Abstract
Background
Although obesity is associated with breast cancer incidence and prognosis, the underlying mechanisms are poorly understood. Identification of obesity-associated epigenetic changes in breast tissue may advance mechanistic understanding of breast cancer initiation and progression. The goal of this study, therefore, was to investigate associations between obesity and gene methylation in breast tumors.
Methods
Using the Illumina GoldenGate Cancer I Panel, we estimated the association between body mass index (BMI) and gene methylation in 345 breast tumor samples from Phase I of the Carolina Breast Cancer Study, a population based case-control study. Multivariable linear regression was used to identify sites that were differentially methylated by BMI. Stratification by tumor estrogen receptor status was also conducted.
Results
In the majority of the 935 probes analyzed (87%), the average beta value increased with obesity (BMI ≥ 30). Obesity was significantly associated with differential methylation (false discovery rate q-value < 0.05) in just 2 gene loci in breast tumor tissue overall and in 21 loci among estrogen receptor (ER)-positive tumors. Obesity was associated with methylation of genes that function in immune response, cell growth, and DNA repair.
Conclusions
Obesity is associated with altered methylation overall, and with hypermethylation among ER-positive tumors in particular, suggesting that obesity may influence the methylation of genes with known relevance to cancer. Some of these differences in methylation by obese status may influences levels of gene expression within breast cells.
Impact
If our results are validated, obesity-associated methylation sites could serve as targets for prevention and treatment research.
Keywords: BMI, obesity, methylation, epigenetics, breast cancer
Introduction
Larger body size, including the body mass index (BMI) defined categories of overweight and obese, are associated with both the incidence and prognosis of breast cancer, though the risk varies by menopausal status and tumor subtype. In premenopausal women, an inverse association has generally been found between BMI and breast cancer incidence (1, 2) while increased BMI has been shown to increase the risk among postmenopausal women (3), with an estimated 3% increase in risk with each one unit gain in BMI (4). There is also evidence that obesity may increase incidence of estrogen receptor (ER)-positive but not ER-negative tumors, though these relationships differ by age and menopausal status (5, 6).
Beyond associations between BMI and breast cancer incidence, obesity is also associated with worse prognosis and outcome in both pre- and postmenopausal women (4). Obese breast cancer patients are more likely to be diagnosed with advanced stage at diagnosis (3), larger tumor size (1), nodal involvement (1), higher grade (4), and higher mitotic cell count (4). They are also at higher risk of experiencing recurrence (7), metastasis (1), and mortality (7).
The biological mechanisms underlying obesity's effects on carcinogenesis are poorly understood. Gene methylation, a mechanism for controlling the expression of genes (8), has the potential to influence the carcinogenic process during both tumor formation and progression. In normal tissues, methylation plays a role in whether cells are growing or senescent and how cells differentiate; if these patterns become abnormal, cancer may result (9). Abnormal methylation can also influence tumor invasiveness and metastasis (10).
Though obesity has been associated with increased methylation in several cancer-related genes (11, 12), previous research has been limited by the use of white blood cells instead of breast tissue (11, 12) or by focusing on only candidate genes (13). We sought to test whether obesity is associated with methylation in a panel of cancer-related genes in the tumor tissue of women diagnosed with breast cancer. Identifying genes that are differentially methylated by BMI can provide evidence for the underlying biological processes through which obesity influences breast carcinogenesis and may facilitate the development of chemoprevention and treatment targets.
Materials and Methods
Study Participants
Data from Phase I of the Carolina Breast Cancer Study (CBCS) was used to assess gene methylation in tumor tissue collected from women with breast cancer. The CBCS study design data collection methods have been previously described (14). Briefly, women diagnosed with breast cancer were recruited with randomized sampling between 1993 and 1996 from the North Carolina Central Cancer Registry. African-American women and women younger than 50 years of age were oversampled to comprise approximately 50% of the study population. Participation in the study was limited to English-speaking women aged 20 to 74 years. Participants were interviewed in-person by trained nurses to collect demographic and breast cancer risk factor data. The University of North Carolina at Chapel Hill Institutional Review Board approved this study.
Tissue Specimens
Only participants with sufficient tumor block material for methylation analyses were included in the current study. CBCS pathologists verified the cancer diagnosis of all participants. The breast tumors were grossly dissected after review by a pathologist, who encircled areas of malignant cellularity. While some intratumoral stroma may be present among tumor cells, this sampling method ensured minimal contamination of our methylation signatures by profiles of adipose, stroma or normal epithelium. As a result, our findings are not likely to be driven by between-sample differences in tumor composition.
Gene Methylation Assessment
The Illumina Cancer Panel I platform (Illumina Inc., San Diego, CA), which consists of 1,505 gene sites with known relevance to cancer (15), measures DNA methylation at cytosine-guanine dinucleotide (CpG) sites. The methylation level of each CpG site on the panel was measured as a beta value, which was calculated by comparing the fluorescent signal ratio of the methylated allele to the sum of the signals from the methylated and unmethylated alleles. The beta values range from 0 to 1, representing the fraction of methylated DNA, with 0 indicating no methylation and 1 indicating complete methylation. Of the 1,505 CpG sites on the platform, 570 were removed from the analysis due to poor performance or because the sites overlapped regions with SNPs or copy number variation, which may render them unreliable (16). The remaining 935 CpG sites (17) were assessed for association with BMI. The input sequence of all probes that were found to be significantly associated with BMI were checked against target sequences in the NCBI Blast database to determine if any of the probes ambiguously mapped to the genome (18). The array data have been deposited in Gene Expression Omnibus under accession number GSE51557.
In a previous study, the demographic and tumor characteristics were compared for those who were eligible for participation in the study and for those whose methylation values were successfully assessed (17). The only characteristic that reached statistical significance (p-value < 0.05) was age, where the tumors of younger women were more likely to be successfully assayed. Age was included as a confounder in our analyses. There were no significant differences in tumor size or clinical stage.
Statistical Analyses
Linear regression, using limma with an empirical Bayes approach (19), was conducted to assess which CpG sites were associated with pre-diagnosis body mass index (BMI). The R statistical package (www.r-project.org/) was used to conduct all regression analyses. Pre-diagnosis BMI was self-reported at the time of interview and was categorized as normal weight (18.5 ≤ BMI < 25), overweight (25 ≤ BMI < 30) or obese (BMI ≥ 30). Comparisons were 2-category contrasts (obese versus normal weight, with overweight women excluded) for regression modeling based on previous studies and our own exploratory analyses that showed that the overweight class was highly heterogeneous with regard to obesity-dependent gene expression (20). Therefore, to enhance our ability to detect obesity-associated methylation, the intermediate overweight phenotype group was excluded. Use of a categorical rather than continuous coding for BMI also minimized potential bias due to underestimation of weight (21).
Statistical analyses were restricted to those of African-American or white race. Race and age (treated as a continuous variable) were considered confounders a priori and were included as covariates in all analyses. Other variables assessed for confounding included menopausal status, age at first full-term birth, parity, alcohol use, smoking status, physical activity, fruit and vegetable intake, income, and education status. Among these variables, only menopausal status was associated with both BMI (p-value <0.05) and gene methylation (false discovery rate (FDR) q-value <0.05), resulting in a final adjustment set that included age, race, and menopausal status. In addition to testing the association between BMI and gene methylation in breast tumor tissue overall, the relationship was also assessed among ER-positive tumors because prior research has shown that this subgroup of tumors may be more stable genomically than other tumor subtypes (22, 23). In a sensitivity analysis restricted to ER-negative tumors, there were no CpG sites associated with obesity in a regression model adjusting for age, race, and menopausal status.
An FDR q-value of <0.05 was used as the statistical significance cut-off for identifying differentially methylated genes.
Though we did not have data on gene expression for the tissue analyzed in this study, we assessed the correlation between methylation and expression in breast tumors that were collected through The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) for the methylation probes that were significantly associated with BMI in our study. The TCGA determined methylation in breast tumors using the Illumina HumanMethylation 450 Beadchip panel (24) while the present study used the Illumina GoldenGate Cancer I Panel. Only seven of the BMI-associated probes in the present study matched directly with probes in the 450 Beadchip panel. For the remaining BMI-associated probes, we identified probes on the 450 Beadchip array that were within 200 base pairs, either up or downstream, of the GoldenGate probe. The correlation between methylation and expression was examined for all of the breast tumor tissue available in the TCGA and also for hormone-receptor positive tumors (the closest approximation to ER-positive tumors for which we had data access).
Results
Methylation Sites Associated with BMI
Of the 517 patients eligible for study participation, 492 had both pre-diagnosis BMI data and methylation data; of these, 345 were either obese or of normal weight (Table 1). The median age for the 492 study participants was 47 years. In unadjusted analyses, 30 CpG sites were differentially methylated by BMI status (obese vs. normal) at a q-value < 0.05; after adjustment for age, race, and menopausal status, two CpG sites remained significantly differentially methylated: SH3BP2_P771_R on the promoter region of the SH3BP2 gene, which regulates transcriptional activity in immune cells, and Xist_seq_80_S95_R on the non-protein coding XIST gene. For both sites, the average methylation beta values were higher in the tissue of obese patients, when compared to those of normal weight.
Table 1.
Selected characteristics of eligible breast cancer cases in the Carolina Breast Cancer Study, Phases 1
| Totala | Normalb | Overweightb | Obeseb | |||||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | |
| Total | 492 | 100% | 195 | 100% | 147 | 100% | 150 | 100% |
| Race | ||||||||
| African-American | 200 | 40.7% | 37 | 19.0% | 61 | 41.5% | 102 | 68.0% |
| White | 282 | 57.3% | 152 | 77.9% | 83 | 56.5% | 47 | 31.3% |
| Other | 10 | 2.0% | 6 | 3.1% | 3 | 2.0% | 1 | 0.7% |
| Menopausal Status | ||||||||
| Premenopausal | 262 | 53.3% | 108 | 55.4% | 78 | 53.1% | 76 | 50.7% |
| Postmenopausal | 230 | 46.7% | 87 | 44.6% | 69 | 46.9% | 74 | 49.3% |
| Alcohol Use | ||||||||
| Never | 141 | 28.7% | 41 | 21.0% | 45 | 30.6% | 55 | 36.7% |
| Ever | 351 | 71.3% | 154 | 79.0% | 102 | 69.4% | 95 | 63.3% |
| Smoking Status | ||||||||
| Never | 251 | 51.0% | 93 | 47.7% | 79 | 53.7% | 79 | 52.7% |
| Former | 146 | 29.7% | 57 | 29.2% | 42 | 28.6% | 47 | 31.3% |
| Current | 95 | 19.3% | 45 | 23.1% | 26 | 17.7% | 24 | 16.0% |
| Income, $ | ||||||||
| <15,000 | 119 | 24.2% | 31 | 17.2% | 30 | 22.6% | 58 | 40.3% |
| 15,000 – 30,000 | 99 | 20.1% | 38 | 21.1% | 27 | 20.3% | 34 | 23.6% |
| 30,000 – 50,000 | 113 | 23.0% | 50 | 27.8% | 33 | 24.8% | 30 | 20.8% |
| ≥50,000 | 126 | 25.6% | 61 | 33.9% | 43 | 32.3% | 22 | 15.3% |
| Educationc | ||||||||
| <High School | 90 | 18.3% | 18 | 9.2% | 26 | 17.7% | 46 | 30.7% |
| High School | 269 | 54.7% | 107 | 54.9% | 87 | 59.2% | 75 | 50.0% |
| College | 133 | 27.0% | 70 | 35.9% | 34 | 23.1% | 29 | 19.3% |
| ER-Positived | ||||||||
| Yes | 285 | 59.6% | 118 | 62.1% | 77 | 54.2% | 90 | 61.6% |
| No | 193 | 40.4% | 72 | 37.9% | 65 | 45.8% | 56 | 38.4% |
| Stagee | ||||||||
| I | 168 | 36.7% | 73 | 38.8% | 49 | 36.8% | 46 | 33.6% |
| II | 238 | 52.0% | 99 | 52.7% | 70 | 52.6% | 69 | 50.4% |
| III & IV | 52 | 11.4% | 16 | 8.5% | 14 | 10.5% | 22 | 16.1% |
Columns may not sum to total due to missing data
Normal weight: 18.5 ≤ BMI < 25; overweight: 25 ≤ BMI < 30; obese: BMI ≥ 30
High School: high school completion (or equivalent), technical or business school, and some college; College: college completion and post-graduate school or professional degree
ER=Estrogen-Receptor
According to AJCC breast tumor staging guidelines
Methylation in ER-Positive Tumors
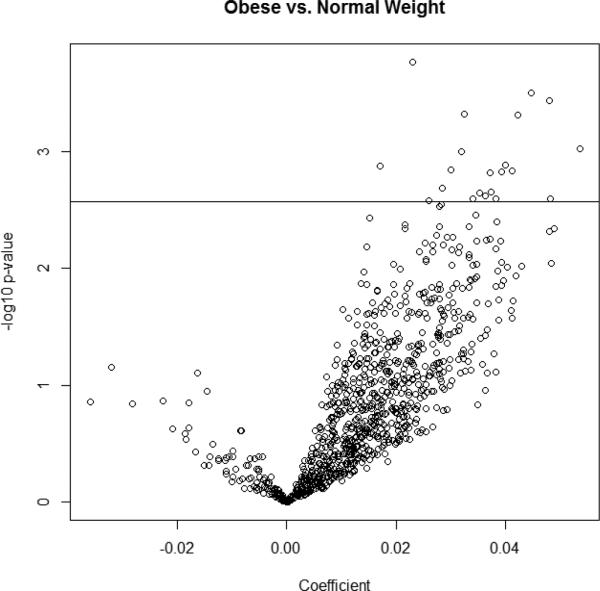
In analyses restricted to ER-positive tumors (n=208), 21 probes were significantly associated with BMI (obese vs. normal) after adjustment for age, race, and menopausal status (Table 2). Twelve of the 21 significant sites were located in CpG islands. Three CpG sites mapped to chromosome 4 (SH3BP2_P771_R, SH3BP2_E18_F, and PKD2_P336_R), three mapped to the X chromosome (Xist_seq_80_S95_R, MCF2_P1024_R, and BTK_P105_F), and two each mapped to chromosome 10 (ERCC6_P698_R, BMPR1A_E88_F), chromosome 11 (TSG101_P257_R, SPI1_E205_F), and chromosome 12 (IGFBP6_E47_F, ARHGDIB_P148_R). Two of the probes (BMPR1A_E88_F and TDGF1_P428_R) mapped to more than one chromosome; as such, these probes should be interpreted with caution. Figure 1 displays a volcano plot, a visual representation of the regression analysis wherein each circle represents a methylation site; circles with positive coefficients signify higher average beta values in the obese compared to those of normal weight, while circles with negative coefficients signify lower beta values in the obese. The majority of the sites were hypermethylated in the obese group, as were all of the probes that were significantly associated with obesity (circles above the horizontal line).
Table 2.
Methylation sites associated with BMI (FDR <.05) in the Carolina Breast Cancer Study among patients with estrogen receptor-positive tumors
| Probe ID | Gene | Gene Description | CHRa | Islandb | Coefficient | p-value | q-value | Normalc | Overweightc,d | Obesec |
|---|---|---|---|---|---|---|---|---|---|---|
| DNMT3B_P352_R | DNMT3B | DNA cytosine-5 methyltransferase 3 beta isoform 2 | 20q11 | N | 0.0230 | 0.00017 | 0.03818 | 15.5% | 17.3% | 17.6% |
| IGFBP6_E47_F | IGFBP6 | Insulin-like growth factor binding protein 6 | 12q13 | N | 0.0447 | 0.00032 | 0.03818 | 37.5% | 40.5% | 43.3% |
| ERCC6_P698_R | ERCC6 | Excision repair cross-complementing rodent repair deficiency, complementation group 6 | 10q11 | Y | 0.0480 | 0.00037 | 0.03818 | 43.1% | 46.8% | 47.3% |
| SH3BP2_P771_Re | SH3BP2 | SH3-domain binding protein 2 | 4p16 | Y | 0.0324 | 0.00048 | 0.03818 | 17.4% | 20.8% | 22.7% |
| PLAGL1_E68_R | PLAGL1 | Pleiomorphic adenoma gene-like 1 isoform 1 | 6q24-25 | Y | 0.0422 | 0.00049 | 0.03818 | 69.8% | 72.5% | 75.4% |
| FZD9_E458_F | FZD9 | Frizzled 9 | 7q11 | Y | 0.0535 | 0.00096 | 0.04640 | 49.2% | 50.0% | 55.4% |
| BMPR1A_E88_F | BMPR1A | Bone morphogenetic protein receptor, type IA precursor | 10q22 | Y | 0.0320 | 0.00101 | 0.04640 | 65.0% | 66.7% | 69.2% |
| TSG101_P257_R | TSG101 | Tumor susceptibility gene 101 | 11p15 | Y | 0.0400 | 0.00132 | 0.04640 | 25.4% | 29.0% | 29.8% |
| RIPK4_E166_F | RIPK4 | receptor-interacting serine-threonine kinase 4 | 21q22 | Y | 0.0171 | 0.00133 | 0.04640 | 7.2% | 9.6% | 10.1% |
| Xist_seq_80_S95_Re | XIST | X inactive specific transcript (non-protein coding) | Xq13 | N | 0.0299 | 0.00143 | 0.04640 | 74.3% | 76.3% | 78.9% |
| TDGF1_P428_R | TDGF1 | Teratocarcinoma-derived growth factor 1 | 3p21 | N | 0.0412 | 0.00147 | 0.04640 | 40.7% | 41.8% | 44.6% |
| MCF2_P1024_R | MCF2 | MCF.2 cell line derived transforming sequence | Xq27 | N | 0.0393 | 0.00151 | 0.04640 | 77.6% | 80.6% | 82.3% |
| SPI1_E205_F | SPI1 | Spleen focus forming virus (SFFV) proviral integration oncogene spi1 | 11p11 | Y | 0.0372 | 0.00154 | 0.04640 | 54.1% | 57.8% | 58.8% |
| SH3BP2_E18_F | SH3BP2 | SH3-domain binding protein 2 | 4p16 | N | 0.0284 | 0.00208 | 0.04934 | 25.3% | 28.6% | 29.9% |
| ABO_E110_F | ABO | ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) | 9q34 | Y | 0.0373 | 0.00225 | 0.04934 | 15.8% | 18.0% | 20.6% |
| CHI3L2_P226_F | CHI3L2 | Chitinase 3-like 2 isoform c | 1p13 | N | 0.0352 | 0.00227 | 0.04934 | 74.8% | 75.2% | 74.6% |
| BTK_P105_F | BTK | Bruton agammaglobulinemia tyrosine kinase | Xq21-22 | N | 0.0363 | 0.00240 | 0.04934 | 39.6% | 42.4% | 44.8% |
| SNURF_P78_F | SNURF | SNRPN upstream reading frame protein | 15q11-12 | Y | 0.0381 | 0.00254 | 0.04934 | 69.5% | 71.0% | 74.3% |
| ARHGDIB_P148_R | ARHGDIB Rho GDP dissociation inhibitor (GDI) beta | 12p12 | N | 0.0340 | 0.00255 | 0.04934 | 44.5% | 45.5% | 47.0% | |
| SEZ6L_P299_F | SEZ6L | Seizure related 6 homolog (mouse)-like precursor | 22q12 | Y | 0.0482 | 0.00255 | 0.04934 | 17.5% | 18.0% | 21.3% |
| PKD2_P336_R | PKD2 | Polycystin 2 | 4q22 | Y | 0.0260 | 0.00265 | 0.04934 | 13.1% | 14.6% | 16.5% |
CHR = Chromsome.
Island indicates whether the probe is located in a CpG Island; N = no; Y = yes.
Unadjusted % Methylation; Normal weight: 18.5 ≤ BMI < 25; overweight: 25 ≤ BMI < 30; obese BMI ≥ 30).
Overweight individuals were not included in the regression analysis, but are included here for descriptive purposes.
Associated with BMI in overall breast tumor tissue regression analysis
Figure 1.
Volcano plot of 935 methylation sites in estrogen receptor-positive tumors. X-axis: Coefficient from model regressing obesity on CpG site, adjusting for age, race, and menopausal status; Y-axis: -log10 of p-value from model regressing obesity on CpG site, adjusting for age, race, and menopausal status. Horizontal line indicates FDR q-value cut-off of 0.05. Each circle represents a CpG site.
The methylation sites that showed the most significant changes in the beta values between obese vs. normal BMI patients were on the DNMT3B gene, which codes for a DNA methyltransferase; the IGFBP6 gene, which codes for an insulin-like growth factor binding protein; the ERCC6 gene, which codes for a protein that functions in excision repair; and the SH3BP2 gene.
Correlation with Gene Expression
The 21 methylation sites that were associated with BMI in the overall or ER-positive analyses were mapped to breast tumors collected by The Cancer Genome Atlas (TCGA) to assess the correlation between gene methylation and expression. Of the 21 sites, 20 matched to probes that were at least within 200 base pairs of the probe used in the TCGA data, and three sites matched to more than one probe (Supplemental Table 1). Approximately 60% of the 20 BMI-associated methylation sites were significantly and inversely associated with gene expression (correlation coefficient ≤ −0.20) in the full TCGA breast tumor tissue data set, and 55% were significantly and inversely associated with expression in the hormone-receptor positive tumors. Though probes that were correlated with gene expression in both the full TCGA dataset and amongst hormone-positive tumors were more likely to be on CpG islands than probes that were not associated with gene expression, the associations were not significant at an alpha = 0.05 level (p-value = 0.36 and .17, respectively).
Discussion
Because methylation is a mechanism for controlling gene expression (8), exploring the relationship between obesity and methylation can provide insight on BMI-associated expression of cancer-related genes. Our regression analyses suggest that BMI is associated with gene methylation in cancer-related genes overall and especially among ER-positive breast tumors. When assessing the correlation between methylation and expression for our BMI-associated methylation sites (from both our overall and ER-positive analyses) in breast tumors collected through The Cancer Genome Atlas, about 60% showed a significant inverse correlation between methylation and expression.
Previous research in breast tissue has demonstrated that BMI is associated with the differential expression of genes involved in pathways related to immune response and insulin-like growth factor availability (20, 25). In our study, BMI was associated with methylation of genes involved not only in immune response and insulin-like growth factor pathways, but also in DNA methylation and DNA repair. Notably, SH3BP2 was the most significant BMI-associated site in our overall analysis, while SH3BP2, IGFBP6, DNMT3, and ERCC6 were the most statistically significant BMI-associated sites among ER-positive tumors. All four of these genes were hypermethylated with obesity. The SH3BP2 gene codes for a protein that acts in the signal transduction of various immune response pathways (26). It has been implicated in responses involving neutrophils (27), B cells (28-30), and T cells (26), and it may function as a tumor suppressor gene (26). Our finding that SH3BP2 is hypermethylated in the tumor tissue of obese breast cancer patients suggests that obesity may confer an abnormal immune response in breast tissue. This is interesting in light of previous research also showing increased infiltration of macrophages in the breast tissue of obese women (20, 31). The IGFBP6 gene codes for a protein that binds with high affinity to insulin-like growth factor-2 (IGF-II) (32), which plays a role in cellular proliferation, differentiation, and migration, and is regulated by the IGFBP6 protein (32). Expression of IGFBP6 may be associated with cancer incidence (32), with studies finding lower expression of the protein in the serum of breast cancer patients when compared to the serum of those with benign breast disease (33, 34). Our finding of hypermethylation of IGFBP6 in obese patients suggests that obesity may lead to reduced expression in breast tissue via gene methylation. BMI may also impact broader mechanisms controlling methylation. For example, DNMT3 is a member of a family of DNA methyltransferases that code for enzymes involved in the methylation of promoter regions of genes (35), and DNMT3 mRNA has been shown to be overexpressed in breast tumor tissue (35, 36) and cell lines (37). Finally, the ERCC6 protein, also known as the Cockayne syndrome complementation group B (CSB) protein, is involved in transcription-coupled nucleotide excision DNA repair (38).
Our study was strengthened by examining the association between BMI and methylation in breast tumor tissue rather than white blood cells (WBCs), as in most previous studies. Nonetheless, prior research has shown that differences in methylation between those with and without cancer may be driven by changes in the distribution of WBC populations (39). Obesity may also be associated with changes in the distribution of WBC populations, resulting in methylation differences by BMI status. In one study of WBCs, methylation was increased in those with higher BMI for three of four differentially methylated CpG sites (MMP9, PM20D1, and PRKG1) (12). Of those sites, only MMP9 was included in the methylation panel used in this study, and it was not associated with obesity either overall or among ER-positive tumors. In another study of methylation in WBCs, BMI was associated with increased methylation of several sites on HIF3A, a gene that encodes for a protein involved in responses to hypoxia (11). However, in the latter study, the magnitude of change was not replicated when the HIF3A loci were examined in adipose tissue, underscoring the importance of tissue specific methylation. There were no CpG sites on the HIF3A gene in the methylation panel used for this study.
Given previous reports of differentially methylated genes associated with BMI in both genome wide and candidate loci studies (11, 12), it is interesting that studies examining global methylation in WBCs are inconclusive regarding the direction of methylation in response to increasing BMI. In a study of global methylation in 85 women, overweight and obese patients had non-statistically significant higher levels of methylation than those with a normal BMI after adjustment for variables including childhood smoking exposure, nulliparity, and age at 1st birth (40). When methylation of the repetitive element, LINE-1, was measured in the peripheral blood of men and women, those with a normal BMI had the lowest mean methylation percentage and those who were obese had the highest methylation percentages, though the results were not statistically significant in crude analyses (41). Other studies reviewed in Terry et al (42) did not find significant correlations between methylation values in the LINE-1 and ALU repetitive elements and BMI (43, 44), and one study examining BMI in women of childbearing age reported less LINE-1 methylation in those with higher BMI (45).
These aforementioned studies suggest that obesity may be weakly associated with an increase in methylation in WBCs, but do not address whether BMI may affect methylation in breast tissue. Given our emphasis on the tumor tissue, the most relevant previous research for our purposes are studies of breast tumor methylation and BMI (13, 46). In one study, Tao et al. examined the association between obesity and methylation in three candidate genes, E-cadherin, p16, and RAR-β(2), in breast tumor tissue, and showed no statistically significant differences in percentage methylation by BMI status in their study population; regression analyses stratified by ER status were not reported (13). Though the three genes examined by Tao et al. (13) were represented on our methylation panel, we were unable to ascertain the exact CpG loci on which methylation values were measured. Given that DNA methylation levels can vary greatly within short base pair distances (47), we were unable to meaningfully compare our gene methylation results with those of Tao et al. In another study, Naudshad et al. examined the association between BMI and methylation in Ec-SOD, RASSF1, BRCA1, and BNIP3; all of the gene loci except for BNIP3 were significantly, increasingly methylated with higher BMI while BNIP3 was significantly, inversely associated with BMI (46). All but the BNIP3 gene examined by Naushad et al. (46) were represented on our methylation panel. However, there were no exact loci matches between the probes available on our methylation panel and the Ec-SOD and BRCA1 sites examined by Naushad et al.; we were unable to determine the location of the RASSF1 probe. Because gene methylation is loci-specific (47), we were unable to compare their reported methylation results to our findings.
Our study is novel in that it assesses the relationship between BMI and methylation in breast tumor tissue using a panel of cancer-associated genes. This enabled us to discern relationships between obesity and methylation that might not be apparent in WBCs or that might be overlooked by focusing on a small number of candidate genes. Nonetheless, because the methylation array was limited to cancer-relevant genes, only a subset of genes that may be differentially methylated by obese status were analyzed. Thus, our results are biased towards methylation sites with established associations with cancer. Further, there is evidence that genes with known relevance to cancer can be either hypermethylated or hypomethylated relative to normal breast tissue (10, 48). However, the goal of our study was to determine the association between obesity and methylation status, regardless of the genes’ hyper- or hypomethylation status relative to normal tissue. Our analysis demonstrates that obesity is associated with the degree of methylation in some cancer-relevant genes, providing insight into which genes should be targeted by research for further exploration.
Though our study was strengthened by assessing the correlation between methylation and expression for our BMI-associated sites, we were unable to assess whether these differences in expression correlated with functional differences. Additionally, we were unable to validate our results in an independent dataset because we were not able to identify public data sources where both methylation and BMI were measured and reported. The Cancer Genome Atlas, for example, does not collect data on BMI. Our study, however, contributes data on both BMI and methylation status to the public domain, and future studies can seek to replicate our findings using these data.
Our analysis was also strengthened by analyzing ER-positive tumors, which have been shown to feature fewer chromosomal aberrations than other tumor subtypes (22, 23). However, there were some limitations associated with our analysis. Although tissue from ER-positive tumors may be more stable than that from other tumor subtypes, in the analysis of any tumor tissue, it is difficult to determine which methylation changes influence the carcinogenic process and which occur as a result of it (49). Furthermore, there may be heterogeneity within ER-positive tumors (i.e. some tumors are luminal A and others luminal B) that may have influenced our power to detect associations between methylation and obesity. A further limitation of the study design was the lack of power to adequately assess the association between BMI and gene methylation by menopausal status. Given the variability in the obesity-breast cancer association by menopausal status (1, 2), it is possible that different or additional genes would be associated with methylation by menopausal status. Finally, the motivating goal of this study was to explore whether there might be epigenetic biomarkers of the effect of obesity on epithelial tumor cells that can be used to direct research of targeted breast cancer therapies. However, a focus on normal breast tissue is also important for future research, as it can give insight into how obesity may influence cancer initiation and promotion.
In summary, our study provides evidence that obesity may be associated with increased gene methylation in breast tumor tissue and may play a role in the expression of genes associated with immune response, insulin-like growth factor availability, and other pathways involved in carcinogenesis. If obesity does act through methylation to control expression of carcinogenesis-related genes, differentially methylated sites could serve as targets for the chemoprevention and treatment of breast cancer in women with obesity as a risk factor.
Supplementary Material
Acknowledgments
Financial Support: This research was funded, in part, by the University Cancer Research Fund of North Carolina, the National Cancer Institute Specialized Program of Research Excellence (SPORE) in Breast Cancer (NIH/ NCI P50-CA58223), the Lineberger Comprehensive Cancer Center Core Grant (NIH/NCI P30-CA16086). Support for K. Conway was provided by a grant from Susan G. Komen for the Cure (#KG081397). Support or W.R. Robinson was provided by the National Cancer Institute (1K01CA172717-01). Support for M.A. Troester was provided by the National Cancer Institute/National Institute of Environmental Health Sciences Breast Cancer and the Environment Research Program (U01-ES019472).
Footnotes
Conflict of Interest: The authors report no conflicts of interest.
References
- 1.Carmichael AR, Bates T. Obesity and breast cancer: a review of the literature. Breast. 2004;13:85–92. doi: 10.1016/j.breast.2003.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Ursin G, Longnecker MP, Haile RW, Greenland S. A meta-analysis of body mass index and risk of premenopausal breast cancer. Epidemiology. 1995;6:137–41. doi: 10.1097/00001648-199503000-00009. [DOI] [PubMed] [Google Scholar]
- 3.White KK, Park SY, Kolonel LN, Henderson BE, Wilkens LR. Body size and breast cancer risk: The multiethnic cohort. Int J Cancer. 2011 doi: 10.1002/ijc.27373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmichael AR. Obesity as a risk factor for development and poor prognosis of breast cancer. BJOG. 2006;113:1160–6. doi: 10.1111/j.1471-0528.2006.01021.x. [DOI] [PubMed] [Google Scholar]
- 5.Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103:250–63. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enger SM, Ross RK, Paganini-Hill A, Carpenter CL, Bernstein L. Body size, physical activity, and breast cancer hormone receptor status: results from two case-control studies. Cancer Epidemiol Biomarkers Prev. 2000;9:681–7. [PubMed] [Google Scholar]
- 7.Loi S, Milne RL, Friedlander ML, McCredie MR, Giles GG, Hopper JL, et al. Obesity and outcomes in premenopausal and postmenopausal breast cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1686–91. doi: 10.1158/1055-9965.EPI-05-0042. [DOI] [PubMed] [Google Scholar]
- 8.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–70. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 10.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–82. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- 11.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014 doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 12.Feinberg AP, Irizarry RA, Fradin D, Aryee MJ, Murakami P, Aspelund T, et al. Personalized epigenomic signatures that are stable over time and covary with body mass index. Sci Transl Med. 2010;2:49ra67. doi: 10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tao MH, Marian C, Nie J, Ambrosone C, Krishnan SS, Edge SB, et al. Body mass and DNA promoter methylation in breast tumors in the Western New York Exposures and Breast Cancer Study. Am J Clin Nutr. 2011;94:831–8. doi: 10.3945/ajcn.110.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moorman PG, Newman B, Millikan RC, Tse CK, Sandler DP. Participation rates in a case- control study: the impact of age, race, and race of interviewer. Ann Epidemiol. 1999;9:188–95. doi: 10.1016/s1047-2797(98)00057-x. [DOI] [PubMed] [Google Scholar]
- 15.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–93. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18:4808–17. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conway K, Edmiston SN, May R, Kuan P, Chu H, Bryant C, et al. DNA methylation profiling in the Carolina Breast Cancer Study defines cancer subclasses differing in clinicopathologic characteristics and survival. Breast Cancer Res. 2014;16:450. doi: 10.1186/s13058-014-0450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2015. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X, Su S, Barnes VA, De Miguel C, Pollock J, Ownby D, et al. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics. 2013;8:522–33. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun X, Casbas-Hernandez P, Bigelow C, Makowski L, Joseph Jerry D, Smith Schneider S, et al. Normal breast tissue of obese women is enriched for macrophage markers and macrophage- associated gene expression. Breast Cancer Res Treat. 2012;131:1003–12. doi: 10.1007/s10549-011-1789-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorber SC, Tremblay M, Moher D, Gorber B. A comparison of direct vs. self-report measures for assessing height, weight and body mass index: a systematic review. Obes Rev. 2007;8:307–26. doi: 10.1111/j.1467-789X.2007.00347.x. [DOI] [PubMed] [Google Scholar]
- 22.Fang M, Toher J, Morgan M, Davison J, Tannenbaum S, Claffey K. Genomic differences between estrogen receptor (ER)-positive and ER-negative human breast carcinoma identified by single nucleotide polymorphism array comparative genome hybridization analysis. Cancer. 2011;117:2024–34. doi: 10.1002/cncr.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smid M, Hoes M, Sieuwerts AM, Sleijfer S, Zhang Y, Wang Y, et al. Patterns and incidence of chromosomal instability and their prognostic relevance in breast cancer subtypes. Breast Cancer Res Treat. 2011;128:23–30. doi: 10.1007/s10549-010-1026-5. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Suga K, Imai K, Eguchi H, Hayashi S, Higashi Y, Nakachi K. Molecular significance of excess body weight in postmenopausal breast cancer patients, in relation to expression of insulin-like growth factor I receptor and insulin-like growth factor II genes. Jpn J Cancer Res. 2001;92:127–34. doi: 10.1111/j.1349-7006.2001.tb01074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deckert M, Rottapel R. The adapter 3BP2: how it plugs into leukocyte signaling. Adv Exp Med Biol. 2006;584:107–14. doi: 10.1007/0-387-34132-3_8. [DOI] [PubMed] [Google Scholar]
- 27.Chen G, Dimitriou I, Milne L, Lang KS, Lang PA, Fine N, et al. The 3BP2 adapter protein is required for chemoattractant-mediated neutrophil activation. J Immunol. 2012;189:2138–50. doi: 10.4049/jimmunol.1103184. [DOI] [PubMed] [Google Scholar]
- 28.Chen G, Dimitriou ID, La Rose J, Ilangumaran S, Yeh WC, Doody G, et al. The 3BP2 adapter protein is required for optimal B-cell activation and thymus-independent type 2 humoral response. Mol Cell Biol. 2007;27:3109–22. doi: 10.1128/MCB.01014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de la Fuente MA, Kumar L, Lu B, Geha RS. 3BP2 deficiency impairs the response of B cells, but not T cells, to antigen receptor ligation. Mol Cell Biol. 2006;26:5214–25. doi: 10.1128/MCB.00087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shukla U, Hatani T, Nakashima K, Ogi K, Sada K. Tyrosine phosphorylation of 3BP2 regulates B cell receptor-mediated activation of NFAT. J Biol Chem. 2009;284:33719–28. doi: 10.1074/jbc.M109.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howe LR, Subbaramaiah K, Hudis CA, Dannenberg AJ. Molecular pathways: adipose inflammation as a mediator of obesity-associated cancer. Clin Cancer Res. 2013;19:6074–83. doi: 10.1158/1078-0432.CCR-12-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bach LA, Fu P, Yang Z. Insulin-like growth factor-binding protein-6 and cancer. Clin Sci (Lond) 2013;124:215–29. doi: 10.1042/CS20120343. [DOI] [PubMed] [Google Scholar]
- 33.Kaulsay KK, Ng EH, Ji CY, Ho GH, Aw TC, Lee KO. Serum IGF-binding protein-6 and prostate specific antigen in breast cancer. Eur J Endocrinol. 1999;140:164–8. doi: 10.1530/eje.0.1400164. [DOI] [PubMed] [Google Scholar]
- 34.Pazaitou-Panayiotou K, Kelesidis T, Kelesidis I, Kaprara A, Blakeman J, Vainas I, et al. Growth hormone-binding protein is directly and IGFBP-3 is inversely associated with risk of female breast cancer. Eur J Endocrinol. 2007;156:187–94. doi: 10.1530/EJE-06-0611. [DOI] [PubMed] [Google Scholar]
- 35.Girault I, Tozlu S, Lidereau R, Bieche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–22. [PubMed] [Google Scholar]
- 36.Veeck J, Esteller M. Breast cancer epigenetics: from DNA methylation to microRNAs. J Mammary Gland Biol Neoplasia. 2010;15:5–17. doi: 10.1007/s10911-010-9165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer. 2008;7(15):4598–7-15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lake RJ, Fan HY. Structure, function and regulation of CSB: a multi-talented gymnast. Mech Ageing Dev. 2013;134:202–11. doi: 10.1016/j.mad.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koestler DC, Marsit CJ, Christensen BC, Accomando W, Langevin SM, Houseman EA, et al. Peripheral blood immune cell methylation profiles are associated with nonhematopoietic cancers. Cancer Epidemiol Biomarkers Prev. 2012;21:1293–302. doi: 10.1158/1055-9965.EPI-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, et al. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev. 2008;17:2306–10. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623–9. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–37. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim KY, Kim DS, Lee SK, Lee IK, Kang JH, Chang YS, et al. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy Koreans. Environ Health Perspect. 2010;118:370–4. doi: 10.1289/ehp.0901131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang FF, Cardarelli R, Carroll J, Zhang S, Fulda KG, Gonzalez K, et al. Physical activity and global genomic DNA methylation in a cancer-free population. Epigenetics. 2011;6:293–9. doi: 10.4161/epi.6.3.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piyathilake C, Badiga S, Johanning G, Alvarez R, Partridge E. Predictors and Health Consequences of Epigenetic Changes Associated with Excess Body Weight in Women of Child- bearing Age. Cancer Epidemiol Biomarkers Prev. 2011;20:719. [Google Scholar]
- 46.Naushad SM, Hussain T, Al-Attas OS, Prayaga A, Digumarti RR, Gottumukkala SR, et al. Molecular insights into the association of obesity with breast cancer risk: relevance to xenobiotic metabolism and CpG island methylation of tumor suppressor genes. Mol Cell Biochem. 2014;392:273–80. doi: 10.1007/s11010-014-2037-z. [DOI] [PubMed] [Google Scholar]
- 47.Bardowell SA, Parker J, Fan C, Crandell J, Perou CM, Swift-Scanlan T. Differential methylation relative to breast cancer subtype and matched normal tissue reveals distinct patterns. Breast Cancer Res Treat. 2013;142:365–80. doi: 10.1007/s10549-013-2738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jackson K, Yu MC, Arakawa K, Fiala E, Youn B, Fiegl H, et al. DNA hypomethylation is prevalent even in low-grade breast cancers. Cancer Biol Ther. 2004;3:1225–31. doi: 10.4161/cbt.3.12.1222. [DOI] [PubMed] [Google Scholar]
- 49.Choi JD, Lee JS. Interplay between Epigenetics and Genetics in Cancer. Genomics Inform. 2013;11:164–73. doi: 10.5808/GI.2013.11.4.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.