Abstract
Background
The transcription factor DOF AFFECTING GERMINATION1 (DAG1) is a repressor of the light-mediated seed germination process. DAG1 acts downstream PHYTOCHROME INTERACTING FACTOR3-LIKE 5 (PIL5), the master repressor, and negatively regulates gibberellin biosynthesis by directly repressing the biosynthetic gene AtGA3ox1. The Dof protein DOF AFFECTING GERMINATION (DAG2) shares a high degree of aminoacidic identity with DAG1. While DAG1 inactivation considerably increases the germination capability of seeds, the dag2 mutant has seeds with a germination potential substantially lower than the wild-type, indicating that these factors may play opposite roles in seed germination.
Results
We show here that DAG2 expression is positively regulated by environmental factors triggering germination, whereas its expression is repressed by PIL5 and DAG1; by Chromatin Immuno Precipitation (ChIP) analysis we prove that DAG1 directly regulates DAG2. In addition, we show that Red light significantly reduces germination of dag2 mutant seeds.
Conclusions
In agreement with the seed germination phenotype of the dag2 mutant previously published, the present data prove that DAG2 is a positive regulator of the light-mediated seed germination process, and particularly reveal that this protein plays its main role downstream of PIL5 and DAG1 in the phytochrome B (phyB)-mediated pathway.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0453-1) contains supplementary material, which is available to authorized users.
Keywords: DAG2, Seed germination, DAG1, Arabidopsis thaliana
Background
The DNA BINDING WITH ONE FINGER (Dof) proteins are a family of plant-specific transcription factors characterised by a single zinc-finger DNA-binding domain. So far Dof proteins have been identified in Chlamydomonas reinharditii, where only one Dof gene is present, in ferns, mosses and in higher plants [1-3].
The number of Dof genes varies depending on the species; bioinformatic analysis of the Arabidopsis and rice genome predicts 36 and 30 Dof genes, respectively [1], while 26 are present in barley [2], 31 in wheat [4], and 28 in sorghum [5]. Members of this family have been found to be involved in the regulation of diverse plant-specific processes. Although the biological role of many Dof proteins has not been clarified yet, a number of them has been shown to be involved in responses to light and phytohormones, as well as in seed development and germination [6-15].
Seed germination is regulated by environmental factors such as light, temperature and nutrients, and by phytohormones, particularly gibberellins (GA) and abscissic acid (ABA) [16]. The effect of light is mediated mainly by the photoreceptor phytochrome B (phyB) [17], and light modulates in opposite ways the levels of GA and ABA, as it induces GA biosynthesis and causes a reduction in ABA levels [18,19]. Among the factors involved in phyB-mediated GA-induced seed germination, the bHLH transcription factor PHYTOCHROME INTERACTING FACTOR 3-LIKE 5 (PIL5) represents the master repressor of this process in Arabidopsis [20].
We have previously shown that inactivation of the Dof proteins DAG1 and DAG2 affects in opposite ways seed germination: dag2 mutant seeds required more light and GA than wild-type seeds to germinate, whereas germination of dag1 seeds was less dependent on these factors [7,8,21].
Recently, we have also pointed out that DAG1 acts as a negative regulator in the phyB-mediated pathway: DAG1 gene expression is reduced in seeds irradiated for 24 hours with Red light, and this reduction is dependent on PIL5; in pil5 mutant seeds DAG1 expression is reduced irrespective of light conditions, indicating that DAG1 acts downstream of PIL5. Moreover, DAG1 negatively regulates GA biosynthesis by directly repressing the GA biosynthetic gene AtGA3ox1 [22]. Very recently we showed that in repressing AtGA3ox1 DAG1 directly interacts with the GA INSENSITIVE (GAI) DELLA protein [23]. Furthermore, we pointed out that DAG1 plays a role also in embryo development, as inactivation of DAG1 results in a significant number of embryo abnormalities [7,24], and simultaneous inactivation of both DAG1 and GAI results in an embryo-lethal phenotype. Here, we provide evidence suggesting that DAG2, opposite to DAG1, functions as a positive regulator in the molecular pathway controlling seed germination, and that it is negatively regulated by DAG1.
Differently from DAG1, DAG2, although it is expressed during embryo development, is not likely to play a role in this process, as dag2 mutant embryos develop similarly to wild-type embryos.
Results
DAG2 inactivation affects phyB-dependent seed germination
We have previously demonstrated that dag2 mutant seeds have a reduced germination potential, as they are substantially more dependent than the wild-type on the stimuli that promote germination [8]. This germination phenotype is opposite to that of dag1 mutant seeds. As we have recently shown that DAG1 is a component of the phyB-mediated pathway controlling seed germination in Arabidopsis [22,23], we set up to verify whether DAG2 is also a component of this regulatory network.
Since seed germination, although promoted mainly by phyB, may be induced also by phyA under very low light fluences [17], we checked whether Red (R) or Far Red (FR) light may control expression of the DAG2 gene. Analysis of wild-type seeds exposed to phyB- or phyA-dependent conditions, according to Oh et al. 2006 [25], revealed that the DAG2 gene is induced by exposure to R light (Figure 1A), whereas DAG2 expression in seeds exposed to FR light was not significantly different than in seeds kept in the dark (Figure 1B). To assess whether DAG2 plays its role under R light, we analysed seed germination under phyB-dependent conditions [22] using the dag2 mutant previously characterised [8], compared to the corresponding wild-type (Ws-4). Germination of dag2 mutant seeds was significantly lower than that of wild-type seeds (30% and 90%, respectively - Figure 1C), thus confirming that DAG2 plays a positive role in seed germination and showing that it acts in the phyB-mediated pathway.
Figure 1.
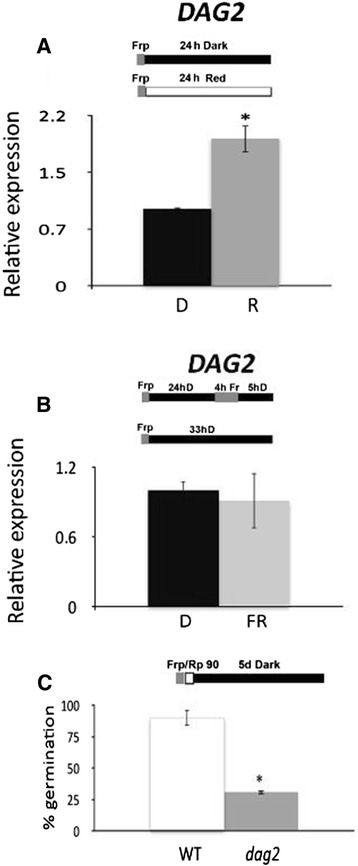
Mutation of DAG2 affects seed germination under R light. Relative expression level of DAG2 in wild-type seeds imbibed 24 hours in the dark (D), or under phyB-dependent conditions, (A), and in the dark or under phyA-dependent conditions (B). Relative expression levels were normalized with that of the UBQ10 (At4g05320) gene, and are presented by the ratio of the corresponding mRNA level in Dark, which was set to 1. Similar results were obtained from three independent experiments, and a typical result is presented with SD values.Germination rates of wild-type and dag2 mutant seeds, grown 5 days under phyB-dependent germination conditions (C). Error bars = SEM. The diagram at top depicts the light treatment scheme for the experiment. FRp, Far Red pulse (40 μmol m−2 s−1); Rp, Red pulse (90 μmol m−2 s−1). Significative differences were analyzed by t-test (*P ≤ 0,05).
Since water uptake is a fundamental requirement for seed germination, we verified whether expression of DAG2 was regulated during imbibition. We performed RT-qPCR assays on wild-type (Ws-4) dry seeds, and on seeds imbibed under White (W) and R light or in the dark for 12 and 24 hrs. Figure 2A shows that, compared to the low amount present in dry seeds, DAG2 expression in seeds was much increased following water uptake in the dark (2 and 4 fold, respectively, at 12 and 24 hrs). Interestingly, the increase in DAG2 mRNA level in seeds exposed to W or R light was even higher, probably due to the effect of both light and imbibition (3.7 and 7.8 fold in W light and 4 and 7-fold in R light, at 12 and 24 hrs, respectively - Figure 2A). GUS histological assays, performed on seeds of the DAG2:GUS transgenic line [8], dry or imbibed 12 hours under W light or in the dark respectively, showed that the DAG2 promoter was active only in the vascular tissue (Figure 2B).
Figure 2.
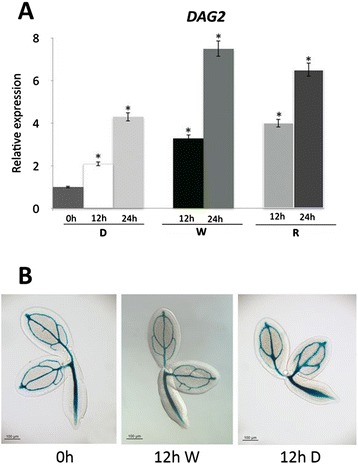
DAG2 expression is induced by imbibition. Relative expression level of DAG2 in wild-type dry seeds (0 h), or imbibed 12 (12 h) or 24 hours (24 h) in the dark (D) or under White (W) or Red (R) light (A). Relative expression levels were normalized with that of the ACTIN2 (At3g18780) gene, and are presented by the ratio of the corresponding mRNA level in dry seeds, which was set to 1. Similar results were obtained from three independent experiments, and a typical result is presented with SD values. Significative differences were analyzed by t-test (*P ≤ 0,05). Histochemical staining of DAG2:GUS dry seeds, or imbibibed 12 hours under W light (W) or in the dark (D) (B).
DAG2 is directly regulated by DAG1
We have previously investigated the genetic interactions between the DAG2 and DAG1 genes by isolating the dag2dag1 double mutant, and showed that DAG1 is epistatic over DAG2 [8]. Since the function of DAG2 appears to be opposite to that of DAG1, we verified whether DAG1 and DAG2 would mutually affect their expression, by performing an RT- qPCR analysis in dag1 and dag2 mutant seeds imbibed for 12 hours in the dark or under R light. As shown in Figure 3A, expression of DAG2 is significantly (approximately 3-fold) increased by lack of DAG1, irrespective of light conditions. Conversely, DAG1 expression level in wild-type and dag2 mutant seeds was comparable, both in the dark and under R light (Figure 3B).
Figure 3.
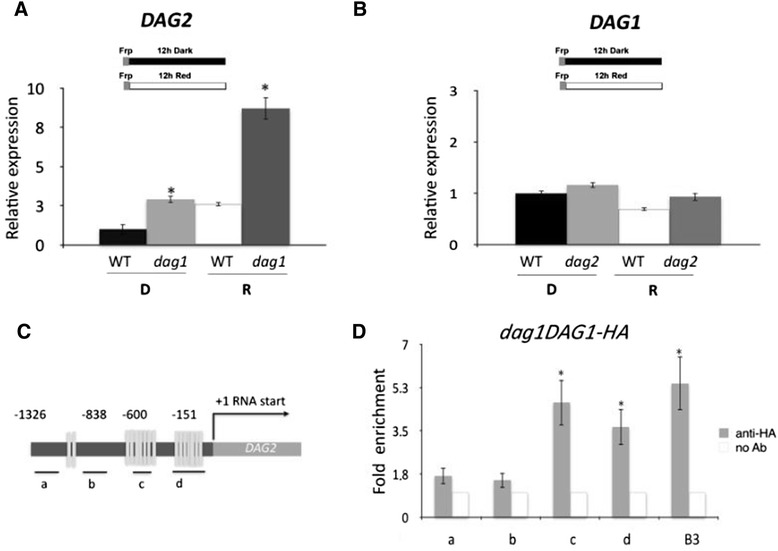
DAG2 is directly regulated by DAG1. Relative expression level of: DAG2 in dag1 mutant and wild-type (WT) seeds (A), and of DAG1 in dag2 mutant and wild-type seeds (B). Seeds were imbibed 12 hours in the dark (D), or under R light (R). Relative expression levels were normalized with that of the UBQ10 gene and are presented by the ratio of the corresponding wild-type mRNA level in D, which was set to 1. Similar results were obtained from three independent experiments, and a typical result is presented with SD values. Significative differences were analyzed by t-test (*P ≤ 0,05). (C) Graphic representation of the DAG2 promoter. Underlying thick lines marked by letters (a, b, c, d) are referred to different promoter fragments used for qPCR, containing 0 (a, b), 4 and 7 Dof sites respectively (c,d). (D) Chromatin from dag1DAG1-HA seeds was immunoprecipitated with anti-HA or without antibody, and the amount of DNA was measured by qPCR. B3 is referred to the positive control, fragment B3 of the AtGA3ox1 promoter bound by DAG1-HA The values of fold enrichment are the average of three independent experiments presented with SD values. Significative fold enrichment was analyzed by t-test (*P ≤ 0,05).
To assess whether DAG1 regulates DAG2 by directly binding to the DAG2 promoter in vivo, we performed chromatin immunoprecipitation (ChIP) assays, utilizing the dag1DAG1-HA line previously reported [22,23]. A scheme of the DAG2 promoter is reported in Figure 3C, showing the positions of the PCR fragments amplified for the ChIP assays, each containing different numbers of Dof binding sites: 0 (a, b), 4 (c) and 7 sites (d). Consistently, anti-HA antibodies revealed that the amplification of fragments c and d were the most efficient, compared to the positive control, the fragment B3 of the AtGA3ox1 promoter bound by DAG1-HA, as previously reported [23]. On the contrary, the signal for fragments a and b was quite faint. No PCR product was present for any of the fragments in the sample precipitated without antibodies as a negative control, and not even for the negative control on wild-type seeds (Figure 3D; Additional file 1: Figure S1). These results indicate that DAG1 negatively regulates DAG2 by directly binding the DAG2 promoter.
PIL5 negatively regulates DAG2 in the Dark
Since DAG1 and DAG2 seem to have opposite roles in the phyB-mediated seed germination pathway, we wondered whether PIL5, which positively regulates DAG1, might negatively control the expression of DAG2. To verify this hypothesis, we analysed the expression of the DAG2 gene in wild-type and pil5 mutant seeds after 12 hours imbibition in the dark or under R light. Interestingly, as shown in Figure 4, the relative amount of DAG2 in pil5 mutant seeds in the dark was significantly higher than in the wild-type, suggesting that PIL5 negatively regulates the expression of DAG2 in the dark. On the other hand, DAG2 expression level in R light does not depend on PIL5, as it is degraded following interaction with phyB.
Figure 4.
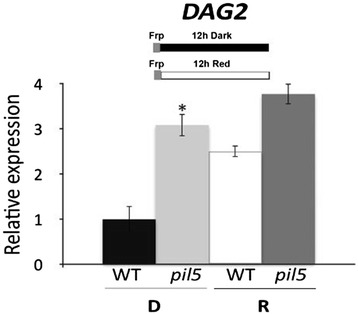
DAG2 expression is repressed by PIL5. Relative expression level of DAG2 in pil5 mutant and wild-type seeds. Seeds were imbibed 12 hours in the dark (D), or under R light (R). Relative expression levels were normalized with that of the UBQ10 gene, and are presented by the ratio of the corresponding wild-type mRNA level in D, which was set to 1. Similar results were obtained from three independent experiments, and a typical result is presented with SD values. Significative differences were analyzed by t-test (*P ≤ 0,05).
The DELLA proteins GAI and RGA are negative regulators of seed germination, acting downstream of PIL5 [26]. In particular, we have recently shown that GAI and DAG1 mutually regulate their expression level and directly interact with each other [23]. Thus, we set to assess whether GAI and/or RGA might control the expression level of DAG2. Analysis of DAG2 expression on gai-t6 and rga28 mutant seeds and on the corresponding Col-0 wild-type seeds, imbibed 12 hours in the dark or under R light, revealed that neither GAI nor RGA control DAG2 expression, as the relative amount of DAG2 mRNA was similar in the gai-t6 and rga28 single mutants compared to the wild-type, under both light conditions (Figure 5A, B). To verify whether DAG2 might regulate expression of these DELLA proteins, we analysed the expression of GAI and RGA in dag2 mutant seeds compared to the wild-type. As shown in Figure 5C, the expression of the RGA gene was significantly increased in dag2 mutant seeds, whereas GAI expression in wild-type and dag2 mutant seeds was not significantly different, both in the dark and under R light (Figure 5D).
Figure 5.
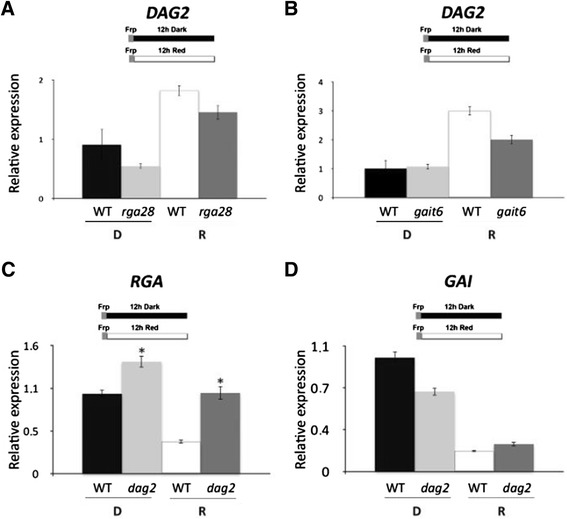
DAG2 expression is regulated by RGA and GAI. Relative expression level of: DAG2 in rga28 (A), and gai-t6 mutant seeds (B), and of RGA (C) or GAI (D) in dag2 mutant seeds, compared to wild-type seeds. Seeds were imbibed 12 hours in the dark (D), or under R light (R). Relative expression levels were normalized with that of PP2A (At1g13320) (A, B), or of UBQ10 (C, D), and are presented by the ratio of the corresponding wild-type mRNA level in D, which was set to 1. Similar results were obtained from three independent experiments, and a typical result is presented with SD values. Significative differences were analyzed by t-test (*P ≤ 0,05).
These results point to DAG2 as a positive component of light-mediated signalling pathway, downstream of PIL5 and in turn controlling the DELLA protein RGA in the phyB signalling pathway.
We then verified whether expression of some factors known to be involved in the phyA-signalling pathway [27] may be affected in dag2 mutant seeds. In particular, we analysed expression of the FR light-regulated ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2 (ATHB2) and PHYTOCHROME INTERACTING FACTOR 3-LIKE 2 (PIL2) genes, of the GA-regulated GA-STIMULATED ARABIDOPSIS 4 and 6 (GASA4 and GASA6), and of the ABA signalling gene ABA INSENSITIVE 4 (ABI4). Our data revealed that under phyA-dependent conditions neither expression of ATHB2 and PIL2, nor that of ABI4 were affected in the dag2 mutant (Additional file 2: Figure S2) whereas expression of both GASA4 and GASA6 was downregulated, thus opening the possibility that DAG2 may also play a role in phyA signalling.
The dag2 mutation alters GA metabolism
It has been shown that phyB controls the ratio of GA and ABA levels during seed germination by altering the expression of different GA and ABA metabolic genes through PIL5 [18,26]. In particular, DAG1 directly represses the GA biosynthetic gene AtGA3ox1 in cooperation with GAI [22,23], and its inactivation affects expression of the ABA metabolic genes ABA1, ABA2 and CYP707A2 [22].
As DAG2 seems to have a role opposite to DAG1 in seed germination, we investigated whether DAG2 would regulate the expression of GA and ABA metabolic genes in germinating seeds. We performed a RT-qPCR analysis of the expression of the GA biosynthetic genes AtGA3ox1, AtGA3ox2 and of the catabolic gene AtGA2ox2 in dag2 and wild-type seeds imbibed 12 hours in the dark or under R light. As shown in Figure 6A, the expression of both GA biosynthetic genes was significantly reduced in dag2 mutant seeds irrespective of light conditions, whereas the catabolic gene AtGA2ox2 was expressed similarly in dag2 and wild-type seeds (Figure 6A).
Figure 6.
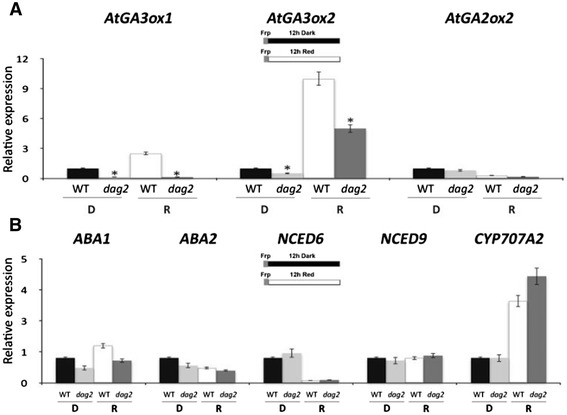
Mutation of the DAG2 gene affects GA biosynthesis. Relative expression level of AtGA3ox1, AtGA3ox2 and AtGA2ox2 (A), and of ABA1, ABA2, NCED6, NCED9 and CYP707A2 (B) in dag2 mutant seeds compared to wild-type seeds. Seeds were imbibed 12 hours in the dark (D), or under R light (R). Relative expression levels were normalized with that of the UBQ10 gene, and are presented by the ratio of the corresponding wild-type mRNA level in D, which was set to 1. Similar results were obtained from three independent experiments, and a typical result is presented with SD values. Significative differences were analyzed by t-test (*P ≤ 0,05).
As for ABA metabolism, we analysed the expression level of the biosynthetic genes ABA1, ABA2, NCED6 and NCED9, and of the catabolic gene CYP707A2, on dag2 and wild-type seeds imbibed 12 hours in the dark or under R light. The expression profile of the biosynthetic genes, as well as of the catabolic gene CYP707A2 did not show significant differences in dag2 and wild-type seeds (Figure 6B).
We have previously shown that the sensitivity of seeds to GA is affected by mutation of the DAG2 gene: a concentration of GA 10-fold higher than for wild-type seeds was needed for dag2 mutant seeds to attain 50% germination [8]. To verify whether GA affect DAG2 expression, we carried out an RT-qPCR analysis on wild-type seeds imbibed 24 hours in the presence of GA or of paclobutrazol, an inhibitor of GA biosynthesis. Since GA metabolism is controlled by the ABA level [18], we also checked DAG2 expression on wild-type seeds imbibed 24 hours in the presence of ABA. The results of this analysis did not show any significant difference in DAG2 transcript levels in all conditions tested, clearly showing that the DAG2 gene is not regulated by GA nor by ABA irrespective of light conditions (Figure 7).
Figure 7.
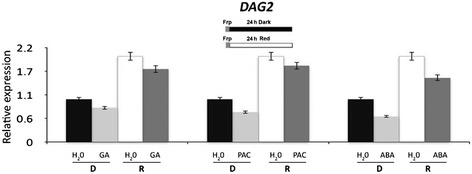
DAG2 expression is not altered by ABA or GA. Relative expression level of DAG2 in wild-type seeds imbibed 24 hours in the presence of GA, of Paclobutrazol, an inhibitor of GA biosynthesis, or of ABA in the dark (D), or under R light (R), compared to seeds imbibed in water as a control (H2O). Relative expression levels were normalized with that of the UBQ10 gene, and are presented by the ratio of the corresponding mRNA level in seeds imbibed in water, which was set to 1. Similar results were obtained from three independent experiments, and a typical result is presented with SD values.
Inactivation of the DAG2 gene does not affect embryo development
We have recently shown that DAG1 is expressed during embryo development, and that lack of DAG1 affects this process [24]. Thus, we set to assess whether also DAG2 is required for embryo development. We first analyzed the expression of DAG2 during embryo development by histochemical GUS analysis of seeds of the DAG2:GUS transgenic line. GUS activity was observed in embryos at the heart, torpedo, and bent-cotyledon stages. Interestingly, GUS staining was extended to all cells at the heart stage, whereas from the torpedo stage on it was restricted to the procambium (Figure 8A). These results were confirmed and extended to later seed development stages by a RT-qPCR analysis on wild-type embryos at 13, 16 and 19 Days After Pollination (DAP), compared to mature seeds, to verify whether the DAG2 gene was expressed also during seed maturation.
Figure 8.
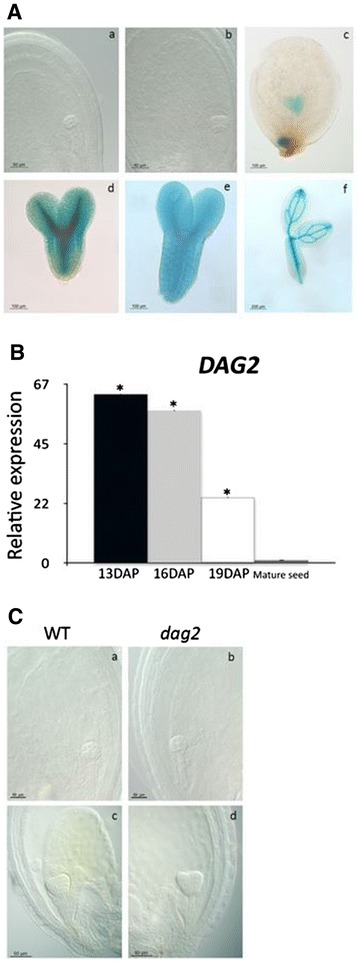
DAG2 inactivation does not affect embryo development. Histochemical staining of DAG2:GUS during embryogenesis, in early globular, globular, heart, late heart, torpedo and mature embryo (A).Relative expression level of DAG2 in wild-type seeds at 13, 16 and 19 Days After Pollination (DAP), and in mature seeds (28 DAR). Relative expression levels were normalized with that of the UBQ10 gene, and are presented by the ratio of the corresponding mRNA level in mature seeds, which was set to 1. Similar results were obtained from three independent experiments, and a typical result is presented with SD values. Significative differences were analyzed by t-test (*P ≤ 0,05) (B). Phenotypes of wild-type (a, c) and dag2 mutant (b, d) embryos, at globular (a, b) and heart stage (c, d) (C).
Expression of DAG2, at 13 and 16 DAP was extremely high (63- and 57-fold the basal level, respectively), and gradually decreased at 19 DAP (24-fold) compared to mature seeds (Figure 8B).
Despite the high expression level of the DAG2 gene during embryo and seed development, microscopic analysis of dag2 mutant embryos did not reveal any noticeable phenotypical alteration (Figure 8C).
Discussion
We had previously shown that the dag2 mutant has seeds which require higher light fluences and higher GA levels than wild-type ones to germinate [8], suggesting a positive role of the Dof transcription factor DAG2 in the regulation of seed germination.
Here, we have expanded our analysis of the function of DAG2 and we confirm the positive role of DAG2 in seed germination and provide molecular and genetic evidences that assign this protein to the phyB/PIL5 pathway.
To date the molecular pathway controlling seed germination has been partially elucidated. In this model PIL5 acts as the master repressor, which inhibits seed germination in the dark partly by activating the expression of the genes encoding the DELLA proteins RGA and GAI - which repress germination acting as negative GA signaling components - and of the transcription factors ABA INSENSITIVE 3 and 5 (ABI3 and ABI5) - which function as positive ABA signaling molecules [26,28]. Other transcription factors acting as repressors have been added in this pathway: the bHLH transcription factor SPATULA (SPT) [29], the C3H-type zinc finger protein SOMNUS (SOM) [30], and the Dof transcription factor DAG1, which we have shown to directly regulate the GA biosynthetic gene AtGA3ox1, with the cooperation of GAI [22,23].
DAG1 and DAG2 share 77% overall aminoacidic identity, with 100% identity in the Dof domain and, based on the opposite germination properties of dag1 and dag2 mutant seeds, we had assumed that the function of these two Dof proteins was opposite. This was also supported by DAG1 overexpression, which caused phenotypes similar to mutation of DAG2 [8]. Consistently, germination of dag2 mutant seeds in phyB-dependent conditions (i.e. under R light) was significantly reduced compared to wild-type seeds, whereas dag1 seeds showed a higher germination frequency [21,22]. In addition, DAG2 expression is induced by exposure to R light, as opposed to DAG1, whose transcript level is lower in R light than in the dark [22].
Analysis of the germination properties of dag2dag1 double mutant seeds revealed that the dag1 mutation is epistatic over the dag2 one [8]. Consistent with these previous reports, here we showed that DAG2 expression is negatively controlled by DAG1, and that DAG1 directly binds the DAG2 promoter as demonstrated by ChIP assay.
This provides molecular support to the genetic evidence of the epistatic relationship between these two Dof proteins shown in previous work [8]. We show here that DAG2 is also repressed by PIL5, since the DAG2 mRNA level is significantly increased in pil5 mutant seeds in the dark but not under R light, where PIL5 is degraded following interaction with phyB in its activated form (Pfr).
Since DAG1 directly interacts with GAI, and cooperates with this DELLA protein in repressing the GA biosynthetic gene AtGA3ox1 [23], and in the light of the opposite role of DAG2 in this molecular pathway, one could hypotesize a relationship of DAG2 with RGA or GAI. Interestingly, our results revealed that expression of RGA, but not of GAI, is significantly affected in dag2 mutant seeds exposed to R light, suggesting that DAG2 may negatively regulate this DELLA gene, whereas expression of DAG2 is not likely to be controlled by both RGA and GAI, as DAG2 transcript levels are similar in rga28 and in gai-t6 mutant seeds compared to wild-type seeds, in both light conditions.
Our expression analysis under phyA-dependent conditions further supports the notion that DAG2 acts in the phytochrome-mediated seed germination. In fact, of the marker genes of the phyA-dependent germination pathway we analyzed, PIL2, ATHB2, and ABI4 remained unaffected in the dag2 mutant, while GASA4 and GASA6 were severely downregulated - consistent with the role of DAG2 in the positive control of GA biosynthesis - opening the interesting possibility that DAG2 participates also in phyA signalling.
It should be noted that GASA4 has been previously characterised as a regulatory protein, induced by GA and involved in seed development and germination, independently of light conditions [31,32].
Phytochromes promote seed germination partly through GA. Red light induces the expression of the two GA anabolic genes GA3-oxidase genes GA3ox1 and GA3ox2, whereas it represses the GA catabolic gene GA2ox2 [33,34]. Consistent with a positive role of DAG2 in seed germination, mutation of the DAG2 gene severely affects expression of both AtGA3ox1 and AtGA3ox2, although it does not alter the expression level of AtGA2ox2. Unlike DAG1, DAG2 does not seem to play its function through regulation of ABA metabolism, as the expression profile of the ABA metabolic genes tested is quite similar in dag2 and wild-type seeds [22].
In recent years, the molecular mechanisms underlying light-mediated seed germination has been partly elucidated; however, it still remains an open question which are the positive regulators of this process. In fact, so far only LONG HYPOCOTYL IN FAR RED1 (HFR1) has been identified as a positive regulator of seed germination: HFR1 acts upstream of PIL5 and interacts directly with PIL5 thus sequestering it to prevent it from binding to its target genes [35]. Interestingly, germination of hfr1 mutant seeds under phyB-dependent germination conditions is very similar to that of dag2 mutant seeds, strengthening the notion that DAG2 is also a positive regulator in the phyB-dependent seed germination pathway.
As previously reported, DAG2 and DAG1 show a very similar expression profile, restricted to the vascular tissue [8], and we showed that during embryo development, DAG1 is expressed from late globular stage [22,24]. We also showed that dag1 mutant embryos displayed abnormal cell divisions at globular stage, altering the radial symmetry of the embryo axis [24].
Here we showed that, in contrast with DAG1, although also DAG2 is expressed during embryo development, its absence does not produce obvious embryo phenotypes.
Conclusions
Our genetic and molecular data indicate that DAG2 is a new positive factor of the phyB/PIL5-mediated seed germination pathway. DAG2 is located downstream PIL5 and DAG1, which directly represses DAG2 expression. Consistent with previous genetic data, DAG2 plays an opposite role to DAG1, although our results indicate that DAG2 acts on GA, but not on ABA, metabolism.
Methods
Plant material and growth conditions
dag2 is the allele described in Gualberti et al. [8] in Ws-4 ecotype.
All Arabidopsis thaliana lines used in this work were grown in a growth chamber at 24/21°C with 16/8-h day/night cycles and light intensity of 300 μmol/m−2 s−1 as previously described [7,22].
Seed germination assays
All seeds used for germination tests were harvested from mature plants grown at the same time, in the same conditions, and stored for the same time (28 Days After Ripening, DAR) under the same conditions. Germination assays were performed according to Gabriele et al. [22]. For phyB-dependent germination experiments, seeds were exposed to a pulse of FR light (40 μmol m−2 s−1), then a pulse of R light (90 μmol m−2 s−1) and subsequently kept in the dark for 5 days. Germination assays were repeated with three seed batches, and one representative experiment is shown. Bars represent the mean ± SEM of three biological repeats (25 seeds per biological repeat). P values were obtained from a Student’s unpaired two-tail t test comparing the mutant with its control (* = p ≤ 0,05).
Expression analysis
For expression analysis, seeds were imbibed for 12 or 24 hours, on five layers of filter paper, soaked with 5 ml water, exposed to a pulse of FR (40 μmol m−2 s−1), then incubated in the dark or under R light (90 μmol m−2 s−1), in the presence of PAC (100 μM) to prevent de-novo GA biosynthesis in response to light [26]. For phyA-dependent conditions, seeds were treated according to Oh et al., 2006 [25]. RNA extraction and RT-qPCR were performed according to Gabriele et al. [22]. Quantification of gene expression was expressed in comparison to the reference gene (See legends of figures), and relative expression ratio was calculated based on the qRT-PCR efficiency (E) for each gene and the crossing point (CP) deviation of our target genes versus a control [36]. The expression analyses were repeated in comparison with a second reference gene (Additional file 3: Figure S3).
Three independent biological replicates were performed, and one representative experiment is reported. Significative differences were analyzed by t-test (*P ≤ 0,05). The primers used for the assays are listed in Additional file 4: Table S1.
ChIP analysis
The dag1DAG1-HA line is the one previously described in Gabriele et al. [22]. ChIP was performed as previously described [22], with 12 hours imbibed seeds. Antibodies against HA tag (Santa Cruz, CA, USA) were used for immunoprecipitation. Equal amounts of starting material and ChIP products were used for qPCR reaction. The primers used are listed in Table S1. Three independent biological replicates were performed. Significative differences were analyzed by t-test (*P ≤ 0,05).
Microscopy and GUS analysis
Analysis of dag2 and wild-type embryos was performed under an Axioskop 2 plus microscope (Zeiss).
The DAG2:GUS line is the one described in Gualberti et al. [8]. Histochemical staining and microscopic analysis were carried out according to Blazquez et al. [37]. Stained embryos (after washing in 70% ethanol) were analysed and photographed under an Axioskop 2 plus microscope (Zeiss).
Availability of supporting data
All the supporting data of this article are included as additional files (Additional files 1, 2 and 3: Figures S1-S3; Additional file 4: Table S1).
Acknowledgments
This work was partially supported by research grants from Ministero dell’Istruzione, Università e Ricerca, Progetti di Ricerca di Interesse Nazionale, and from Sapienza Università di Roma to PC, and from Istituto Pasteur Fondazione Cenci Bolognetti to PV.
Abbreviations
- DOF
DNA Binding With One Finger
- DAG1
Dof AFFECTING GERMINATION 1
- DAG2
Dof AFFECTING GERMINATION 2
- phyB
Phytochrome B
- PIL5
PHYTOCHROME INTERACTING FACTOR3-LIKE 5
- GAI
GA INSENSITIVE
- RGA
REPRESSOR OF ga1-3
- ABI3
ABA INSENSITIVE 3
- ABI5
ABA INSENSITIVE 5
- SPT
SPATULA
- SOM
SOMNUS
- HFR1
LONG HYPOCOTYL IN FAR RED1
- ATHB2
ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2
- GASA4
6, GA-STIMULATED ARABIDOPSIS 4, 6
- PIL2
PHYTOCHROME INTERACTING FACTOR3-LIKE 2
- ABI4
ABA Insensitive 4,ABA, Abscissic Acid
- GA
Gibberellins
- PAC
Paclobutrazol
- ChIP
Chromatin Immuno Precipitation
- RT-qPCR
quantitative reverse transcriptase-polymerase chain reaction
- W light
White light
- R light
Red light
- D
Dark
- FR Light
Far Red Light
- DAP
Days After Pollination
- DAR
Days After Ripening
- GUS
β-glucuronidase
- HA
Heme Agglutinin
Additional files
ChIP analysis of wild-type (WS) seeds immunoprecipitated with anti-HA antibody or without antibody.
Relative expression levels of ATHB2, PIL2, GASA4, GASA6 and ABI4 in wild-type (WT) and dag2 mutant seeds. Relative expression levels were normalized with that of the UBQ10 gene.
Expression analysis with a second reference gene. Relative expression levels of DAG2 in wild-type seeds in the dark (D), Red (R) (A), or Far Red (FR) (B) light, in dry seeds (0h), or imbibed 12 (12h), 24 hours (24h) in the dark, under White (W) or Red light (C), normalized with the PP2A gene. Relative expression levels of DAG2 in dag1 (D), pil5 (F), rga28 (G), gai-t6 (H) seeds compared to WT. Relative expression levels of DAG1 (E), RGA (I), GAI (L), in dag2 seeds compared to WT. The expression levels were normalized with PP2A (D, F, E, I, L), or with eIF1α (At5g60390) (G, H). Relative expression levels of GA (M) or ABA (N) metabolic genes in dag2 seeds compared to WT, normalized with PP2A gene. Relative expression levels of DAG2 in WT seeds imbibed in the presence of ABA, or GA, or PAC (O), in WT embryos at 13, 16, 19 DAP (P), normalized with PP2A gene. Relative expression levels of ATHB2, PIL2, GASA4, GASA6 and ABI4 in dag2 seeds compared to WT (Q), normalized with PP2A.
List of the primers used for expression analyses and for the ChIP assays.
Footnotes
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
PV designed the research. SS, AB and GS contributed to the experimental design and to analysis of the results. SS, AB, RL, VR, DC, EM performed the experiments. All authors analyzed and discussed the data. SS prepared the figures and PV wrote the article. PC supervised the research and the writing of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Silvia Santopolo, Email: silvia.santopolo@hotmail.it.
Alessandra Boccaccini, Email: boccaccini_alessandra@hotmail.com.
Riccardo Lorrai, Email: lorrai_riccardo@hotmail.it.
Veronica Ruta, Email: v.nika@hotmail.it.
Davide Capauto, Email: davidecs@hotmail.it.
Emanuele Minutello, Email: kitelooper@gmail.com.
Giovanna Serino, Email: giovanna.serino@uniroma1.it.
Paolo Costantino, Email: paolo.costantino@uniroma1.it.
Paola Vittorioso, Email: paola.vittorioso@uniroma1.it.
References
- 1.Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol. 2003;3:17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreno-Risueno MA, Martinez M, Vicente-Carbajosa J, Carbonero P. The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol Genet Genomics. 2007;277(4):379–90. doi: 10.1007/s00438-006-0186-9. [DOI] [PubMed] [Google Scholar]
- 3.Hernando-Amado S, Gonzalez-Calle V, Carbonero P, Barrero-Sicilia C. The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol. 2012;12:202. doi: 10.1186/1471-2229-12-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw LM, McIntyre CL, Gresshoff PM, Xue GP. Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct Integr Genomics. 2009;9(4):485–98. doi: 10.1007/s10142-009-0130-2. [DOI] [PubMed] [Google Scholar]
- 5.Kushwaha H, Gupta S, Singh VK, Rastogi S, Yadav D. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol Biol Rep. 2011;38(8):5037–53. doi: 10.1007/s11033-010-0650-9. [DOI] [PubMed] [Google Scholar]
- 6.Yanagisawa S, Sheen J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell. 1998;10(1):75–89. doi: 10.1105/tpc.10.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 2000;14(1):28–33. [PMC free article] [PubMed] [Google Scholar]
- 8.Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C, et al. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell. 2002;14(6):1253–63. doi: 10.1105/tpc.010491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isabel-LaMoneda I, Diaz I, Martinez M, Mena M, Carbonero P. SAD: a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. Plant J. 2003;33(2):329–40. doi: 10.1046/j.1365-313X.2003.01628.x. [DOI] [PubMed] [Google Scholar]
- 10.Park DH, Lim PO, Kim JS, Cho DS, Hong SH, Nam HG. The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 2003;34(2):161–71. doi: 10.1046/j.1365-313X.2003.01710.x. [DOI] [PubMed] [Google Scholar]
- 11.Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;9(5732):3–7. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- 12.Ward JM, Cufr CA, Denzel MA, Neff MM. The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell. 2005;17(2):475–85. doi: 10.1105/tpc.104.027722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwamoto M, Higo K, Takano M. Circadian clock- and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant Cell Environ. 2009;32(5):592–603. doi: 10.1111/j.1365-3040.2009.01954.x. [DOI] [PubMed] [Google Scholar]
- 14.Rueda-Romero P, Barrero-Sicilia C, Gomez-Cadenas A, Carbonero P, Onate-Sanchez L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J Exp Bot. 2012;63(5):1937–49. doi: 10.1093/jxb/err388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguero M, Atif RM, Ochatt S, Thompson RD. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant science : an international journal of experimental plant biology. 2013;209:32–45. doi: 10.1016/j.plantsci.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Bewley JD. Seed Germination and Dormancy. Plant Cell. 1997;9(7):1055–66. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinomura T, Nagatani A, Chory J, Furuya M. The Induction of Seed Germination in Arabidopsis thaliana Is Regulated Principally by Phytochrome B and Secondarily by Phytochrome A. Plant Physiol. 1994;104(2):363–71. doi: 10.1104/pp.104.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, et al. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006;48(3):354–66. doi: 10.1111/j.1365-313X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi S, Sun T, Kawaide H, Kamiya Y. The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 1998;116(4):1271–8. doi: 10.1104/pp.116.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh E, Kim J, Park E, Kim JI, Kang C, Choi G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004;16(11):3045–58. doi: 10.1105/tpc.104.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papi M, Sabatini S, Altamura MM, Hennig L, Schafer E, Costantino P, et al. Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol. 2002;128(2):411–7. doi: 10.1104/pp.010488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 2010;61(2):312–23. doi: 10.1111/j.1365-313X.2009.04055.x. [DOI] [PubMed] [Google Scholar]
- 23.Boccaccini A, Santopolo S, Capauto D, Lorrai R, Minutello E, Serino G, Costantino P, Vittorioso P. The DOF Protein DAG1 and the DELLA Protein GAI Cooperate in Negatively Regulating the AtGA3ox1 Gene. Mol Plant. 2014;7(9):1486–9. doi: 10.1093/mp/ssu046. [DOI] [PubMed] [Google Scholar]
- 24.Boccaccini A, Santopolo S, Capauto D, Lorrai R, Minutello E, Belcram K, et al. Independent and interactive effects of DOF affecting germination 1 (DAG1) and the Della proteins GA insensitive (GAI) and Repressor of. BMC Plant Biol. 2014;14(1):200. doi: 10.1186/s12870-014-0200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006;47(1):124–39. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- 26.Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, et al. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;19(4):1192–208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ibarra SE, Auge G, Sanchez RA, Botto JF. Transcriptional programs related to phytochrome A function in Arabidopsis seed germination. Mol Plant. 2013;6(4):1261–73. doi: 10.1093/mp/sst001. [DOI] [PubMed] [Google Scholar]
- 28.Park J, Lee N, Kim W, Lim S, Choi G. ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell. 2011;23(4):1404–15. doi: 10.1105/tpc.110.080721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol. 2005;15(22):1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, et al. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell. 2008;20(5):1260–77. doi: 10.1105/tpc.108.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M. Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol. 1998;36(6):871–83. doi: 10.1023/A:1005938624418. [DOI] [PubMed] [Google Scholar]
- 32.Roxrud I, Lid SE, Fletcher JC, Schmidt ED, Opsahl-Sorteberg HG. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007;48(3):471–83. doi: 10.1093/pcp/pcm016. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T. Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell. 1998;10(12):2115–26. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamauchi Y, Takeda-Kamiya N, Hanada A, Ogawa M, Kuwahara A, Seo M, et al. Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 2007;48(3):555–61. doi: 10.1093/pcp/pcm023. [DOI] [PubMed] [Google Scholar]
- 35.Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell. 2013;25(10):3770–84. doi: 10.1105/tpc.113.117424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blazquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124(19):3835–44. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]


