Abstract
This contribution describes the trapping of the hydroperoxyl radical at a pH of 4 during turnover of wild-type oxalate decarboxylase and its T165V mutant using the spin trap BMPO. Radicals were detected and identified by a combination of EPR and mass spectrometry. Superoxide, or its conjugate acid, the hydroperoxyl radical, is expected as an intermediate in the decarboxylation and oxidation reactions of the oxalate monoanion both of which are promoted by oxalate decarboxylase. Another intermediate, the carbon dioxide radical anion was also observed. The quantitative yields of superoxide trapping is similar in the wild type and the mutant while it is significantly different for the trapping of the carbon dioxide radical anion. This suggests that the two radicals are released from different sites of the protein.
Keywords: Oxalate decarboxylase, spin-trapping, BMPO, superoxide, carbon dioxide radical
Introduction
Oxalic acid is one of the most common naturally occurring dicarboxylic acids and is toxic to most mammals [1]. Approximately 60% of urinary tract stones contain calcium oxalate [2]. Excessive consumption of dietary plants rich in oxalic acid (e.g., rhubarb, chard, beet, spinach, purslane, etc.) may lead to hyperoxaluria and hypocalcaemia in humans and animals [3–5]. Elevated concentrations of oxalic acid in humans can lead to a variety of disorders including pyridoxine deficiency, cardiomyopathy, cardiac conductance, hyperoxaluria, calcium oxalate stones, renal failure, and at higher concentrations, mortality [6–9].
Oxalate decarboxylase (OxDC) is one of only three categories of enzymes known in plants, fungi, and bacteria that degrade oxalic acid [10]. OxDC is utilized by soil bacteria and fungi and plays an important role in the biogeochemical carbon cycle facilitating the oxalate-carbonate pathway [11]. It accumulates in the cell wall of Bacillus subtilis and can be secreted by many basidiomycetous and ascomycetous fungi [11, 12]. OxDC has garned attention due to its potentially numerous applications ranging from remediation of oxalate scaling in the wood and paper industry [13–16], bioengineering of crop plants for fungal resistance and lower oxalate content [17, 18], diagnostics and sensing of oxalate [19–21], bioengineering of probiotic gut bacteria to release OxDC in the intestine [22], and as a dietary supplement for the degradation of excess oxalate in the stomach [23, 24]. Despite these efforts, significant questions remain about the details of the enzymatic mechanism of OxDC, and in particular about how modifications of the enzyme can direct its chemistry away from decarboxylase to oxidase activities [25].
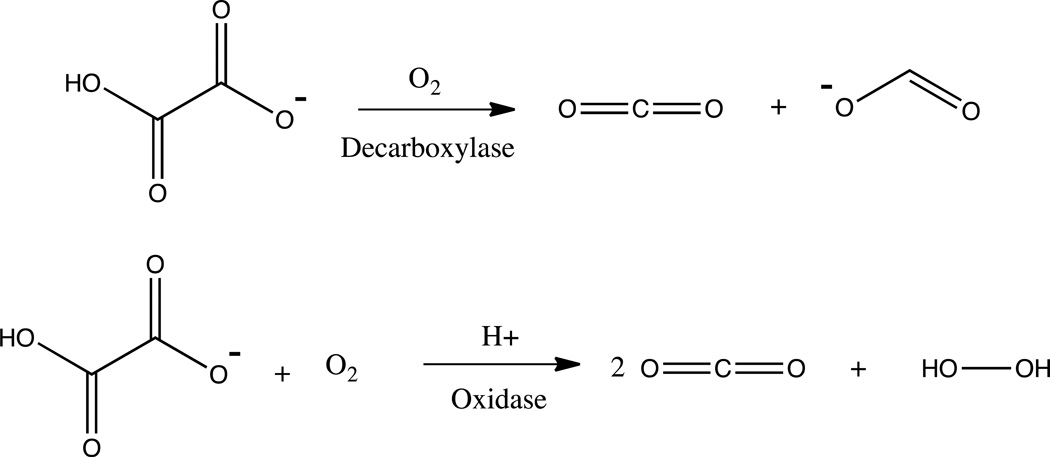
OxDC isolated from Bacillus subtilis is a bicupin enzyme that contains a Mn ion in each of its two cupin folds [26–28]. The cupin superfamily of enzymes are characterized by a series of conserved residues that form β-barrel folds, that typically support the binding of metal ions [29–31]. OxDC shows an impressive enzymatic rate enhancement of 2.5×1013 at its optimum pH of 4.2 [32]. A very interesting but poorly understood aspect of this enzyme is the apparent bifurcation of its chemistry. Wild type (WT) OxDC primarily acts as a decarboxylase producing carbon dioxide and formate (99.8% of all turnovers) while it acts as an oxidase in about 0.2% of all turnover events producing hydrogen peroxide and carbon dioxide (see Scheme 1) [10, 33].
Scheme 1.
(Top) Decarboxylase reaction and (bottom) oxalate oxidase reaction. (Double Column Image)
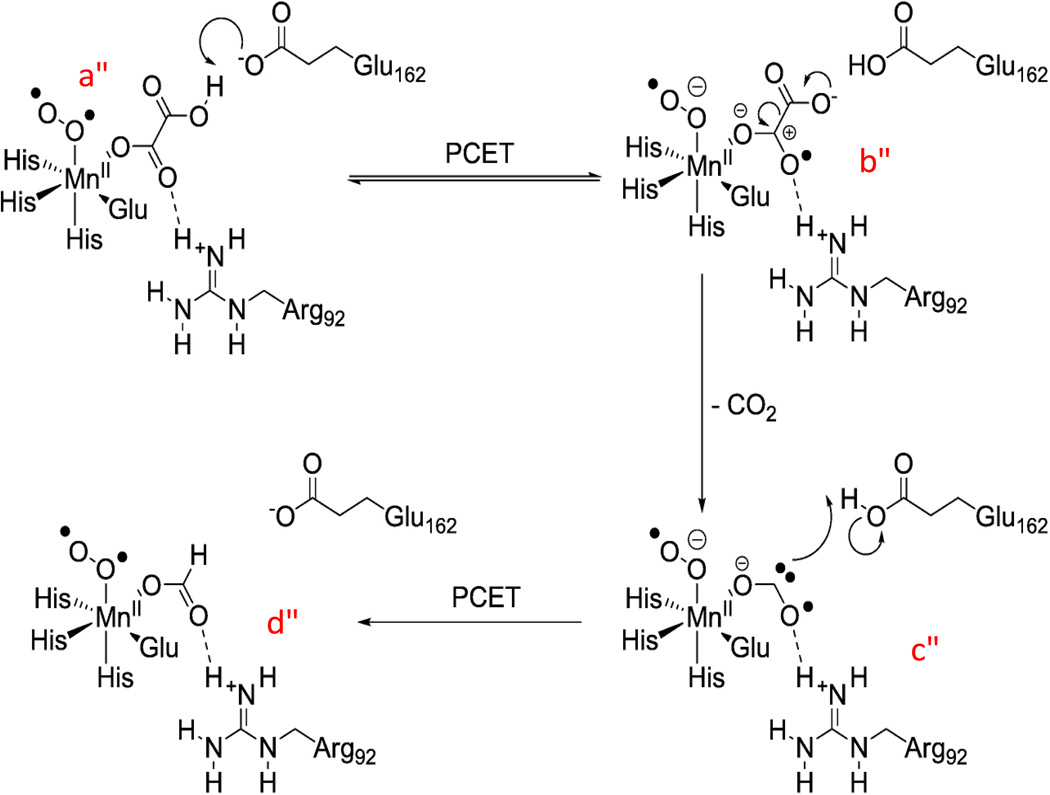
Although the primary reaction catalyzed by OxDC is a redox-neutral disproportionation reaction and does not consume dioxygen, it requires dioxygen for turnover as a co-catalyst [10, 33–36]. Current mechanistic proposals suggest that dioxygen is bound to one of two Mn ions and acts as a transient electron sink to destabilize the carbon-carbon bond in oxalate resulting in a bound superoxide radical. After decarboxylation has taken place superoxide acts as an electron source to reduce the resulting carbon dioxide anion radical (see Scheme 2) [37]. This mechanism requires oxygen to cycle through its 1-electron reduced state as superoxide which would likely be protonated in the pH range in which the enzyme is active [38]. Alternative proposals suggest one of the Mn ions undergoing a redox cycle between its +2 and +3 oxidation states allowing the associated dioxygen to remain a hydroperoxyl radical throughout the reaction [25, 27, 39].
Scheme 2.
Proposed mechanism for the decarboxylation mechanism in OxDC. The formation of superoxide is suggested to occur after dioxygen binds to Mn(II) (a), or alternatively, after the initial proton-coupled electron transfer process (PCET) during enzyme turnover. (Permission granted for reproduction by Elsevier license: 3236061008715) (Double Column Image)
Since 0.2% of all turnovers result in oxalate oxidation with production of hydrogen peroxide (see Scheme 1, bottom) [33], it is instructive to consider the structurally related mono- and bi-cupin oxalate oxidase (OxOx) enzymes [40], which may be evolutionarily related to OxDC [29, 30]. Both OxDC and OxOx enzymes require manganese, dioxygen, and oxalate, and both belong to the cupin superfamily [10]. OxOx also has the same conserved residues coordinating manganese within its own active site, consisting of one glutamate and three histidines [26–28, 40]. Sequence alignment studies of both Bacillus subtilis OxDC and Ceriporiopsis subvermispora OxOx identified a major structural difference between these two enzymes in the existence of an N-terminal active site flexible lid in OxDC, consisting of a serine-glutamate-arginine-serine-threonine sequence at positions 161–165 [41]. Site-directed mutagenesis of this lid region can transform OxDC into an oxidase [41]. For decarboxylation to occur, the presence of a protonating group, tentatively assigned to be glutamate-162 in OxDC, appears to be necessary [25, 41]. The lack of such a group in OxOx may lead to a peroxycarbonate intermediate, which decays under acidic conditions to form hydrogen peroxide and carbon dioxide [27, 28, 41, 42]. However, direct experimental evidence for a peroxycarbonate intermediate is still missing, and other mechanistic proposals exist in the literature [43].
Results from spin-trapping experiments on the T165V OxDC mutant suggested that the destabilization of the closed conformation of this lid region leads to increased loss of the intermediate carbon dioxide radical anion into solution where it reacts diffusion-controlled with dioxygen to produce superoxide and eventually hydrogen peroxide (i.e., the same products expected for oxidase activity) [37]. Even in WT OxDC the CO2•− radical intermediate can be trapped by appropriate spin traps suggesting that the 0.2% oxidase activity might be simply due to loss of this intermediate from the active site [37, 41]. Spin trapping of OxOx under turnover conditions also yields a CO2•− radical adduct with both PBN and DMPO spin traps [44, 45]. It is interesting to note that in the OxOx mutant A241E designed to mimic OxDC the spin trapping yield dropped to negligible levels suggesting that the introduction of glutamate somehow allowed the protein to keep its intermediate radical sequestered even though it did not show any decarboxylase activity just like the wild type OxOx enzyme [45].
In any case, when OxDC loses its intermediate CO2•− radical anion the initial oxidation step has already taken place and the protein is now fixed with an electron on either one of its Mn ions or bound superoxide. This should lead to enzyme inhibition unless the enzyme can rid itself of the reducing equivalent, for example by losing a superoxide anion, or its protonated counterpart (i.e., the hydrogen peroxyl radical). One would therefore expect to find evidence for superoxide in the solution under turnover conditions. However, Burrell et al. were unable to observe trapped superoxide in either WT OxDC or their SENS161–164DASN active-site lid mutant using POBN and DMPO as spin traps [41]. Experimental detection of O2•− at low pH is made difficult by the acid-catalyzed disproportionation of superoxide which occurs at very fast rates [46]. Yet, the trapping efficiency of the hydroperoxyl radical with cyclic nitrones (such as DMPO or BMPO) increases with decreasing pH, which mitigates the problem [47, 48].
A common spin trap used for identification of superoxide is 5,5-dimethyl-pyrroline N-oxide (DMPO) [47]. However, DMPO tends to react rather slowly with superoxide making it necessary to utilize high spin trap concentrations to outcompete the disproportionation reaction [47]. Moreover, it decays quickly with a half-life of less than a minute at pH7 [49], and its superoxide adduct can decay into a hydroxyl adduct necessitating cumbersome control experiments [50–52]. To avoid these problems the spin trap 5-tert-butoxycarbonyl 5-methyl-1-pyrroline N-oxide (BMPO) was used in the present study. It features an extended half-life of the persistent superoxide adduct of t1/2 = 23 min, facilitated by its bulky side groups protecting the nitroxide radical [53, 54]. Since the reaction is carried out at pH4.0 for OxDC, there will be a mixture of O2•− and HO2• with the equilibrium favoring the hydroperoxyl radical [55, 56].
Previous studies have utilized spin trapping in combination with mass spectrometry to confirm the identity of spin trapped radical adducts with both DMPO and BMPO spin traps [57–62]. Here, we report for the first time on the detection and identification of a hydroperoxyl-adduct of BMPO when OxDC reacts with oxalate under turnover conditions.
Materials and Methods
Chemicals
The following chemicals were purchased from Fisher Scientific (ACS Grade): potassium acetate, glycerol, citric acid, ethylenediaminetetraacetic acid (EDTA), and potassium phosphate. Potassium oxalate monohydrate and diethethylenetriamine-pentaacetic acid (DTPA) were purchased from Sigma-Aldrich. 5-tert-butoxycarbonyl 5-methyl-1-pyrroline N-oxide (BMPO) was obtained from Applied Bioanalytical Labs (Bradenton, FL). All solutions were prepared in HPLC grade water obtained from Fisher Scientific. Stock solutions of 1 M acetate or citrate buffer were used to buffer the solutions at pH4.0 at a final buffer concentration of 50 mM. Potassium oxalate stock solutions were prepared at 0.5 M and the pH was adjusted to match that of the pH of the buffering solution. A 2 M BMPO stock solution was also prepared, along with a 4 mM stock of DTPA.
Enzyme Preparation
Expression and purification of recombinant His6-tagged Bacillus subtilis wild-type and T165V mutant OxDC was carried out following previously published procedures [27, 28, 33, 60, 63, 64]. Final preparation of the enzyme required a series of dialyses to transfer the protein into storage buffer (50 mM Tris-Cl, 0.5 M NaCl pH8.5), as well as to remove excess imidazole. Residual imidazole in the enzyme preparation leads to aggregation in subsequent concentration steps. To remove dissolved metals from the preparation, Chelex 100 resin (Bio-Rad, Hercules CA) was added to the enzyme after the serial dialysis steps. The solution was shaken for approximately one hour following removal of the resin. The enzyme solution was then concentrated using Amicon Centriprep YM-30 centrifugal filter units (EMD Millipore, Billerica, MA). Concentrated enzyme samples (approximately 40 mg/mL) were stored as 200 µL aliquots in Eppendorf tubes at −80°C until used for experiments. Enzyme activity was determined using an endpoint-stopped assay, coupling the formation of formate to the reduction of nicotinamide adenine dinucleotide (NAD+) [33, 65, 66]. An additional assay was performed to confirm activity prior to experimentation, the oxidation of o-phenyldiamine to 2,3-diaminophenazine which gives a pale yellow color indicative of active enzyme [33].
Electron Paramagnetic Resonance Spin Trapping
Experiments were performed on a Bruker ELEXSYS E580 CW/Pulsed or a Bruker ELEXSYS-II E500 CW X-band spectrometer equipped with a super high-Q cavity (ER 4123SHQE). Reactions were carried out in mini-Eppendorf tubes with the following final concentrations: 50 mM potassium acetate or citric acid buffer adjusted to a pH of 4.0, 50 mM potassium oxalate pH-adjusted to match the buffer pH, 20 µM DTPA, 100 mM BMPO. Enzyme was added last to the reaction mixture resulting in a final concentration of 5 mg/mL. The total reaction volume was 100 µL and also contained 35 µL distilled water generated by a Thermo Scientific Barnstead Nanopure Model 7134 and 20 µL glycerol. Glycerol was added to prevent precipitation of the enzyme upon lowering the pH. The solution was mixed using a vortexer (Fisher Scientific Deluxe Vortex Mixer) and then immediately transferred to a quartz capillary (1×3 mm ID×OD), which was sealed at the bottom with Cha-seal (Kimble Chase Life Sciences, Vineland NJ). All spectra were collected at room temperature, with the following instrumental parameters: 100 kHz modulation frequency, 1 G modulation amplitude, 20.48 ms time constant, 40.96 ms conversion time, 10 dB microwave attenuation, 60 dB receiver gain, and 1024 data points per spectrum. Simulation of the EPR spectra was carried out using the EasySpin toolbox for MATLAB™ [67]. Individual spectra for the carbon dioxide radical anion adduct and the hydroperoxyl adduct were fitted first to the spectra where they dominate (T165V and WT, respectively), followed by a weighted fit of both components which provided the concentrations after comparison to a calibration curve using 4-hydroxy TEMPO as the calibration standard. All experiments were performed in triplicate for error analysis.
Mass spectrometry
All experiments were performed on an Advion Expression compact mass spectrometer (Advion, Ithaca, NY). The instrument is equipped with an electrospray ionization source, single quadrupole mass analyzer, and an electron multiplier detector. Solutions were prepared similar to those for EPR except no glycerol was used, as its low vapor pressure dramatically reduces electrospray nebulization efficiency, and thus ionization efficiency. Before injection into the mass spectrometer, the enzyme was precipitated from solution by mixing with acetonitrile, followed by centrifugation to pellet out unwanted protein [68]. The supernatant was diluted in acetonitrile/water/formic acid (50:50:1) to a final volume of 500 µL. The solution was infused at a flow rate of 20 µL min−1, nebulized with nitrogen gas, and introduced into the mass spectrometer using positive mode electrospray ionization. Ion source conditions were optimized to promote high signal intensity of the protonated radical-bound spin trap. Due to the inherently low concentrations of analytes in this study, mass spectra were recorded at 1 Hz and were allowed to average for 5 minutes prior to analysis by Mass Express™ (Advion, Ithaca, NY). The samples used for the MS experiments were tested for the presence of the radical adduct by EPR before injection.
Europium tetracycline Hydrogen Peroxide Fluorescence Assay
The Europium tetracycline (EuTc) Hydrogen Peroxide assay kit was obtained from Active Motif (Carlsbad, CA). All experiments were carried out with a Horiba Jobin Yvon FluoroMax-3. Reaction mixtures were prepared, run and quenched in a 1.5 mL eppendorf and then transferred to a 5 mm square cuvette. The sample temperature during the reaction and measurement was 25° C. The assay was carried out as specified by Active Motif. The EuTc powder sample was used as provided with no further purification and dissolved in 100 mL of DI-H2O. A standard curve was prepared using 30% hydrogen peroxide adjusted to final concentrations of 200 µM, 160 µM, 100 µM, 40 µM, 20 µM, and 10 µM. Enzyme reaction mixtures were prepared in 100 µL total volume as follows: 25 µL 0.2 M acetate buffer pH 4.0, 10 µL 0.5 M potassium oxalate pH 4.0, 44 µL DI-H2O, and 1 µL enzyme. The reactions were allowed to run for 3 min and were then quenched with 900 µL 0.1 M HEPES buffer pH 7.0. The reaction product was mixed in a 1:1 volume ratio with the prepared EuTc solution and allowed to incubate for 10 minutes prior to the fluorescence measurement. Samples were excited at a λexcitation of 400 nm, and measured at λemission of 617 nm with a bandwidth of 10 nm.
Results and Discussion
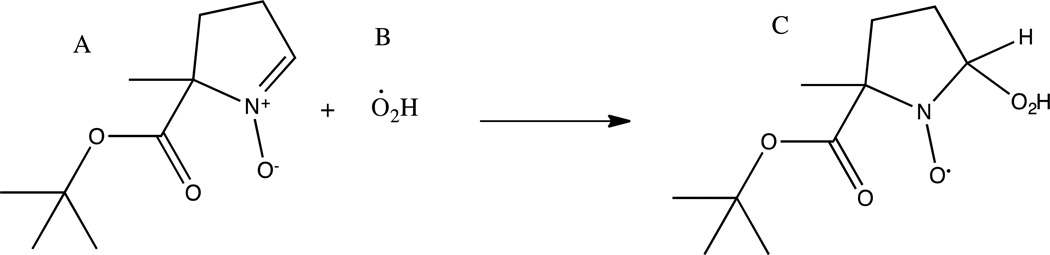
Spin trapping studies were carried out on wild type B. subtilis OxDC and its T165V mutant in an effort to identify free radicals produced during the enzymatic mechanism. Scheme 3 depicts the reaction that occurs when hydroperoxyl reacts with BMPO, forming a persistent nitroxide radical adduct. The reaction is carried out at an optimum pH of 4.0 for OxDC, below the pKa of superoxide (4.88), which shifts the equilibrium to favor the protonated form (species B in Scheme 3).
Scheme 3.
5-tert-butoxycarbonyl 5-methyl-1-pyrroline N-oxide (BMPO, A). Upon incubation with BMPO in the enzyme reaction mixture, hydroperoxyl radical (B) binds to the nitroxide ring yielding a persistent radical adduct (C). (Double Column Image)
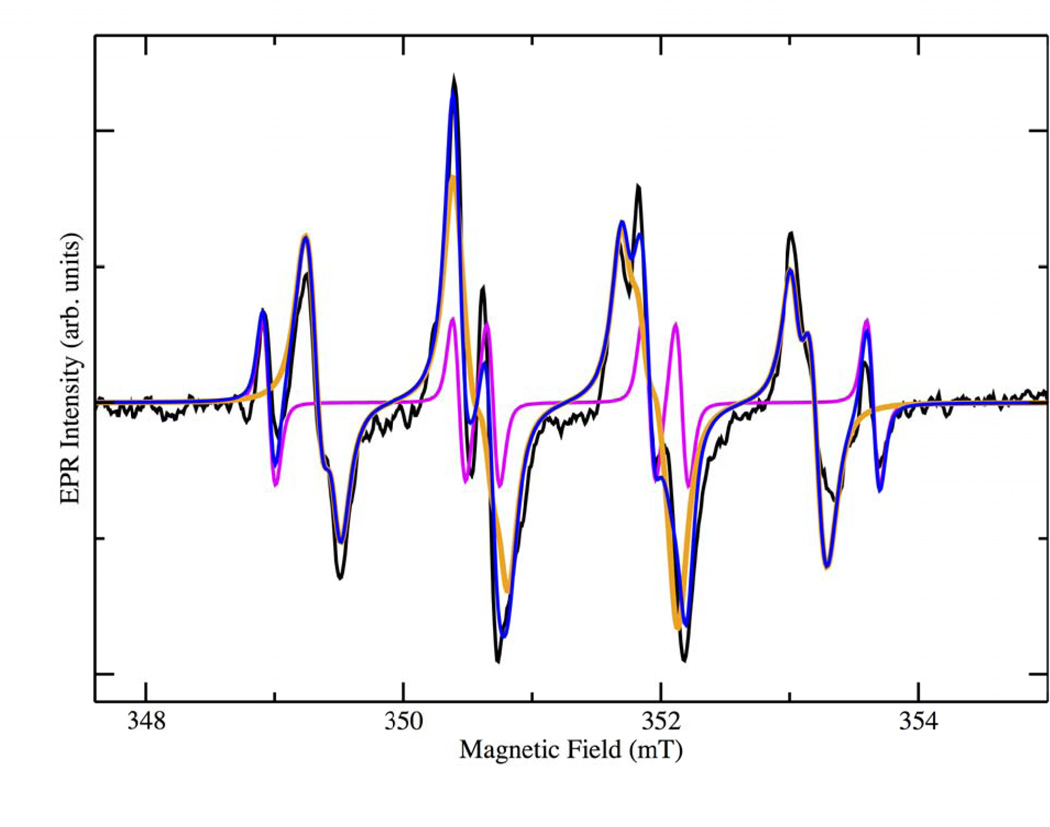
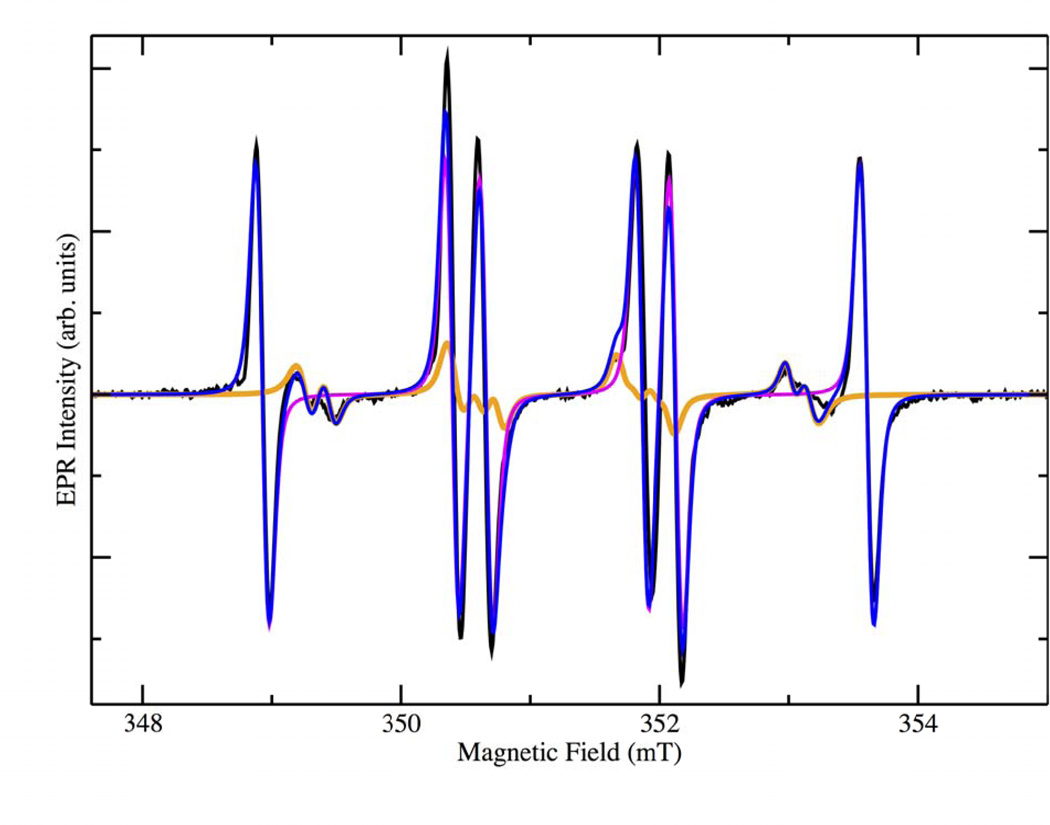
The resulting X-band EPR spectrum is shown in the black trace in Figure 1. It is apparent that contributions are present from more than one species. BMPO radical adducts usually show two slightly different spectra due to the presence of two diastereomers [54]. An additional contribution from a carbon based radical adduct was also observed and was necessary to produce a satisfactory simulation of the spectrum (magenta trace in Figure 1). Table 1 shows the magnetic parameters used in the simulation together with their corresponding spectral weights. The production of a CO2•− adduct was expected and had been previously reported as being derived from oxalate [37]. Unexpectedly, the major contributor to the spectrum was the hydroperoxyl adduct with a combined spectral weight of 90.1% for the two diastereomers, while the carbon dioxide anion radical contributed only 9.9% according to the simulated spectra. The hydroperoxyl radical had evaded detection in our earlier work because PBN was used, which is not a very efficient trap for superoxide/hydroperoxyl. Simulations were initially performed with literature values for the expected radical adducts, followed by an iterative fitting approach producing the converged values reported here. The hyperfine coupling constants for the hydroperoxyl adduct match well with those in the literature suggesting that it is produced during the enzymatic catalysis, as a transient intermediate [53, 54].
Figure 1.
X-band EPR spectra of the BMPO-radical adducts produced from the reactions between oxalate and WT-OxDC in 50 mM citrate buffer pH 4.0 (black). The orange and magenta spectra represent simulations of the superoxide and carbon dioxide anion radical adduct, respectively. Blue: sum of the simulations of the two spin adducts. (Double Column Image, Color Reproduction online/print)
Table 1.
Spectral parameters of BMPO-radical adducts for WT-OxDC, including hyperfine coupling, g-value, and individual spectral weights based on the simulated spectra. (Single Column Image)
| Spin-Trapped Adduct |
g-value | aH(mT) | aN(mT) | Spectral Weight (Relative Percentage) |
|---|---|---|---|---|
| [BMPO-OOH]• (Conformer I) | 2.005 | 1.207 | 1.346 | 61.33 |
| [BMPO-OOH]• Conformer II) | 2.005 | 0.956 | 1.318 | 28.78 |
| [BMPO-CO2−]• | 2.005 | 1.744 | 1.483 | 9.89 |
In order to confirm that the radical adduct formed was indeed derived from superoxide, mass spectrometric experiments were carried out on the reaction mixture. Time resolved spin trapping was conducted on wild type, and T165V OxDC mutants to identify the lifetime of the trapped species, as well as to confirm the time frame for trapping (see supplementary data, figures S1, S2, respectively).
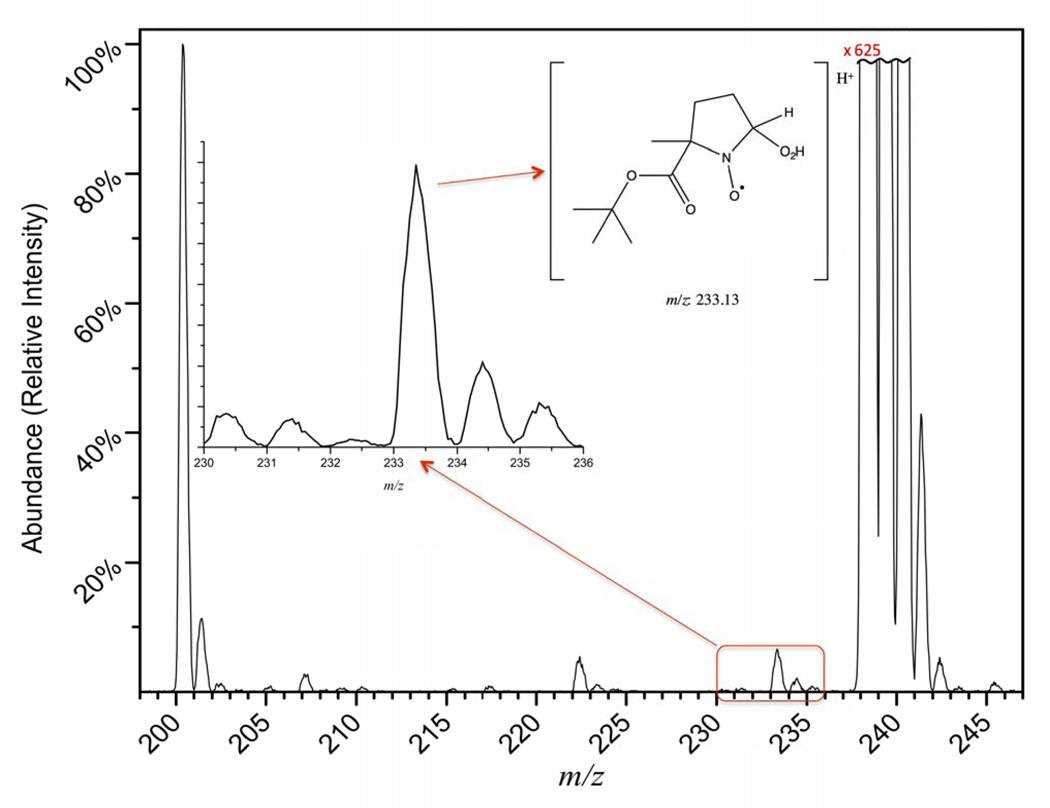
The MS analysis of the enzyme reaction mixture in the presence of BMPO provides confirmation that superoxide is produced during turnover. In Figure 2, peaks at m/z 200, 222, 233, 238, and 245 represent the protonated spin trap, sodiated spin trap, superoxide bound radical adduct, and potassiated spin trap, and a protonated form of the carbon dioxide anion radical adduct, respectively. From the zoomed portion of the figure, indicated by the red box, it is evident that there is a peak at the expected mass-to-charge ratio for the BMPO-hydroperoxyl radical adduct. It is also clear from the mass spectrum that BMPO favors metallation, given that both sodium and potassium bound spin traps are present. The peak at m/z 245 also provides evidence for the second trapped radical. In control experiments, when enzyme is not added to the reaction mixture, the masses representative of the spin trapped adducts are no longer observed (Figure S4). The metal bound forms of the spin trap (m/z 222 and m/z 238) are also drastically reduced due to the lack of salts in the sample that come with the enzyme preparation. The mass spectral analysis in combination with the EPR experiments provides clear evidence that both the CO2•− radical anion and superoxide are produced by the protein during the enzymatic catalysis of oxalate. With our experimental scheme, it is not possible to distinguish whether the observed superoxide was generated during the proton-coupled electron transfer process in the protein or by diffusion-controlled reaction of CO2•− with dissolved dioxygen in the solution. Either way, superoxide is expected as an intermediate and a byproduct of the small oxidase activity seen in OxDC.
Figure 2.
ESI-Q-MS analysis of BMPO-superoxide reaction mixture. Mass spectral features include: [BMPO+H]+ (m/z 200), [BMPO+Na]+ (m/z 222), [BMPO+HO2+H]+ (m/z 233), [BMPO+K]+ (m/z 238), [BMPO+HCO2•+H]+ (m/z 245). The inset gives an expanded view of the m/z range for the radical-bound spin trap. Spectra were normalized to the intensity of the protonated spin trap ([BMPO+H]+). The potassiated spin trap (m/z 238) is approximately 625 times more intense and off the scale. (Double Column Image, Color Reproduction online/print)
All tentative mechanisms for OxDC discussed in the literature to date suggest oxalate activation by one-electron transfer as the first step with dioxygen being the driving oxidative force leading to superoxide or hydroperoxyl bound to Mn(II) [25]. If this takes place in the active site, presumably the N-terminal Mn-binding pocket, a peroxycarbonate species may form leading to oxidase activity which could explain the 0.2% oxidase activity in OxDC [27]. A similar mechanism has been proposed for oxalate oxidase by Opaleye et al. although the peroxo compound suggested there still includes oxalate [69]. More recently, Whittaker et al. favored an oxidase mechanism for OxOx that bypasses peroxycarbonate and generates superoxide or hydroperoxyl through inner or outer sphere electron transfer based on competition experiments with superoxide dismutase [43]. This mechanism allows for the loss of hydroperoxyl from the protein and is consistent with our findings of a hydroperoxyl adduct to BMPO.
At issue is whether superoxide/hydroperoxyl is released into the solution under turnover conditions. BMPO is too bulky to fit into the active site to react with a closely held superoxide radical. If superoxide and the CO2•− intermediate are generated in the same active site pocket, formation of a peroxycarbonate species is the logical next step and one would not expect to observe either CO2•− or hydroperoxyl in solution [27]. On the other hand, release of the carbon dioxide radical anion into solution, e.g., through loss of control of this intermediate by the protein, may automatically lead to superoxide production in solution since this radical will react with oxygen [37]. However, this is not likely to happen under our experimental conditions since the high concentration of spin trap will outcompete oxygen for the reaction with CO2•−. Yet, it may explain the 0.2% oxidase activity in WT OxDC, not as a separate enzymatic pathway but rather as the result of loss of control over the decarboxylase pathway [37]. This raises the question what happens to the enzyme when it loses the CO2•− intermediate since at that point it contains an extra electron, most likely on a hydroperoxyl radical bound to one of the Mn(II) ions. The simplest explanation would be the loss of superoxide in the form of a hydroperoxyl radical with the needed extra proton being donated by the solution [43].
It has been previously shown that the lid mutants T165V and SENS161–164DASN exhibit increased CO2•− yields by spin trapping experiments using α-phenyl N-tertiary-butyl nitrone (PBN) and α-(4-Pyridyl N-oxide)-N-tert-butylnitrone (POBN) [37, 41]. If the observed superoxide arises only from the reaction of escaped CO2•− radical anion with dissolved dioxygen, the trapping rates of superoxide should also increase in these mutants. Spin trapping and mass spectrometry was therefore performed on OxDC T165V in the presence of BMPO. The experimental results together with simulations of the EPR spectra are shown in Figure 3 while Table 2 shows the magnetic parameters used together with the corresponding spectral weights.
Figure 3.
X-band EPR spectra of the BMPO-radical adducts produced from the reactions between oxalate and OxDC mutant T165V in 50 mM citrate buffer pH 4.0 (black). The orange and magenta spectra represent simulations of the superoxide and carbon dioxide anion radical adduct, respectively. Blue: sum of the simulations of the two spin adducts. (Double Column Image, Color Reproduction online/print)
Table 2.
Spectral parameters of BMPO-radical adducts for T165V OxDC, mutant, including hyperfine coupling, g-value, and individual spectral weights based off simulated spectra. (Single Column Image)
| Spin-Trapped Adduct |
g-value | aH(mT) | aN(mT) | Spectral Weight (Relative Percentage) |
|---|---|---|---|---|
| [BMPO-OOH]• (Conformer I) | 2.005 | 1.208 | 1.359 | 24.22 |
| [BMPO-OOH]• Conformer II) | 2.005 | 0.953 | 1.316 | 9.91 |
| [BMPO-CO2−]• | 2.005 | 1.749 | 1.484 | 65.87 |
As can be seen in figure 3, the spectrum is dominated by the carbon dioxide anion radical adduct. Identification of this radical was confirmed by comparison with synthetic CO2•− radical produced via a Fenton reaction in the presence of formic acid and the spin trap BMPO as previously described [37], and mass spectrometry experiments (see supplementary information, figure S3. In order to better compare the relative yields of the two radical adducts, quantitative EPR was performed on the WT and the T165V mutant under the same experimental conditions (example spectra in figures 1 and 3 with quantitative analysis in figure 4).
Figure 4.
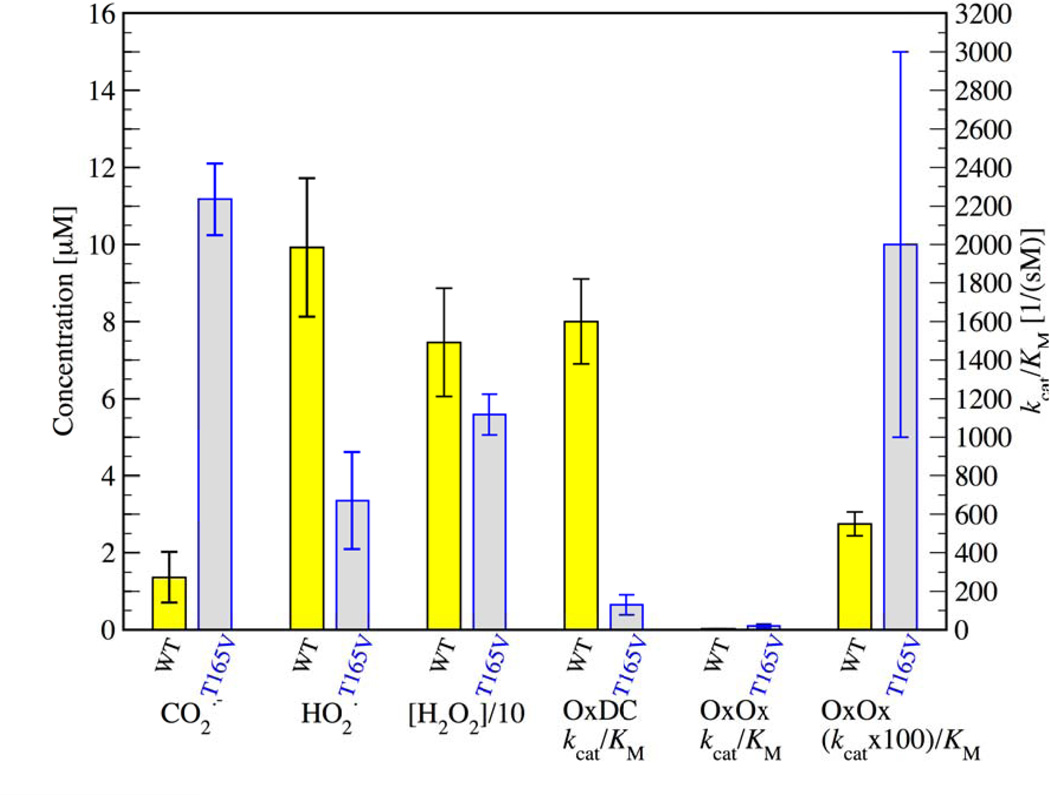
From left to right for WT (yellow) and T165V (gray): concentrations of the BMPO radical adduct for the CO2•− and the O2•− radical in reactions carried out as described in the text, concentrations of H2O2 as determined by the EuTc assay, and published values for the decarboxylase and oxidase activities in terms of observed kcat/KM. Please note that values for the activities refer to the y-axis on the right while concentrations refer to the y-axis on the left. Please note that the H2O2 concentrations were divided by a factor of ten in order to fit on the same ordinate axis as the trapped radicals. Since the oxidase activity is so much lower than the decarboxylase activity for both WT and T165V mutant, the last pair of bars to the right has been multiplied by a factor of 100. Oxidase activities were measured as described in [70] where the oxygen consumption was determined as a function of oxalate concentration using a luminescent oxygen sensor. All experiments were performed in triplicate with the error bars indicating the standard deviation of the experiments. (Double Column Image, Color Reproduction online/print)
It is instructive to compare the ratio of the yields of both radical adducts with the reported decarboxylase and oxidase activities of WT OxDC and mutant T165V in order to put the spin trapping results into perspective. the kcat/KM for decarboxylase activity in the T165V mutant is approximately ten times lower than in WT [64]. Saylor et al. ascribe this to a hindered motion of the flexible SENS161–164 lid, giving it a preference for the open conformation which allows the intermediate CO2•− radical to escape into the surrounding solution at a higher rate [64]. Figure 4 shows that the rate of CO2•− radical escape is approximately ten times higher in T165V than in WT. This is reflected in the ten times lower decarboxylase activity of the mutant (see 4th column pair in figure 4 where the OxDC activities show the inverse relationship compared to the 1st column pair). The intermediate CO2•− radical simply diffuses away from the active site at a much higher rate in the mutant and is therefore not available for conversion to formate. The experimental observation of high CO2•− radical adduct yields is clearly related to reduced decarboxylase activity.
A similar inverse relationship between radical adduct yields and activities is seen between the 2nd and 4th (or 5th for 100× magnification) column pair in figure 4 showing the hydroperoxyl adduct yields and the OxOx activities of the two strains although the differences between the two strains is smaller and the error bars are generally larger due to the fact that oxidase activity is still about two orders of magnitude slower than decarboxylase activity. When using the EuTc fluorescence assay (3rd column pair in figure 4 and supplementary figure 5) we find a slightly smaller yield of H2O2 production in T165V of 56 ± 5 µM compared to 75 ± 14 µM in WT. This is still consistent with the spin trapping experiments because the EuTc assay measures total yield of H2O2 while the spin trapping experiment only determines the trapping yield of escaped superoxide that was generated within the protein.
Our observations can be qualitatively explained by considering two different sources of superoxide during catalytic turn-over: (A) Intra-protein superoxide production through an oxidase mechanism, for example the one suggested for oxalate oxidase by Whittaker [43], and subsequent release of hydroperoxyl radicals into solution. This should lead to superoxide release from the protein either directly from the active site or from a different oxygen binding site of the protein, e.g., the C-terminal bound Mn, and results in the observation of trapped hydroperoxyl radical in the presence of BMPO. The trapped radical can therefore be considered as a signature of ‘true’ oxidase activity taking place within the protein. (B) Extra-protein superoxide production through the reaction of the CO2•− radical with dissolved dioxygen will also happen (see Saylor et al. [64]). In fact it is expected to be enhanced in T165V because of the large amount of released CO2•− radical. In the absence of spin trap this ‘extra-protein’ superoxide will dismutate to oxygen and H2O2 which is seen in the EuTc assay as well as in the oxygen consumption experiments [70]. It masks the true OxOx activity in these assays by allowing for additional production of hydrogen peroxide and additional consumption of dioxygen.
While a more quantitative analysis of the relative yields of hydrogen peroxide from intra- and extra-protein sources will have to await the outcome of competition experiments in which oxidase activity is measured in the presence of spin trap, we wish to point out in this communication that this is the first report of superoxide production in OxDC (or OxOx) under turnover conditions. Our experiments were performed at pH4 where superoxide dismutates with essentially diffusion-controlled kinetics [71]. The trapping rate of superoxide with BMPO is only 0.24 M−1s−1 at pH7.4 [72]. However, trapping yields with many cyclic nitrones increase dramatically in the presence of hydroperoxyl radical at low pH [73], possibly also with superoxide when the spin trap can be protonated [48]. Our EPR and mass spectral results confirm the trapping of the hydroperoxyl radical unequivocally, suggesting that the trapped hydroperoxyl is formed by the protein during the reaction and released into solution. This might help the protein to get rid of the excess electron from oxalate when it acts as an oxidase. The T165V mutation has a dramatic effect on the trapping yield of the carbon dioxide radical anion but much less so and in the opposite direction for the trapping yield of superoxide. This strongly suggests that both species are released from different sites of the protein. Perhaps superoxide gets produced at the C-terminal Mn binding site which would necessitate long-range electron transfer between the two Mn-ions. Alternatively, dioxygen may bind at a distal position of the protein that is solvent accessible and facilitates electron transfer to remove excess reducing equivalents from the protein.
Conclusions
The combination of EPR spin trapping and mass spectrometry experiments have demonstrated the presence of hydroperoxyl radicals in OxDC preparations under turnover conditions. To our knowledge this is the first experimental report of a superoxide-derived radical adduct observed in either OxDC or OxOx at the rather low pH of 4.0. Our experiments also confirm prior observation of the carbon dioxide radical anion intermediate during turn-over. Spin trapping experiments conducted on the OxDC lid mutant T165V suggest that the mutant has a three-fold smaller rate of superoxide production in the protein than WT while its carbon dioxide radical anion yield is increased ten-fold. This unequal effect of the site mutation on radical release suggests that the two radicals are produced at different locations in the protein.
Supplementary Material
Highlights.
Superoxide is released by oxalate decarboxylase under turn-over conditions at pH4.
Identification of radicals by combined EPR spin trapping and mass spectrometry.
Superoxide is released from OxDC at a site different from the active site.
Acknowledgements
Funding for this work was provided by the National Science Foundation under grant CHE-121440, National Institutes of Health under grant DK061666, and the mass spectrometer was purchased from funds by the National Science Foundation under grant CHE-0845450 and an RET supplement under grant MCB-1158000. A plasmid containing the gene encoding C-terminally, His-tagged wild type Bacillus subtilis OxDC was generously provided by Dr. Stephen Bornemann (John Innes Centre, Norwich, UK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller C, Kennington L, Cooney R, Kohjimoto Y, Cao LC, Honeyman T, Pullman J, Jonassen J, Scheid C. Oxalate Toxicity in Renal Epithelial Cells: Characteristics of Apoptosis and Necrosis. Toxicology and Applied Pharmacology. 2000;162:132–141. doi: 10.1006/taap.1999.8835. [DOI] [PubMed] [Google Scholar]
- 2.Frassetto L, Kohlstadt I. Treatment and Prevention of Kidney Stones: An Update. Am. Fam. Physician. 2011;84:1234–1242. [PubMed] [Google Scholar]
- 3.Libert B, Franceschi VR. Oxalate in crop plants. J. Agric. Food. Chem. 1987;35:926–938. [Google Scholar]
- 4.Noonan SCSGP. Oxalate content of foods and its effect on humans. Asia Pacific Journal of Clinical Nutrition. 1999;8:64–74. [PubMed] [Google Scholar]
- 5.Rahman MM, Abdullah RB, Wan Khadijah WE. A review of oxalate poisoning in domestic animals: tolerance and performance aspects. J. Anim. Physiol. Anim. Nutr. 2013;97:605–614. doi: 10.1111/j.1439-0396.2012.01309.x. [DOI] [PubMed] [Google Scholar]
- 6.Hodgkinson A. Oxalic acid in biology and medicine. New York: Academic Press, London; 1977. [Google Scholar]
- 7.Elder TD, Wyngaarden JB. The Biosysnthesis and Turnover of Oxalate in Normal and Hyperoxaluric Subjects. The Journal of Clinical Investigation. 1960;39:1337–1344. doi: 10.1172/JCI104151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sidhu H, Schmidt ME, Cornelius JG, Thamilselvan S, Khan SR, Hesse A, Peck AB. Direct correlation between hyperoxaluria/oxalate stone disease and the absence of the gastrointestinal tract-dwelling bacterium Oxalobacter formigenes: possible prevention by gut recolonization or enzyme replacement therapy. J. Am. Soc. Nephrol. 1999;14(Suppl):S334–S340. [PubMed] [Google Scholar]
- 9.Sidhu H, Allison M, Peck AB. Identification and Classification of Oxalobacter formigenes Strains by Using Oligonucleotide Probes and Primers. J. Clin. Microbiol. 1997;35:350–353. doi: 10.1128/jcm.35.2.350-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svedružić DJS, Toyota CG, Reinhardt LA, Ricagno S, Linqvist Y, Richards NGJ. The enzymes of oxalate metabolism: unexpected structures and mechanisms. Archives of Biochemistry and Biophysics. 2005;433:176–192. doi: 10.1016/j.abb.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 11.Martin G, Guggiari M, Bravo D, Zopfi J, Cailleau G, Aragno M, Job D, Verrecchia E, Junier P. Fungi, bacteria and soil pH: the oxalate–carbonate pathway as a model for metabolic interaction. Environ. Microbiol. 2012;14:2960–2970. doi: 10.1111/j.1462-2920.2012.02862.x. [DOI] [PubMed] [Google Scholar]
- 12.Mäkelä MR, Hildén K, Hatakka A, Lundell TK. Oxalate decarboxylase of the white-rot fungus Dichomitus squalens demonstrates a novel enzyme primary structure and non-induced expression on wood and in liquid cultures. Microbiology. 2009;155:2726–2738. doi: 10.1099/mic.0.028860-0. [DOI] [PubMed] [Google Scholar]
- 13.Hart PW, Rudie AW. Mineral scale management, Part I. Case studies. TAPPI Journal. 2006;5:22–27. [Google Scholar]
- 14.Rudie AW, Hart PW. Mineral Scale Management Part II. Fundamental Chemistry. TAPPI Journal. 2006;5:17–23. [Google Scholar]
- 15.Winestrand S, Gandla ML, Hong F, Chen QZ, Jönsson LJ. Oxalate decarboxylase of Trametes versicolor: biochemical characterization and performance in bleaching filtrates from the pulp and paper industry. J. Chem. Technol. Biotechnol. 2012;87:1600–1606. [Google Scholar]
- 16.Mäkelä MRHK, Lundell TK. Oxalate decarboxylase: biotechnological update and prevalence of the enzyme in filamentous fungi. Applied Microbiology and Biotechnology. 2010;87:801. doi: 10.1007/s00253-010-2650-z. [DOI] [PubMed] [Google Scholar]
- 17.Chakraborty N, Ghosh R, Ghosh S, Narula K, Tayal R, Datta A, Chakraborty S. Reduction of Oxalate Levels in Tomato Fruit and Consequent Metabolic Remodeling Following Overexpression of a Fungal Oxalate Decarboxylase. Plant Physiol. 2013;162:364–378. doi: 10.1104/pp.112.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dias BBA, Cunha WG, Morais LS, Vianna GR, Rech EL, de Capdeville G, Aragão FJL. Expression of an oxalate decarboxylase gene from Flammulina sp. in transgenic lettuce (Lactuca sativa) plants and resistance to Sclerotinia sclerotiorum. Plant Physiol. 2006;55:187–193. [Google Scholar]
- 19.Langman LJ, Allen LC. An Enzymatic Method for Oxalate Automated Using The Hitachi 911 Analyzer. Clin. Biochem. 1998;31:429–432. doi: 10.1016/s0009-9120(98)00029-0. [DOI] [PubMed] [Google Scholar]
- 20.Pundir CS, Sharma M. Oxalate biosesor: a review. J. Sci. & Ind. Res. 2010;69:489–494. [Google Scholar]
- 21.Hesse A, Bongartz D, Heynck H, Berg W. Measurement of Urinary Oxalic Acid: A Comparison of Five Methods. Clin. Biochem. 1996;29:467–472. doi: 10.1016/0009-9120(96)00067-7. [DOI] [PubMed] [Google Scholar]
- 22.Sasikumar P, Gomathi S, Anbazhagan K, Selvam GS. Secretion of Biologically Active Heterologous Oxalate Decarboxylase (OxdC) in Lactobacillus plantarum WCFS1 Using Homologous Signal Peptides. Biomed. Res. Int. 2013;2013:9. doi: 10.1155/2013/280432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowley AB, Poage DW, Dean RR, Meschter CL, Ghoddusi M, Li QS, Sidhu H. 14-Day Repeat-Dose Oral Toxicity Evaluation of Oxazyme in Rats and Dogs. International Journal of Toxicology. 2010;29:20. doi: 10.1177/1091581809353611. [DOI] [PubMed] [Google Scholar]
- 24.Grujic D, Salido EC, Shenoy BC, Langman CB, McGrath ME, Patel RJ, Rashid A, Mandapati S, Jung CW, Margolin AL. Hyperoxaluria Is Reduced and Nephrocalcinosis Prevented with an Oxalate-Degrading Enzyme in Mice with Hyperoxaluria. Am. J. Nephrol. 2009;29:86–93. doi: 10.1159/000151395. [DOI] [PubMed] [Google Scholar]
- 25.Svedružić DLY, Reinhardt LA, Wroclawska E, Cleland WW, Richards NGJ. Investigating the roles of putative active site residues in the oxalate decarboxylase from Bacillus subtilis. Archives of Biochemistry and Biophysics. 2007;464:36–47. doi: 10.1016/j.abb.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anand R, Dorrestein PC, Kinsland C, Begley TP, Ealick SE. Structure of Oxalate Decarboxylase from Bacillus subtilis at 1.75 Å Resolution. Biochemistry. 2002;41:7659–7669. doi: 10.1021/bi0200965. [DOI] [PubMed] [Google Scholar]
- 27.Just VJ, Burrell MR, Bowater L, McRobbie I, Stevenson CEM, Lawson DM, Bornemann S. The identity of the active site of oxalate decarboxylase and the importance of the stability of active-site lid conformations. Biochemical Journal. 2007;407:397–406. doi: 10.1042/BJ20070708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Just VJ, Stevenson CEM, Bowater L, Tanner A, Lawson DM, Bornemann S. A Closed Conformation of Bacillus subtilis Oxalate Decarboxylase OxdC Provides Evidence for the True Identity of the Active Site. Journal of Biological Chemistry. 2004;279:19867–19874. doi: 10.1074/jbc.M313820200. [DOI] [PubMed] [Google Scholar]
- 29.Dunwell JM, Gane PJ. Microbial Relatives of Seed Storage Proteins: Conservation of Motifs in a Functionally Diverse Superfamily of Enzymes. J Mol Evol. 1998;46:147–154. doi: 10.1007/pl00006289. [DOI] [PubMed] [Google Scholar]
- 30.Khuri S, Bakker FT, Dunwell JM. Phylogeny, Function, and Evolution of the Cupins, a Structurally Conserved, Functionally Diverse Superfamily of Proteins. Mol. Biol. Evol. 2001;18:593–605. doi: 10.1093/oxfordjournals.molbev.a003840. [DOI] [PubMed] [Google Scholar]
- 31.Dunwell J, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Wolfenden R, Lewis CA, Yuan Y. Kinetic Challenges Facing Oxalate, Malonate, Acetoacetate, and Oxaloacetate Decarboxylases. J. Am. Chem. Soc. 2011;133:5683–5685. doi: 10.1021/ja111457h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanner A, Bowater L, Fairhurst SA, Bornemann S. Oxalate Decarboxylase Requires Manganese and Dioxygen for Activity - Overexpression and Characterization of Bacillus subtilis YvrK and YoaN. Journal of Biological Chemistry. 2001;276:43627–43634. doi: 10.1074/jbc.M107202200. [DOI] [PubMed] [Google Scholar]
- 34.Emiliani E, Bekes P. Enzymatic Oxalate Decarboxylation in Aspergillus niger. Archives of Biochemistry and Biophysics. 1964;105:488–493. doi: 10.1016/0003-9861(64)90040-2. [DOI] [PubMed] [Google Scholar]
- 35.Shimazono H. Oxalic Acid Decarboxylase, A new Enyme from the Mycelium of Wood Destroying Fungi. Journal of Biochemistry. 1955;42:321–340. [Google Scholar]
- 36.Reinhardt LA, Svedružić D, Chang CH, Cleland WW, Richards NGJ. Heavy Atom Isotope Effects on the Reaction Catalyzed by the Oxalate Decarboxylase from Bacillus subtilis. Journal of the American Chemical Society. 2003;125:1244–1252. doi: 10.1021/ja0286977. [DOI] [PubMed] [Google Scholar]
- 37.Imaram WSBT, Centonze CP, Richards NGJ, Angerhofer A. EPR spin trapping of an oxalate-derived free radical in the oxalate decarboxylase reaction. Free Rad. Biol. & Med. 2011;50:1009–1015. doi: 10.1016/j.freeradbiomed.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bielski BHJ. Reevaluation of the Spectral and Kinetic Properties of HO2 and O2- Free Radicals. Photochem. Photobiol. 1978;28:645–649. [Google Scholar]
- 39.Begley TP, Ealick SE. Enzymatic reactions involving novel mechanisms of carbanion stabilization. Curr. Opin. Chem. Biol. 2004;8:508–515. doi: 10.1016/j.cbpa.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Woo E-J, Dunwell JM, Goodenough PW, Marvier AC, Pickersgill RW. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Mol. Biol. 2000;7:1036–1040. doi: 10.1038/80954. [DOI] [PubMed] [Google Scholar]
- 41.Burrell MWJVJ, Bowater L, Fairhurst SA, Requena L, Lawson DM, Bornemann S. Oxalate Decarboxylase and Oxalate Oxidase Activities Can Be Interchanged with a Specificity Switch of up to 282 000 by Mutating an Active Site Lid. Biochemistry. 2007;46:12327–12336. doi: 10.1021/bi700947s. [DOI] [PubMed] [Google Scholar]
- 42.Escutia MR, Bowater L, Edwards A, Bottrill AR, Burrell MR, Polanco R, Vicuña R, Bornemann S. Cloning and Sequencing of Two Ceriporiopsis subvermispora Bicupin Oxalate Oxidase Allelic Isoforms: Implications for the Reaction Specificity of Oxalate Oxidases and Decarboxylases. Appl. Environ. Microbiol. 2005;71:3608–3616. doi: 10.1128/AEM.71.7.3608-3616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whittaker MM, Pan H-Y, Yukl ET, Whittaker JW. Burst Kinetics and Redox Transformations of the Active Site Manganese Ion in Oxalate Oxidase: Implications for the Catalytic Mechanism. J. Biol. Chem. 2007;282:7011–7023. doi: 10.1074/jbc.M609374200. [DOI] [PubMed] [Google Scholar]
- 44.Whittaker M, Whittaker J. Characterization of recombinant barley oxalate oxidase expressed by Pichia pastoris. J. Biol. Inorg. Chem. 2002;7:136–145. doi: 10.1007/s007750100281. [DOI] [PubMed] [Google Scholar]
- 45.Moomaw EW, Hoffer E, Moussatche P, Salerno JC, Grant M, Immelman B, Uberto R, Ozarowski A, Angerhofer A. Kinetic and Spectroscopic Studies of Bicupin Oxalate Oxidase and Putative Active Site Mutants. PLoS ONE. 2013;8:e57933. doi: 10.1371/journal.pone.0057933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bielski BHJ, Allen AO. Mechanism of the Disproportionation of Superoxide Radicals. J. Phys. Chem. 1977;81:1048–1050. [Google Scholar]
- 47.Finkelstein E, Rosen GM, Rauckman EJ. Spin Trapping. Kinetics of the Reaction of Superoxide and Hydroxyl Radicals with Nitrones. J. Am. Chem. Soc. 1980;102:4994–4999. [Google Scholar]
- 48.Villamena FA, Merle JK, Hadad CM, Zweier JL. Superoxide Radical Anion Adduct of 5,5-Dimethyl-1-pyrroline N-Oxide (DMPO). 1. The Thermodynamics of Formation and Its Acidity. J. Phys. Chem. A. 2005;109:6083–6088. doi: 10.1021/jp052431f. [DOI] [PubMed] [Google Scholar]
- 49.Buettner GR, Oberley LW. Considerations in the Spin Trapping of Superoxide and Hydroxyl Radical in Aqueous Systems Using 5,5-Dimethyl-1-pyrroline-1-oxide. Biochem. Biophys. Res. Commun. 1978;83:69–74. doi: 10.1016/0006-291x(78)90398-4. [DOI] [PubMed] [Google Scholar]
- 50.Villamena FA, Zweier JL. Detection of Reactive Oxygen and Nitrogen Species by EPR Spin Trapping. Antioxid. Redox Signaling. 2004;6:619–629. doi: 10.1089/152308604773934387. [DOI] [PubMed] [Google Scholar]
- 51.Buettner GR, Mason RP. Spin-Trapping Methods for Detecting Superoxide and Hydroxyl Free Radicals in Vitro and in Vivo. In: Packer LGAN, editor. Methods in Enzymology. Academic Press; 1990. pp. 127–133. [DOI] [PubMed] [Google Scholar]
- 52.Finkelstein E, Rosen GM, Rauckman EJ. Production of Hydroxyl Radical by Decomposition of Superoxide Spin-Trapped Adducts. Mol. Pharmacology. 1982;21:262–265. [PubMed] [Google Scholar]
- 53.Khan N, Wilmot CM, Rosen GM, Demidenko E, Sun J, Joseph J, Hara J, Kalyanaraman B, Swartz HM. Spin Traps: in Vitro Toxicity and Stability of Radical Adducts. Free Rad. Biol. & Med. 2003;34:1473–1481. doi: 10.1016/s0891-5849(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 54.Zhao H, Joseph J, Zhang H, Karoui H, Kalyanaraman B. Synthesis and Biochemical Applications of a Solid Cyclic Nitrone Spin Trap: a Relatively Superior Trap for Detecting Superoxide Anions and Glutathiyl Radicals. Free Rad. Biol. & Med. 2001;31:599–606. doi: 10.1016/s0891-5849(01)00619-0. [DOI] [PubMed] [Google Scholar]
- 55.Rosen GM, Britigan BE, Halpern HJ, Pou S. Free Radicals: Biology And Detection By Spin Trapping. 198 Madison Avenue, New York, New York 10016: Oxford University Press; 1999. [Google Scholar]
- 56.Bielski BHJ, Cabelli DE, Arudi RL, Ross AB. Reactivity of HO2/O2-Radicals in Aqueous Solution. J. Phys. Chem. Ref. Data. 1985;14:1041–1100. [Google Scholar]
- 57.Iwahashi H, Parker CE, Mason RP, Tomer KB. Combined Liquid Chromatography/Electron Paramagnetic Resonance Spectrometry/Electrospray Ionization Mass Spectrometry for Radical Identification. Analytical Chemistry. 1992;64:2244–2252. doi: 10.1021/ac00043a011. [DOI] [PubMed] [Google Scholar]
- 58.Qian SYKMB, Guo Q, Mason RP. A novel protocol to identify and quantify all spin trapped free radicals from in vitro/in vivo interaction of HO and DMSO: LC/ESR, LC/MS, and dual spin trapping combinations. Free Rad. Biol. & Med. 2005;38:125–135. doi: 10.1016/j.freeradbiomed.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 59.Domingues P, Domingues MRM, Amado FML, Ferrer-Correia AJ. Detection and Characterization of Hydroxyl Radical Adducts by Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2001;12:1214–1219. doi: 10.1016/S1044-0305(01)00310-5. [DOI] [PubMed] [Google Scholar]
- 60.Imaram W, Gersch C, Kim KM, Johnson RJ, Henderson GN, Angerhofer A. Radicals in the reaction between peroxynitrite and uric acid identified by electron spin resonance spectroscopy and liquid chromatography mass spectrometry. Free Rad. Biol. & Med. 2010;49:275–281. doi: 10.1016/j.freeradbiomed.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo Q, Qian SY, Mason RP. Separation and Identification of DMPO Adducts of Oxygen-Centered Radicals Formed from Organic Hydroperoxides by HPLC-ESR, ESI-MS and MS/MS. J. Am. Soc. Mass Spectrom. 2003;14:862–871. doi: 10.1016/S1044-0305(03)00336-2. [DOI] [PubMed] [Google Scholar]
- 62.Huang C-H, Shan G-Q, Mao L, Kalyanaraman B, Qin H, Ren F-R, Zhu B-Z. The first purification and unequivocal characterization of the radical form of the carbon-centered quinone ketoxy radical adduct. Chem. Commun. 2013;49:6436–6438. doi: 10.1039/c3cc42732c. [DOI] [PubMed] [Google Scholar]
- 63.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 64.Saylor BT, Reinhardt LA, Lu Z, Shukla MS, Nguyen L, Cleland WW, Angerhofer A, Allen KN, Richards NGJ. A Structural Element That Facilitates Proton-Coupled Electron Transfer in Oxalate Decarboxylase. Biochemistry. 2012;51:2911–2920. doi: 10.1021/bi300001q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moomaw EW, Angerhofer A, Moussatche P, Ozarowski A, García-Rubio I, Richards NGJ. Metal Dependence of Oxalate Decarboxylase Activity. Biochemistry. 2009;48:6116–6125. doi: 10.1021/bi801856k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schütte H, Flossdorf J, Sahm H, Kula M-R. Purification and Properties of Formaldehyde Dehydrogenase and Formate Dehydrogenase from Candida boidinii. Eur. J. Biochem. 1976;62:151–160. doi: 10.1111/j.1432-1033.1976.tb10108.x. [DOI] [PubMed] [Google Scholar]
- 67.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Res. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 68.Polson C, Sarkar P, Incledon B, Raguvaran V, Grant R. Optimization of protein precipitation based upon effectiveness of protein removal and ionization effect in liquid chromatography–tandem mass spectrometry. J. Chromat. B. 2003;785:263–275. doi: 10.1016/s1570-0232(02)00914-5. [DOI] [PubMed] [Google Scholar]
- 69.Opaleye O, Rose R-S, Whittaker MM, Woo E-J, Whittaker JW, Pickersgill RW. Structural and Spectroscopic Studies Shed Light on the Mechanism of Oxalate Oxidase. J. Biol. Chem. 2006;281:6428–6433. doi: 10.1074/jbc.M510256200. [DOI] [PubMed] [Google Scholar]
- 70.Molina LMGTJ, Twahir UT, Moomaw EW, Angerhofer A. Kinetic Studies of Oxalate Decarboxylase and Oxalate Oxidase using a Luminescent Oxygen Sensor. Journal of Biochemical Technology. 2014 Oct; Submitted. [Google Scholar]
- 71.Ozawa T, Hanaki A. Reactions of Superoxide with Water and with Hydrogen Peroxide. Chem. Pharm. Bull. 1981;29:926–928. [Google Scholar]
- 72.Keszler A, Kalyanaraman B, Hogg N. Comparative Investigation of Superoxide Trapping by Cyclic Nitrone Spin Traps: the Use of Singular Value Decomposition and Multiple Linear Regression Analysis. Free Radic. Biol. & Med. 2003;35:1149–1157. doi: 10.1016/s0891-5849(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 73.Allouch A, Lauricella RP, Tuccio BN. Effect of pH on superoxide/hydroperoxyl radical trapping by nitrones: an EPR/kinetic study. Molec. Phys. 2007;105:2017–2024. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.