, exhibited at least “moderate” synergism. Lastly, we also found that pre-treatment with ALW-II-41-27 and HG-6-64-1 restored VEM sensitivity in resistant A375 (A375-P) and SK-mel-28 (SK-mel-28-P) cells (Fig. 4H, I).
EphA2 inhibition abolishes AKT and ERK phosphorylation and induces apoptosis
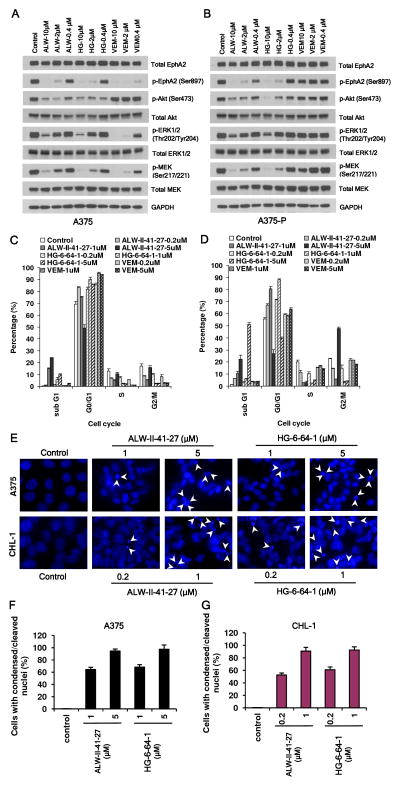
The physiological effects of ALW-II-41-27 and HG-6-64-1 on EphA2/AKT and EphA2/MEK/ERK signaling were then investigated. At 2 μM, both agents effectively inhibited EphA2Ser897 and AKTSer473 phosphorylation; at 10 μM, efficient inhibition of MEK and partial inhibition of ERK were also achieved. ALW-II-41-27 and HG-6-64-1 extinguished MAPK and AKT signaling in both VEM- sensitive (A375) and VEM-resistant (A375-P) lines while VEM abrogated only MAPK, but not AKT, signaling and then only in the sensitive cells. VEM also inhibited p-EphA2Ser897 in the sensitive A375 line (Fig. 5A,B).
Fig. 5. EphA2 inhibitors inhibit phosphorylation of AKT and ERK, arrest cell cycle at G0/G1 and induce apoptosis in both VEM sensitive and resistant cells.
ALW-II-41-27 and HG-6-64-1 abrogated EphA2/AKT and EphA2/MAPK signaling in both VEM sensitive A375 (A) and A375-P (B) cells while VEM inhibited only the MAPK pathway and only in the parental A375 line. Similarly, ALW-II-41-27 and HG-6-64-1 caused both G0/G1 arrest and sub-G1 fractionation in both VEM sensitive A375 (C) and A375-P (D) cells. However, VEM only induced G0/G1 arrest and exclusively in the A375 sensitive line. Error bars indicate percentage of cells ± SEM (n=2). (E) ALW-II-41-27 and HG-6-64-1 caused nuclear condensation and fragmentation in both A375 (VEM sensitive) and CHL-1 (VEM resistant) cells. The nuclei were stained with Hoechst, and analyzed using a fluorescent microscope (the condensed/fragmented nuclei are indicated by arrowheads). (F,G) Quantification results for the nuclei staining in (E).
Since both EphA2/AKT and EphA2/MEK/ERK pathways play critical roles in promoting proliferation and inhibiting apoptosis, we next assessed the impact of the EphA2 compounds on the cell cycle and on the induction of apoptosis. FACS analysis showed that treatment with either ALW-II-41-27 or HG-6-64-1 caused cell cycle arrest at G0/G1 and induced apoptosis at higher doses in both VEM-sensitive and resistant cell lines (Fig. 5C,D); morphologically, the nuclei of cells treated with either ALW-II-41-27 or HG-6-64-1 exhibited a condensed and fragmented morphology, characteristic of apoptosis (Fig. 5E–G). In contrast, VEM was effective in triggering G0/G1 arrest but not apoptosis and only in sensitive cells (Fig. 5C,D). These data suggest that EphA2 blockade abolishes EphA2/AKT as well as EphA2/MEK/ERK signaling and induces apoptosis independent of VEM responsiveness.
EphA2 inhibitors suppress in vivo tumor growth of both VEM sensitive and resistant melanomas
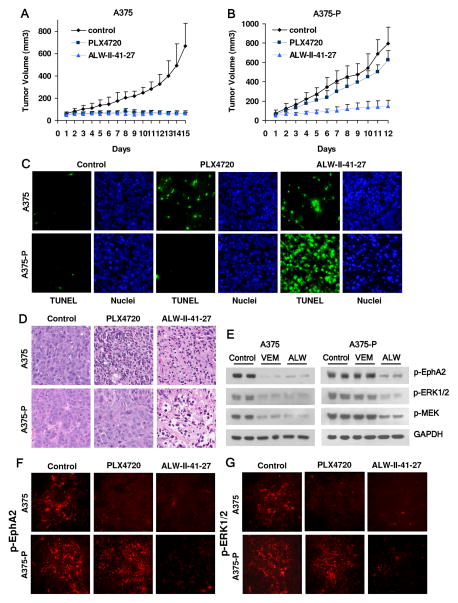
Lastly, we evaluated the efficacy of ALW-II-41-27 against A375 xenografts in mice. ALW-II-41-27 dosed at 30 mg/kg by i.p. significantly suppressed growth of both VEM-sensitive A375 and VEM-resistant A375-P tumors in vivo compared to the vehicle control (P<0.001). In contrast, PLX4720 only inhibited the growth of the parental A375 (P<0.001) tumors but not the resistant A375-P (P=N.S.) tumors in vivo (Fig. 6A,B, fig. S5A,B). Compared to control groups, ALW-II-41-27 reduced A375 and A375-P tumor volumes by 79.7% and 81.6%, respectively, while PLX4720 reduced the tumor volumes by 78.6 and 16.9%, respectively (fig. S5C). Administration of ALW-II-41-27 or PLX4720 was well tolerated by healthy mice without significant signs of overt toxicity or weight loss (P>0.05) (fig. S5D, E).
Fig. 6. EphA2 inhibitor ALW-II-41-27 suppresses in vivo tumor growth of both VEM sensitive and resistant melanomas.
Administration of ALW-II-41-27 inhibited growth of both A375 and A375-P tumors in vivo. After inoculation of 4×107 A375 (A) or A375-P (B) cells, ALW- II-41-27 was administered twice-a-day by intraperitoneal injection (30 mg/kg) while PLX-4720 was administered once a day by intraperitoneal injection (30 mg/kg). The tumor volumes were measured every day by caliper. Error bars indicate mean tumor volume ± SEM (n=6 animals each arm). ALW-II-41-27 administration induced significant apoptosis of both A375 and A375-P tumor cells, which was readily apparent with the TUNEL assay (green in C) and hematoxylin/eosin staining (D). (E) ALW-II-41-27 suppressed phosphorylation of EphA2 and ERK in both A375 and A375-P tumors as demonstrated by western blot analysis. Immunohistochemistry showed that both ALW-II-41-27 and PLX4720 abrogated phosphorylation of EphA2 (F) and ERK1/2 (G) in the A375 tumors while only ALW-II-41-27 suppressed these signaling events in the A375-P tumors.
Consistent with the in vitro data, analysis of both A375 and A375-P tumors from ALW-II-41-27-treated and PLX4720-treated animals showed widespread apoptosis as indicated by TUNEL staining, nuclear fragmentation (Fig. 6C) and both apoptosis and necrosis on hematoxylin/eosin staining (Fig. 6D). PLX4720 induced significant apoptotic changes only in A375 tumors (Fig. 6C, D). Notably, ALW-II-41-27 caused more dramatic apoptosis in the VEM-resistant A375-P tumors compared to the A375 tumor (Fig. 6C, D) further supporting a stronger EphA2 dependency for the VEM- resistant tumor cells. Consistent with the in vitro observations, administration of ALW-II-41-27 caused significant suppression of EphA2/MEK/ERK signaling in both A375 and A375-P tumors while VEM suppressed EphA2/MEK/ERK signaling only in the parental A375 line (Fig. 6E–G). Moreover, since EphA2 influences migration and invasion, we assessed for tumor metastasis in our animal experiments and failed to observe any gross metastatic deposits in the visceral organs (data not shown). Taken together, these experiments provide in vivo evidence of feasibility for EphA2 inhibition in the setting of therapeutic resistance.
DISCUSSION
EphA2 is overexpressed in many human malignancies including melanomas, is associated with a worse prognosis especially among glioblastomas and esophageal carcinomas and is a promoter of proliferation and invasiveness in various cancer cell lines (10, 19, 21–30). Previous studies from our laboratory showed that EphA2 is an essential survival factor in melanoma (10). In this manuscript, we provide several lines of evidence to suggest that EphA2 is also a novel mediator of vemurafenib resistance and potential pharmacologic target in melanoma. First, both total and phosphorylated EphA2 are upregulated in VEM-resistant cells compared to VEM-sensitive cells while EphA2 levels are also increased in relapse specimens compared to pre-treatment specimens from clinical trials. Next, overexpression of EphA2 in VEM-sensitive cells significantly increased VEM resistance while depletion of EphA2 in both naive and VEM-resistant cells can dramatically confer or restore VEM sensitivity. Third, we found that the natural EphA2-selective ligand, ephrinA1, reduced EphA2 signaling and enhanced VEM responsiveness. Lastly, VEM resistant cells appear to be more “addicted” to EphA2 in its survival and migration phenotype compared to its VEM sensitive counterparts.
EphA2 joins the cannon of RTKs known to be upregulated with BRAFi resistance- these include PDGFRβ (12), PDGFRα (13), IGF-1R (14), FGFR3 (15) and EGFR (17,18). Like other RTKs, higher p-EphA2Ser897 correlates with a resistant phenotype. It is interesting to note that the VEM-resistant cells appear to have acquired a greater dependence on EphA2 in terms of migration and colony formation capacity (Fig. 3). This could also explain why EphA2 inhibitors are effective as single agents in the resistant cells even in the absence of additional VEM though there is clear synergism between anti-EphA2 and anti-MAPK compounds. In the animal studies, VEM-resistant A375-P tumors exhibited greater levels of apoptosis compared to VEM-sensitive A375 tumors when treated with ALW-II-41-27. Thus, secondary VEM resistance may come with a heightened “addiction” to EphA2, which could reflect biologic changes induced by the increased EphA2 or a fundamental change in the nature of the resistant cells or both. Unlike other receptor systems which are activated by soluble ligands, interaction between ephrinA1 and EphA2 suppresses EphA2Ser897 phosphorylation and sensitizes cells to VEM. Thus, the mechanism by which EphA2 and other RTK signaling is sustained in the setting of acquired resistance is not known and a focus of ongoing investigation.
With the myriad of resistance lesions reported, it is in fact possible that drug selection recruits multiple simultaneous mechanisms to bypass growth blockade. These can be concurrent in the same cell or scattered across clones within a single population. We have observed a modest increase in pY-AXL in two of the EphA2-upregulated resistant lines though functional studies suggest that the increased AXL signaling is not significantly contributing to VEM resistance (manuscript submitted). Furthermore, Wagle et al. recently performed whole exome sequencing of paired pre-treatment and post-relapse specimens and delineated a constellation of genetic changes associated with gain-of-resistance to combination dabrafenib+trametinib (40). However, it is not yet possible to parse out the contribution of individual changes to the overall pharmacophenotype. In fact, a BRAF splice variant in one of the EphA2-augmented relapse tumor specimens (patient #22) was identified in that study (40) thereby supporting the idea that long-term drug selection may elicit multiple concurrent resistance lesions.
Earlier studies have employed co-targeting of both MAPK and PI-3K/AKT pathways to overcome RTK-mediated VEM resistance (12, 13). However, we set out to demonstrate that EphA2 can be directly antagonized by generating first-in-class small molecule inhibitors of EphA2. These agents proved efficacious in overcoming VEM resistance both in vitro and in vivo. There have been previous inroads made into anti-EphA2 therapy (41). The early use of soluble EphA2-Fc molecules demonstrated significant anti-tumor activity in animal studies (42, 43). In 2008, two 2,5-dimethylpyrrolyl benzoic acid derivatives were shown to competitively inhibit the ephrin-Eph interaction by interacting with the ligand-binding pocket of EphA4 and EphA2 (44); despite this antagonism, the disruption in ephrin binding did not lead to cellular toxicity. Several studies have also shown pre-clinical responses using immune-based strategies that target EphA2 (45). Moreover, depletion of EphA2 using systemic siRNA has also been successfully attempted in animal models (41). However, none of these approaches to date hold the same promise as a small molecule kinase inhibitor that is selective against EphA2. ALW-II-41-27 and HG-6-64-1 were discovered using a rationally-designed kinase-directed library and are believed to bind to the ‘DFG-out’ inactive kinase (type II) conformation based on similarity to structurally related compounds that have been crystallized with EphA3 and EphA7. Like other type II inhibitors that are clinically approved, such as imatinib, nilotinib and sorafenib, ALW-II-41-27 and HG-6-64-1, possess a spectrum of kinase targets in addition to EphA2 (fig. S2; manuscript submitted). While the biologic effects are undoubtedly related to the entire constellation of intracellular targets, our molecular and pharmacologic results support the idea that EphA2 is a critical target for both ALW-II-41-27 and HG-6-64-1 and that EphA2 may be successfully leveraged as a novel pharmacologic target in VEM-resistant melanomas.
Although there are important findings revealed by these studies, there are also limitations. First, a phenotypic selection in vitro, such as induction of VEM resistance, is likely to identify compensatory mechanisms based in part by the nutrient and growth factors conditions in which these experiments were performed. Moreover, pharmacologic selection, either in vitro or in vivo, may galvanize multiple resistance mediators as alluded to above. This could explain why VEM responsiveness is not fully reproduced or restored despite significant alterations in EphA2 levels. We are attempting to improve the gene/drug response specification by isolating individual subclones for analysis. Second, although ALW-II-41-27 and HG-64-1 potently inhibit EphA2 biochemically and in cells, both compounds possess additional targets which most likely contribute to their pharmacology. Efforts are underway to further refine the structure in order to enhance EphA2 specificity and to identify mutations in EphA2 that confer resistance to the inhibitors. Thus, future iterations of these compounds may in fact demonstrate even greater potency. Third, our studies do not address the mechanisms by which EphA2, or possibly other RTKs, becomes activated. Given our current understanding of HGF/MET in VEM resistance (15), autocrine and paracrine loops are likely to play a role. On the other hand, these RTKs are clearly vulnerable to pharmacologic targeting, as evidenced by our results.
In summary, we have identified EphA2 induction as a mechanism of VEM resistance and successfully provided a first-pass pharmacologic solution to inhibit this RTK. Our results confirm that small molecule inhibition of EphA2 can be an effective means to neutralize VEM resistance. These studies thus offer a new strategy to treat patients with melanomas that are either primarily insensitive to VEM or those are have developed resistance against BRAF inhibition.
MATERIALS AND METHODS
Materials
PLX4032/vemurafenib, PLX4720 and AZD6244 were purchased from Chembridge (San Diego, CA). ALW-II-41-27 and HG-6-64-1 were provided by Dr. Nathanael Gray’s lab at Dana-Farber Cancer Institute, Harvard Medical School (Boston, MA). The purity of all compounds was >95%. Hairpin shRNAs targeting EphA2 were purchased from Open Biosystems, Huntsville, AL. The recombinant ephrin-A1 and ephrin-B1 were purchased from R&D systems, Minneapolis, MN. Mouse polyclonal anti-EphA2 (C-20) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-phospho-EphA2 (Ser897), anti-phospho-Erk(1/2) (Thr202/Tyr204), anti-phospho-MEK (Ser217/221), anti-phospho-Akt (Ser473), anti-total MEK, anti-GAPDH and the secondary HRP-conjugated antibodies were purchased from Cell Signaling Technologies (Beverly, MA). All other reagents and chemicals were purchased from Sigma (St. Louis, MO).
Cells and cell culture
Human melanoma cell lines (A375, SK-mel-28, UACC903, MGH-MC-1, K4, WM239, WM1158, WM164, G-mel, CHL-1, SK-mel-119) were developed in-house, purchased from the American Type Culture Collection (Rockville, MD) or gifts from Meenhard Herlyn (Wistar Institute, Philadelphia, PA). Vemurafenib resistant melanoma cell lines (A375-P, SK-Mel 28-P, UACC903-P, MGH-MC-1-P, K4-P, WM239-P and WM1158-P) were obtained by sequentially exposing of cells to escalating doses of VEM in 2014. DMEM medium with 10% fetal bovine serum (FBS), supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine was used for routine culturing of cells; VEM-resistant cells were cultured in VEM-free media for 10–14 days prior to re-testing VEM sensitivity; this drug holiday period had no effect on the VEM resistance (data not shown). All cells were incubated in a humidified atmosphere of 95% air and 5% CO2 at 37°C. Tumor specimens from patients were obtained prior to treatment with BRAFi (vemurafenib) or BRAFi+MEKi (dabrafenib+trametinib) and post-relapse as indicated in Table S1. Acquisition of tissue was covered under a protocol approved by the DF/Harvard Cancer Center (legacy #11-181; Boston, U.S.) in accordance with the Declaration of Helsinki.
Cell line authentication
Cell lines were verified in 2014 by matching mutational profiles and/or copy number variants (CNVs) obtained in our laboratory with those independently reported in COSMIC (A375, Gmel, SKmel28, CHL-1) or with those published by at least two independent laboratories (WM1158, SK-mel-119, WM239, UACC903).
Data on PLX4720 sensitivity (PLX activity area) and EPHA2 expression for the melanoma lines were obtained through the Cancer Cell Line Encyclopedia (CCLE) website (http://www.broadinstitute.org/ccle/home). Statistical analysis was performed using GraphPad Prism 6 in 2014.
Cell viability assay
Melanoma cells were seeded into 96-well white plates at a density of 5–10×103 cells per well in 100 μl media. The compounds were added at the designated concentrations and incubated for 72 hr. A luminescence-based commercial kit (CellTiter-Glo, Promega, Madison, WI) was used to measure cell viability. Briefly, 30 μl of cell lysis/ATP detection reagent was added to each well, incubated on a shaking platform for 10 min at room temperature, and the luminescence was measured with a plate reader (Molecular Devices, Sunnyvale, CA). Cellular GI50 values were determined using CompuSyn software.
Cellular colony formation assay
Melanoma cells were seeded in 6-well plates at a density of 500–1000 cells/well. The media was changed every other day and the colonies were counted at day 14 after staining with 0.1% crystal violet.
Cell migration assays
The 24-transwell Boyden chambers (Costar, Bedford, MA) with a polystyrene membrane (6.5 mm diameter, 10 μm thickness, and 8 μm pore size) were used. Melanoma cells were seeded in the compartment of well in serum-free media (5×104 cells/well) with or without compounds. The upper lower compartment was supplied with 600 μl serum-free media supplemented with 20 μg/ml fibronectin. Cells were treated for 8-hr, then were fixed and stained with 0.1% crystal violet. The non-migrating cells on the upper surface of the membrane were removed, and the migrated cells on the lower side were photographed with a microscope (Nikon, Japan) in five random fields. For quantitation, the migrated crystal violet-stained cells were lysed with 10% acetic acid, and colorimetric determination was made at 595 nm.
Biochemical profiling compounds inhibition of EphA2
SelectScreen™ Kinase Profiling Service (Life Technologies, Grand Island, NY) was used to profile potency of compounds in vitro. In general, the assay was conducted in 50 mM HEPES (pH 7.5), 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA, 2 mM DTT, and 100 μM ATP for 1 hr. The biochemical IC50 was calculated according to 10-point compound titration (0.5 nM to 10 μM).
Real time PCR
RNA was isolated from melanoma patient samples using RNA easy kit (Qiagen, Valencia, CA). cDNA was generated from 1 μg of RNA using First Strand Ready-To-Go beads (GE Healthcare Life Science, Pittsburgh, PA). Real-time PCR was performed in triplicates using EPHA2 and (Invitrogen, Carlsbad, CA) and LightCycler TaqMan Master Kit (Roche GUSB TaqMan primers Applied Sciences, Indianapolis, IN) on a Roche LC480 qRT-PCR machine. EPHA2 cDNA levels in human samples were normalized to GUSB, and are presented as relative units to pre-treatment levels. Data acquisition and analysis was performed according to Manufacturer’s instructions.
Plasmid construction and transfection
The eukaryotic expression plasmid of human EPHA2 gene was generated by cDNA cloning followed by sequence confirmation. The cell lines were transfected with 4 μg of the EphA2 plasmid or control vector using Lipofectamine (Invitrogen, Carlsbad, CA) according to manufacturer’s protocol. The cells were selected with G418 (1 mg/ml) for stable incorporation. The stable clones were collected and protein expression levels were confirmed with western blotting.
EphA2 gene silencing
Hairpin shRNAs targeting EphA2 were purchased from Open Biosystems (Huntsville, AL). The shRNA plasmids were mixed with the lentivirus packaging plasmids (Invitrogen, Carlsbad, CA) in the ratio according to the manufacturer’s protocol. They were then mixed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and transfected into HEK 293T cells for 48 hr to generate virus. Then the viral supernatants were filtered and used for target cell infection in the presence of 8 μg/ml Polybrene (American Bioanalytical, Natick, MA), followed by selection with puromycin to get stable populations of cells.
Western blotting
Cells were lysed in 20 mM HEPES, pH 7.5, 150 mM NaCl, 1% NP-40, 10 mM tetrasodium pyrophosphate, 100 mM NaF, 17.5 mM 1-glycerophosphate buffer supplemented with Complete Mini Protease Inhibitor Cocktail tablets (Roche Applied Sciences, Indianapolis, IN). Samples separated by SDS-PAGE were transferred to nitrocellulose membranes, blocked with 5% bovine serum albumin (w/v) at room temperature for 1 hr, and incubated with primary antibodies (1:1000 dilution) at 4°C overnight. After incubation with secondary antibody (1:3000 dilution) at room temperature for 1 hr, the membranes were developed with chemiluminescence ECL reagent (LumiGold, SignaGen Laboratories, Rockville, MD) and exposed to Hyperfilm MP (GE Healthcare Life Science, Pittsburgh, PA). Tumor samples were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40, supplemented with Complete Mini protease inhibitors) and equal amounts of protein were subjected to western blotting analysis.
Fluorescent immunocytochemistry
Cells were seeded on coverslips, followed by treatments with compounds. Then cells were fixed with 4% polyformaldehyde for 30 min, permeabilized with 0.1% Triton X-100 for 20 min, blocked with 5% normal serum for 30 min and incubated with Hoechst for 10 min. All images were obtained using Olympus BX51 microscope.
Flow cytomery analysis
Melanoma cells were seeded in 6-well plates (5×105 cells/well) with or without compounds treatment. Then cells were harvested, fixed with 70% ethanol and stained with propidium iodide (PI, 5 mg/l) in the presence of RNase (1 g/l), 1 g/l sodium citrate and 0.5% Triton X-100 (v/v) in the dark for 30 min. Cells were collected for apoptosis analysis using FacsCalibur (BD Biosciences, San Jose, CA). The percentage of hypodiploidy was analyzed using ModFIT LT software (Verity Software).
Immunohistochemistry (IH)
Tumor tissues were fixed in phosphate-buffered formalin, embed in paraffin, cut in 4 μm thickness, and applied to slides. The slides were deparaffinized in xylenes using three changes for 5 min each, and hydrated gradually through graded alcohols: 100% ethanol twice for 10 min each, 95% ethanol twice for 10 min each, and then deionized water for 1 min with stirring. For antigen unmasking, slides were placed in a container, covered with 10 mM sodium citrate buffer, pH 6.0, and heated in a convection steamer for 1 hr. The slides were washed in deionized water three times for 2 min each, blocked with 5% normal goat blocking serum for 30 min, incubated with primary antibodies for 1 hr, and incubated with a Rhodamine-conjugated secondary antibody for 30 min, and then incubated with Hoechst for 10 min. The slides were analyzed and photographed using a DeadEnd™ Fluorometric TUNEL fluorescent microscope. TUNEL staining was performed using a kit according to manufacturer’s instructions (Promega, Madison, WI)
Subcutaneous xenograft tumor growth in vivo
Nud/Nud mice were purchase from in-house colonies at MGH, and housed in a BL2 lab at MGH. All animal experiments were carried out in accordance with protocols approved by MGH Animal Care and Use committees. A375 and A375-P cells were cultured in vitro in DMEM medium and then re-suspended in PBS (4×107 cells/ml). The melanoma cells were injected subcutaneously (4×106 cells/100 μl/mouse). When the tumor volumes into axillary regions of nude/nude mice reached ~60 mm3 (about one week), the mice were randomized to control group and various treatment groups (n=6 per group), including ALW-II-41-27 (30 mg/kg, twice/day, vehicle: 10% 1-methyl-2-pyrrolidinone and 90% PEG 300) and PLX4720 (30 mg/kg, vehicle: DMSO) groups. Tumors were measured every day for ~2 weeks with a microcaliper. The body weight was measured every day with a scale. The tumor volumes were calculated with the formula: . After administration with vehicle, ALW-II-41-27 or PLX4720 via intraperitoneal injection for ~2 weeks, the mice were euthanized using carbon dioxide (CO2) and tumors were harvested. Tumor cell apoptosis and EphA2 related signaling were analyzed using immunohistochemistry and western blotting.
Statistics
Student’s t test and analysis of variance (ANOVA) were performed using StatView (SAS Institute, Cary, NC). The data shown are representatives of at least two independent experiments with similar results, and the data points represent the mean of at least triplicate measurements with error bars corresponding to standard deviation.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
In this study, we show that resistance to selective BRAF inhibitors can be mediated by the receptor tyrosine kinase, EphA2. Furthermore, direct targeting of EphA2 can successfully suppress melanoma growth and mitigate therapeutic resistance.
Acknowledgments
GRANT SUPPORT
This work was supported in part by the National Institutes of Health (K24 CA149202, P01CA163222, 5T32AR007098 and R01 CA173469), the American Skin Association (to H.T.) and the generous donors to the MGH Millennium Melanoma Fund.
Abbreviations list
- MAPK
mitogen-activated protein kinase
- RTK
receptor tyrosine kinases
- Eph
erythropoietin-producing hepatocellular
- BRAFi’s
BRAF inhibitors
- VEM
vemurafenib
- CCLE
Cancer Cell Line Encyclopedia
- CI
combination index
Footnotes
Conflict of interest disclosure statement:
The authors disclose no potential conflicts of interest
References
- 1.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauschild A, Agarwala SS, Trefzer U, Hogg D, Robert C, Hersey P, et al. Results of a phase III, randomized, placebo-controlled study of sorafenib in combination with carboplatin and paclitaxel as second-line treatment in patients with unresectable stage III or stage IV melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:2823–30. doi: 10.1200/JCO.2007.15.7636. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev. 2012;26:1131–55. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 9.Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41:1127–32. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udayakumar D, Zhang G, Ji Z, Njauw CN, Mroz P, Tsao H. Epha2 is a critical oncogene in melanoma. Oncogene. 2011;30:4921–9. doi: 10.1038/onc.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlegel J, Sambade MJ, Sather S, Moschos SJ, Tan AC, Winges A, et al. MERTK receptor tyrosine kinase is a therapeutic target in melanoma. J Clin Invest. 2013;123:2257–67. doi: 10.1172/JCI67816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabbatino F1, Wang Y, Wang X, Flaherty KT, Yu L, Pepin D, et al. PDGFRα up-regulation mediated by sonic hedgehog pathway activation leads to BRAF inhibitor resistance in melanoma cells with BRAF mutation. Oncotarget. 2014;5:1926–41. doi: 10.18632/oncotarget.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yadav V, Zhang X, Liu J, Estrem S, Li S, Gong XQ, et al. Reactivation of mitogen- activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. The Journal of biological chemistry. 2012;287:28087–98. doi: 10.1074/jbc.M112.377218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girotti MR, Marais R. Deja Vu: EGF receptors drive resistance to BRAF inhibitors. Cancer discovery. 2013;3:487–90. doi: 10.1158/2159-8290.CD-13-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girotti MR, Pedersen M, Sanchez-Laorden B, Viros A, Turajlic S, Niculescu-Duvaz D, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer discovery. 2013;3:158–67. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaught D, Chen J, Brantley-Sieders DM. Regulation of mammary gland branching morphogenesis by EphA2 receptor tyrosine kinase. Molecular biology of the cell. 2009;20:2572–81. doi: 10.1091/mbc.E08-04-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang G, Brantley-Sieders DM, Vaught D, Yu J, Xie L, Wells S, et al. Elevation of receptor tyrosine kinase EphA2 mediates resistance to trastuzumab therapy. Cancer research. 2010;70:299–308. doi: 10.1158/0008-5472.CAN-09-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz JC, Robertson EJ. The expression of the receptor-protein tyrosine kinase gene, eck, is highly restricted during early mouse development. Mechanisms of development. 1994;46:87–100. doi: 10.1016/0925-4773(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 22.Moore-Scott BA, Opoka R, Lin SC, Kordich JJ, Wells JM. Identification of molecular markers that are expressed in discrete anterior-posterior domains of the endoderm from the gastrula stage to mid-gestation. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236:1997–2003. doi: 10.1002/dvdy.21204. [DOI] [PubMed] [Google Scholar]
- 23.Ganju P, Shigemoto K, Brennan J, Entwistle A, Reith AD. The Eck receptor tyrosine kinase is implicated in pattern formation during gastrulation, hindbrain segmentation and limb development. Oncogene. 1994;9:1613–24. [PubMed] [Google Scholar]
- 24.Connor RJ, Menzel P, Pasquale EB. Expression and tyrosine phosphorylation of Eph receptors suggest multiple mechanisms in patterning of the visual system. Developmental biology. 1998;193:21–35. doi: 10.1006/dbio.1997.8786. [DOI] [PubMed] [Google Scholar]
- 25.Andres AC, Reid HH, Zurcher G, Blaschke RJ, Albrecht D, Ziemiecki A. Expression of two novel eph-related receptor protein tyrosine kinases in mammary gland development and carcinogenesis. Oncogene. 1994;9:1461–7. [PubMed] [Google Scholar]
- 26.Easty DJ, Guthrie BA, Maung K, Farr CJ, Lindberg RA, Toso RJ, et al. Protein B61 as a new growth factor: expression of B61 and up-regulation of its receptor epithelial cell kinase during melanoma progression. Cancer research. 1995;55:2528–32. [PubMed] [Google Scholar]
- 27.Easty DJ, Hill SP, Hsu MY, Fallowfield ME, Florenes VA, Herlyn M, et al. Up-regulation of ephrin-A1 during melanoma progression. Int J Cancer. 1999;84:494–501. doi: 10.1002/(sici)1097-0215(19991022)84:5<494::aid-ijc8>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 28.Hendrix MJ, Seftor EA, Kirschmann DA, Quaranta V, Seftor RE. Remodeling of the microenvironment by aggressive melanoma tumor cells. Ann N Y Acad Sci. 2003;995:151–61. doi: 10.1111/j.1749-6632.2003.tb03218.x. [DOI] [PubMed] [Google Scholar]
- 29.Kamat AA, Coffey D, Merritt WM, Nugent E, Urbauer D, Lin YG, et al. EphA2 overexpression is associated with lack of hormone receptor expression and poor outcome in endometrial cancer. Cancer. 2009;115:2684–92. doi: 10.1002/cncr.24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zelinski DP, Zantek ND, Stewart JC, Irizarry AR, Kinch MS. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer research. 2001;61:2301–6. [PubMed] [Google Scholar]
- 31.Zhuang G1, Brantley-Sieders DM, Vaught D, Yu J, Xie L, Wells S, et al. Elevation of receptor tyrosine kinase EphA2 mediates resistance to trastuzumab therapy. Cancer Res. 2010;70:299–308. doi: 10.1158/0008-5472.CAN-09-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menges CW, McCance DJ. Constitutive activation of the Raf-MAPK pathway causes negative feedback inhibition of Ras-PI3K-AKT and cellular arrest through the EphA2 receptor. Oncogene. 2008;27:2934–40. doi: 10.1038/sj.onc.1210957. [DOI] [PubMed] [Google Scholar]
- 33.Beauchamp A, Debinski W. Ephs and ephrins in cancer: ephrin-A1 signalling. Semin Cell Dev Biol. 2012;23:109–15. doi: 10.1016/j.semcdb.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang WB, Brantley-Sieders DM, Parker MA, Reith AD, Chen J. A kinase-dependent role for EphA2 receptor in promoting tumor growth and metastasis. Oncogene. 2005;24:7859–68. doi: 10.1038/sj.onc.1208937. [DOI] [PubMed] [Google Scholar]
- 35.Miao H, Burnett E, Kinch M, Simon E, Wang B. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nature cell biology. 2000;2:62–9. doi: 10.1038/35000008. [DOI] [PubMed] [Google Scholar]
- 36.Kinch MS, Carles-Kinch K. Overexpression and functional alterations of the EphA2 tyrosine kinase in cancer. Clin Exp Metastasis. 2003;20:59–68. doi: 10.1023/a:1022546620495. [DOI] [PubMed] [Google Scholar]
- 37.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5:1136–44. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 38.Miao H, Li DQ, Mukherjee A, Guo H, Petty A, Cutter J, et al. EphA2 mediates ligand- dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer cell. 2009;16:9–20. doi: 10.1016/j.ccr.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, et al. Comprehensive analysis of kinase inhibitor selectivity. Nature biotechnology. 2011;29:1046–51. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 40.Wagle N1, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–8. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tandon M, Vemula SV, Mittal SK. Emerging strategies for EphA2 receptor targeting for cancer therapeutics. Expert opinion on therapeutic targets. 2011;15:31–51. doi: 10.1517/14728222.2011.538682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brantley DM, Cheng N, Thompson EJ, Lin Q, Brekken RA, Thorpe PE, et al. Soluble Eph A receptors inhibit tumor angiogenesis and progression in vivo. Oncogene. 2002;21:7011–26. doi: 10.1038/sj.onc.1205679. [DOI] [PubMed] [Google Scholar]
- 43.Dobrzanski P, Hunter K, Jones-Bolin S, Chang H, Robinson C, Pritchard S, et al. Antiangiogenic and antitumor efficacy of EphA2 receptor antagonist. Cancer research. 2004;64:910–9. doi: 10.1158/0008-5472.can-3430-2. [DOI] [PubMed] [Google Scholar]
- 44.Noberini R, Koolpe M, Peddibhotla S, Dahl R, Su Y, Cosford ND, et al. Small molecules can selectively inhibit ephrin binding to the EphA4 and EphA2 receptors. The Journal of biological chemistry. 2008;283:29461–72. doi: 10.1074/jbc.M804103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi S, Tatsumi T, Takehara T, Sakamori R, Uemura A, Mizushima T, et al. Immunotherapy of murine colon cancer using receptor tyrosine kinase EphA2-derived peptide- pulsed dendritic cell vaccines. Cancer. 2007;110:1469–77. doi: 10.1002/cncr.22958. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.