Abstract
L-selectin is constitutively expressed on leukocytes and mediates their interaction with endothelial cells during inflammation. Previous studies on the association of soluble L-selectin (sL-selectin) with cardiovascular disease (CVD) are inconsistent. Genetic variants associated with sL-selectin levels may be a better surrogate of levels over a lifetime. We explored the association of genetic variants and sL-selectin levels in a race/ethnicity stratified random sample of 2,403 participants in the Multi-Ethnic Study of Atherosclerosis (MESA). Through a genome-wide analysis with additive linear regression models, we found that rs12938 on the SELL gene accounted for a significant portion of the protein level variance across all four races/ethnicities. To evaluate potential additional associations, elastic net models were used for variants located in the SELL/SELP/SELE genetic region and an additional two SNPs, rs3917768 and rs4987361, were associated with sL-selectin levels in African Americans. These variants accounted for a portion of protein variance that ranged from 4% in Hispanic to 14% in African Americans. To investigate the relationship of these variants with CVD, 6,317 subjects were used. No significant association was found between any of the identified SNPs and carotid intima-media thickness or presence of carotid plaque using linear and logistic regression, respectively. Similarly no significant results were found for coronary artery calcium or coronary heart disease events. In conclusion, we found that variants within the SELL gene are associated with sL-selectin levels. Despite accounting for a significant portion of the protein level variance, none of the variants was associated with clinical or subclinical CVD.
Keywords: atherosclerosis, cardiovascular disease, genetic epidemiology, single nucleotide polymorphism (SNP)
Introduction
Cardiovascular disease (CVD) is the main cause of morbidity and mortality in the United States (Go et al. 2014). Inflammation plays a pivotal role in atherosclerosis; in particular, the adhesion of leukocyte to endothelial cells represents an early step of plaque formation (Libby 2002; Ross 1999; Williams and Tabas 2002). L-selectin is constitutively expressed on leukocytes and has an essential role on the initial steps of their adhesion to the endothelium during inflammation (Wedepohl et al. 2012). Given its activity during early recruitment of leukocytes at inflammatory foci, previous in vivo studies suggested a potential role of L-selectin in the development and progression of subclinical atherosclerosis (Eriksson et al. 2001; Galkina et al. 2006). L-selectin is enzymatically cleaved from the leukocyte surface (Humbria et al. 1994).
Endoproteolytic cleavage regulates both homeostatic and activation-induced changes in cell surface L-selectin density during inflammation. In particular, blocking L-selectin shedding resulted in enhanced cell-bound L-selectin expression and increase neutrophils migration to inflamed areas (Venturi et al. 2003). On the contrary, there is some evidence that soluble L-selectin can competitively block other selectins receptors reducing leukocyte-endothelium interactions. Relatively high levels of soluble L-selectin (sL-selectin) were observed in the general population, suggesting a possible role of the shedding process in the physiologic surface-bound protein turnover (Ponthieux et al. 2004). In addition, previous studies observed a long half-life for sL-selectin in animal models, suggesting that levels may remain elevated for many hours after the protein has been shed (Tu et al. 2002).
Only a few case-control studies have assessed the association of sL-selectin with CVD, with results being inconsistent. Within the Multi-Ethnic Study of Atherosclerosis (MESA) cohort, we found no association between plasma or serum sL-selectin levels and subclinical or clinical CVD (Berardi et al. 2014). However, genetic variants associated with the protein levels may be a better surrogate of levels of sL-selectin over time, and exploring the association of these variants with clinical and subclinical CVD could shed light on the importance of this protein in the pathogenesis of CVD. This approach was previously used for P-selectin; in particular, multiple variants of the SELP gene were shown to be associated with circulating P-selectin levels in community-based samples (Lee et al. 2008; Reiner et al. 2008). Similarly, several variants of the ICAM1 gene have been associated with soluble ICAM-1 levels (Bielinski et al. 2008; Bielinski et al. 2011).
Little is currently known about the genetic determinants of sL-selectin levels, with previous association studies limited to two relatively small studies evaluating candidate single nucleotide polymorphisms (SNP) in the local region of the L-selectin protein coding gene (SELL). Wei et al. identified a significant association with SELL missense SNP rs2229569 (C>T; p.P226S) in a Chinese population, determining the SNP to also be associated with ischemic stroke (Wei et al. 2011). Russell et al. identified significant associations with SELL missense variant rs1131498 (previously denoted rs3177980; T>C, p.F206L) and 3′ untranslated region (UTR) SNP rs12938 (T>C) in a study of systemic lupus erythematosus (Russell et al. 2005). Hajilooi et al. additionally identified rs1131498 to be associated with coronary heart disease (CHD) in an Iranian population (Hajilooi et al. 2006). We sought to identify potential genetic surrogates of circulating sL-selectin levels in a large multi-ethnic population using comprehensive genetic data heavily enriched for protein coding variation. We investigated the importance of genetic variants in determining the levels of circulating sL-selectin and the association of these variants with clinical and subclinical CVD.
Materials and methods
Multi-Ethnic Study of Atherosclerosis (MESA) population
The Multi-Ethnic Study of Atherosclerosis (MESA) is a multi-center population-based prospective cohort study initiated in July 2000 to investigate subclinical cardiovascular endpoints in 6,814 African, non-Hispanic white, Chinese, and Hispanic American men and women. MESA participants were examined at one of six field centers located in Baltimore, MD; Chicago, IL; Forsyth County, NC; Los Angeles County, CA; Northern Manhattan, NY; and Saint Paul, MN. Detailed methods have been described elsewhere.(Bild et al. 2002)
At each visit, information on demographics, cardiovascular risk factors, past medical history and co-morbidities, social history, family history, and medications was collected through a combination of self-administered questionnaires and interview-administered questionnaires. Height was measured while participants were standing without shoes, heels together against a vertical mounted ruler. BMI was calculated as weight (kg)/height2 (m2). Resting seated blood pressure was measured three times using an automated oscillometric method (Dinamap), and the average of the second and third readings are used in analyses. Hypertension was defined according to the Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure (JNC 7) guidelines as systolic blood pressure ≥140 mmHg, diastolic blood pressure ≥90 mmHg, or use of anti-hypertensive medications (Chobanian et al. 2003). Diabetes was defined as any participant who self-reported a physician diagnosis, used diabetic medication, had a fasting glucose ≥126 mg/dL, or a non-fasting glucose of ≥200 mg/dL. Serum glucose was assayed by a hexokinase/glucose-6-phosphate dehydrogenase method. Triglycerides were measured in plasma by a glycerol blanked enzymatic method, and cholesterol was measured in plasma using a cholesterol oxidase method. HDL cholesterol was measured by the cholesterol oxidase method after precipitation of non-HDL-cholesterol with magnesium/dextran. LDL-cholesterol was calculated in specimens having a triglyceride <400 mg/dL via the Friedewald equation.
Genetic association analysis sample
To identify genetic variants associated with circulating sL-selectin levels, a race/ethnicity stratified random sample of 2,880 individuals was used. At Exam 2, the first follow-up visit after enrollment (2002–2004), serum samples were available for 2,441 participants in the random sample. Of those, 38 individuals were excluded; 34 due to the occurrence of CVD prior to Exam 2, 1 due to cognitive impairment, and 3 due to inconsistencies between their self-reported race/ethnicity and the ethnic group that was actively enrolled by the field center. sL-selectin and DNA samples for the genetic analysis were therefore available in 2,403 participants, about 600 of each race/ethnicity.
Blood samples were obtained from fasting participants as previously described (Bild et al. 2002). Serum was obtained allowing blood samples to clot at room temperature for 40 minutes. Samples were centrifuged at 4°C at 2,000g × 15 minutes or 3,000g × 10 minutes for a total of 30,000 g-minutes, serum was aliquoted and stored frozen at −70°C. A single aliquot was thawed at room temperature and circulating sL-selectin was measured immediately by a quantitative sandwich enzyme-linked immunosorbent assay (ELISA) using the Human L-selectin/CD62L Immunoassay kit (R&D Systems, Minneapolis, MN). The inter-assay coefficient of variation of the MESA laboratory was 6.7% at a mean concentration of 943 ng/mL for an in-house serum pooled control and 7.9% at a mean concentration of 866 ng/mL for a lyophilized control. The manufacturer’s minimum detection level is 0.3 ng/mL.
Sample to test the association of SNPs with cardiovascular outcomes
The subset of the Exam 1 (2000–2002) population that gave informed consent for DNA studies (n = 6,317; 1,634 African, 764 Chinese, 2,491 non-Hispanic white and 1,428 Hispanic Americans) was used to test the association of identified variants with clinical and subclinical CVD. Subclinical endpoints of interest were common carotid intima-media thickness (IMT) and presence of carotid plaque measured via ultrasound, and coronary artery calcium (CAC) measured via computed tomography. Standard protocols were used in each field center as previously described (Carr et al. 2005; Polak et al. 2013). In addition, incidence of CHD, defined as myocardial infarction (MI), resuscitated cardiac arrest, angina, and CHD death. Event ascertainment methods are detailed elsewhere (Folsom et al. 2008). In brief, public files (death certificates), medical records from hospitalizations, autopsy reports, and phone interviews from participants at 9–12 month intervals, and in some instances, interviews or questionnaires from their physicians, relatives, or friends were used. Information was reviewed by two independent reviewers for adjudication of an event.
Genetic data
The genetic data consist of four genotype panels: Exome chip,(Huyghe et al. 2013) Cardio-Metabochip (Voight et al. 2012), i-Select.HG18 (IBC) (Keating et al. 2008), and MESA Candidate Gene (Illumina Golden Gate assay). The MESA Candidate Gene panel was run on a random sample of 720 participants from each race/ethnic group. All other SNP panels were genotyped using all MESA participants who consented for genetic studies (n = 6,323). Overall 417,752 SNPs in African Americans; 418,460 SNPs in Chinese; 417,771 SNPs in non-Hispanic whites; and 417,770 SNPs in Hispanics passed quality control procedures. All four panels had quality control performed on their genotype data prior to the merge. The data were merged in several steps using PLINK v1.07 (Purcell et al. 2007).
Statistical analysis
Participant characteristics were summarized using mean, standard deviation, and select percentiles for continuous variables, and number and percent for categorical variables within each race/ethnicity. These were compared across races/ethnicities using analysis of variance (Kruskal-Wallis) for continuous variables and the chi-square test (exact) for categorical variables. For this study a two-stage analysis approach was applied. First, the association of sL-selectin values with genome-wide genetic variants was assessed using linear regression under an additive genetic model using PLINK v1.07. Population stratification was assessed using STRUCTURE and using EigenStrat for participants with genome-wide SNP data (Patterson et al. 2006). Appropriate principal components (PCs) were included as covariates to adjust for population stratification.
Following the genome-wide approach, we identified one genetic variant that was significant across all four races/ethnicities. To assess potential additional independent associations in the region of the originally identified SNP, we applied race/ethnicity-specific elastic net models using the glmnet package in R (Friedman et al. 2010). The elastic net is a penalized regression approach that combines LASSO and ridge regression to simultaneously perform parameter shrinkage and variable selection. For our models, the mixing parameter α was set equal to 0.95 to accommodate linkage disequilibrium (LD), while the penalty parameter λ was selected based upon 10-fold cross-validation of the mean-squared prediction error. Age, sex, and the originally identified SNP were included as unpenalized covariates, with the previously used PCs capturing population stratification excluded from this and all further regression analyses due to the poor correlation between global and local ancestry (Qin et al. 2010). SNPs were modeled under an additive genetic model and missing genotypes imputed with mean observed values. Imputation was necessary for the penalized regression; however, the overall number of imputed SNPs was minimal as we used a SNP call rate threshold of at least 90%.
The association of genetic variants identified in the two-stage analysis with subclinical outcomes was assessed using linear regression for IMT, logistic regression for presence of carotid plaque, and the Tobit model (Fornage et al. 2004) for CAC score. The Tobit model accounts for the large percentage of zero measurements found in the CAC distribution. The association of genetic variants with time-to-CHD was assessed using Cox proportional hazard models. All regression models were stratified by race/ethnicity.
Results
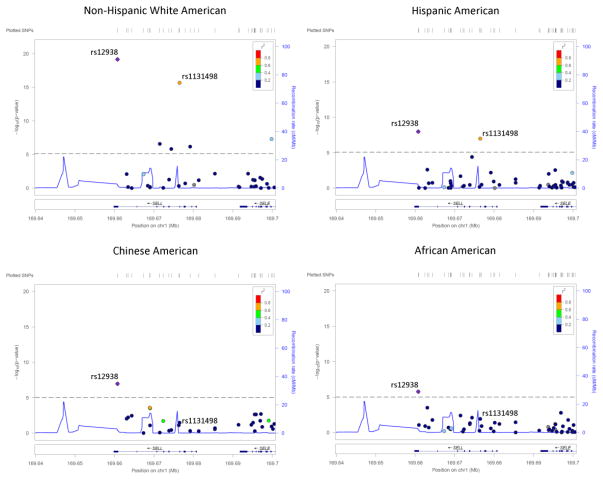
For the first step of the analysis, the subset of the MESA population including 2,403 individuals was used (Supplemental Figure S1); sample characteristics are presented in Supplemental Table S1. Genetic variants significantly (p < 10−5) associated with sL-selectin levels for each race/ethnicity after adjustment for age, sex, and principal components are listed in Supplemental Table S2. The majority of significantly associated SNPs are located in the genes encoding for L-selectin (SELL), P-selectin (SELP), and E-selectin (SELE). One common SNP in SELL, rs12938, was significantly associated in all four races/ethnicities (Figure 1).
Figure 1.
LocusZoom plots of the SNP associations with soluble L-selectin in proximity to SELL polymorphism rs12938 by race/ethnicity. Color of each SNP is indicative of linkage disequilbrium with rs12938. Significance threshold (1e-05) is indicated by dashed gray line. SELL polymorpism rs1131498 (previously reported to be associated with CHD) is additionally labeled.
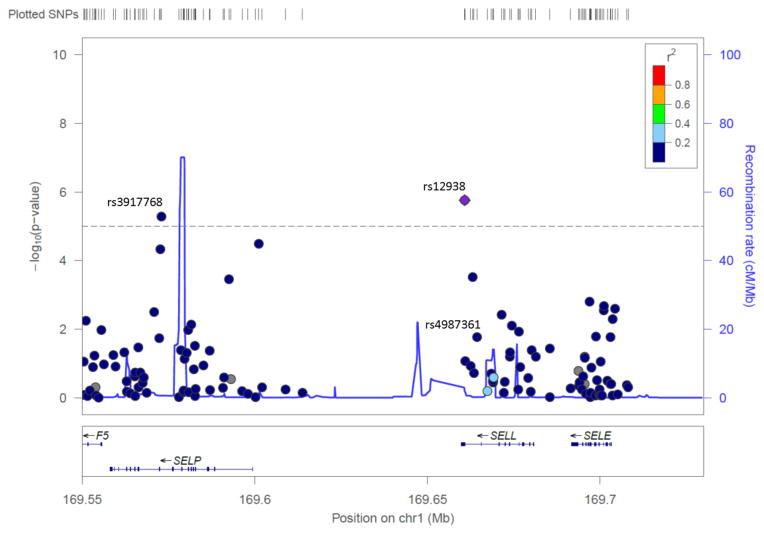
In order to evaluate potential additional associations, in the second step of the analysis, variants located in the SELL/SELP/SELE region were assessed using elastic net models in each race/ethnicity. SNP rs12938 was the only variant that remained in the model for Chinese, non-Hispanic white, and Hispanic Americans. For African Americans, rs12938, rs3917768, and rs4987361 remained in the model (Table 1, Figure 2). The C allele of SNP rs12938 was associated with significantly lower sL-selectin levels and the SNP explained an additional 6%, 12% and 4% of the total protein variance after adjusting for age and sex in Chinese, non-Hispanic white, and Hispanic Americans, respectively. Among African Americans, the three SNPs associated with sL-selectin levels accounted for an additional 14% of the total protein variance after adjusting for age and sex.
Table 1.
Genetic variants on SELL/SELP/SELE associated with soluble L-selectin (sL-selectin) using elastic net models
| Race/Ethnic Group (a Genotype) | n | MAF | Mean (SD) sL-selectin (ng/mL) | p valueb | Protein variance explained by SNP(s)c | ||
|---|---|---|---|---|---|---|---|
| African American | |||||||
| rs12938 (TT, TC, CC) | 544 | 0.31 | 893 (190) | 857 (191) | 745 (178) | 4.43E-07 | |
| rs3917768 (AA, AC, CC) | 544 | 0.26 | 892 (196) | 842 (186) | 769 (176) | 2.35E-05 | 14% |
| rs4987361 (GG, CG, CC) | 544 | 0.27 | 878 (204) | 854 (186) | 820 (151) | 0.008 | |
| Chinese American | |||||||
| rs12938 (TT, TC, CC) | 598 | 0.07 | 848 (171) | 740 (167) | 627 (123) | 8.85E-08 | 6% |
| Non-Hispanic white American | |||||||
| rs12938 (TT, TC, CC) | 619 | 0.29 | 1017 (203) | 919 (205) | 754 (153) | 6.88E-20 | 12% |
| Hispanic American | |||||||
| rs12938 (TT, TC, CC) | 589 | 0.22 | 936 (187) | 856 (187) | 815 (163) | 1.02E-07 | 4% |
MAF minor allele frequency, SD standard deviation, SNPs single nucleotide polymorphisms
The minor allele for each SNP is rs12938 (T), rs3917768 (C), and rs4987361 (C) based on RefSNP.
A = adenine, G = guanine, C = cytosine, and T = thymine
p value derived from linear regression assuming additivity, adjusting for age and sex
Additional protein variance explained after accounting for age and sex
The mean standard deviation (SD) for sL-selectin was 864 (194), 833 (175), 955 (215), and 903 (190) for African, Chinese, Non-Hispanic white, and Hispanic Americans, respectively
Figure 2.
LocusZoom plot of SELL/SELP/SELE genetic region for the African American cohort, with rs12938 the reference SNP. Additional SNPs selected by the elastic net model (rs, rs) are also labeled.
The association of these genetic variants with clinical and subclinical CVD was assessed among Exam 1 participants who gave informed consent for genotype analysis (93%). Table 2 summarizes the population characteristics, as well as the prevalence subclinical disease and incident events over a median follow-up of 10.1 years. As previously described, within this population, significant differences in the prevalence of traditional cardiovascular risk factors across races/ethnicities were observed. In addition, subclinical CVD was more prevalent among non-Hispanic whites. Within the Exam 1 population, no association was found between SNPs associated with sL-selectin and clinical or subclinical CVD. In non-Hispanic white, Chinese, and Hispanic Americans, none of the investigated outcomes was associated with the SNP rs12938, after adjustment of the significance threshold using the Bonferroni method (accounting for four outcomes: 0.05/4 = 0.0125), as shown in Table 3. Similar results were found for rs12938, rs3917768 and rs4987361 within African Americans (Table 4).
Table 2.
Exam 1 characteristics by race/ethnicity for participants with genotype data (count, percent, mean and standard deviation)
| Characteristics | Non-Hispanic white American | Chinese American | African American | Hispanic American | p value |
|---|---|---|---|---|---|
| n (%) | 2491 (39) | 764 (12) | 1634 (26) | 1428 (23) | |
| Age, years | 63 (10) | 62 (10) | 62 (10) | 61 (10) | 0.0019 |
| Sex, n (%) female | 1297 (52) | 387 (51) | 880 (54) | 736 (52) | 0.43 |
| Body mass index, kg/m2 | 28 (5.1) | 24 (3.3) | 30 (5.9) | 29 (5.1) | <0.0001 |
| Systolic blood pressure, mmHg | 123 (20) | 125 (22) | 132 (22) | 127 (22) | <0.0001 |
| Diastolic blood pressure, mmHg | 70 (9.9) | 72 (10) | 74 (10) | 72 (10) | <0.0001 |
| Hypertension, n (%) Yes | 964 (39) | 289 (38) | 965 (59) | 596 (42) | <0.0001 |
| Diabetes mellitus, n (%) Yes | 149 (6) | 101 (13) | 275 (17) | 252 (18) | <0.0001 |
| Total cholesterol, mg/dl | 196 (35) | 192 (31) | 189 (36) | 198 (38) | <0.0001 |
| HDL cholesterol, mg/dl | 52 (15) | 49 (12) | 52 (15) | 47 (13) | <0.0001 |
| LDL cholesterol, mg/dl | 117 (30) | 115 (29) | 116 (33) | 120 (33) | 0.02 |
| Triglycerides, mg/dl | 133 (90) | 143 (84) | 105 (70) | 159 (102) | <0.0001 |
| Antilipidemic therapy, % Yes | 453 (18) | 109 (14) | 259 (16) | 190 (13) | 0.0004 |
| Current smoker, n (%) Yes | 286 (12) | 43 (6) | 300 (18) | 193 (14) | <0.0001 |
| Current use of alcohol, n (%) yes | 1772 (72) | 234 (31) | 803 (50) | 667 (47) | <0.0001 |
| CAC > 0, n (%) yes | 1412 (57) | 388 (51) | 719 (44) | 653 (46) | <0.0001 |
| CAC Categories, Agatston score | <0.0001 | ||||
| < 50, n (%) | 1581 (64) | 538 (70) | 1226 (75) | 1054 (74) | |
| 50–149, n (%) | 275 (11) | 105 (14) | 160 (10) | 153 (11) | |
| 150–399, n (%) | 308 (12) | 70 (9) | 118 (7) | 105 (7) | |
| > 400, n (%) | 327 (13) | 51 (7) | 130 (8) | 116 (8) | |
| IMT, mm | 0.7 (0.2) | 0.7 (0.2) | 0.7 (0.2) | 0.7 (0.2) | <0.0001 |
| Carotid plaque, n (%) Yes | 1139 (47) | 203 (27) | 701 (44) | 554 (40) | <0.0001 |
| CHD events (all), n (%) thru 2012 | 188 (8) | 34 (5) | 100 (6) | 91 (6) |
CAC coronary artery calcium, CHD coronary heart disease, HDL high-density lipoprotein, IMT: intima-media thickness, LDL low-density lipoprotein
Table 3.
Association of SELL rs12938 and subclinical and clinical cardiovascular disease in Non-Hispanic white, Chinese, and Hispanic Americans (Exam 1 data)
| Non-Hispanic white American | Chinese American | Hispanic American | ||||
|---|---|---|---|---|---|---|
| CAC, Agatston Score | Beta (S.E.) | p value | Beta (S.E.) | p value | Beta (S.E.) | p value |
| Model 1 | −19 (21) | 0.37 | −61 (47) | 0.19 | −12 (34) | 0.71 |
| Model 2 | −27 (21) | 0.21 | −81 (47) | 0.09 | −5 (34) | 0.89 |
| IMT, mm | ||||||
| Model 1 | −0.01 (0.01) | 0.02 | −0.005 (0.02) | 0.80 | 0.002 (0.01) | 0.80 |
| Model 2 | −0.01 (0.01) | 0.02 | −0.01 (0.02) | 0.74 | 0.003 (0.01) | 0.73 |
|
| ||||||
| Presence of Plaque | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value |
|
| ||||||
| Model 1 | 0.94 (0.82–1.08) | 0.39 | 0.72 (0.44–1.19) | 0.20 | 0.81 (0.66–0.99) | 0.04 |
| Model 2 | 0.92 (0.80–1.06) | 0.24 | 0.67 (0.39–1.13) | 0.13 | 0.82 (0.66–1.01) | 0.06 |
|
| ||||||
| Coronary Heart Disease | HR (95% CI) | p value | HR (95% CI) | p value | HR (95% CI) | p value |
|
| ||||||
| Model 1 | 1.0 (0.80–1.26) | 0.99 | 0.60 (0.19–1.87) | 0.37 | 1.30 (0.93–1.80) | 0.12 |
| Model 2 | 0.98 (0.77–1.24) | 0.84 | 0.58 (0.18–1.82) | 0.35 | 1.31 (0.93–1.85) | 0.12 |
CI confidence interval, CAC coronary artery calcium, IMT intima-media thickness, OR odds ratio, SE standard error
An additive genetic model was fit modeling 0, 1, or 2 copies of the minor allele (C)
Model 1 = age and sex
Model 2 = age, sex, body mass index (BMI), smoking and alcohol use status, LDL and HDL cholesterol, triglycerides, and hypertension and diabetes status
Table 4.
Association of SELL rs12938, rs3917768 and rs4987361 with subclinical and clinical cardiovascular disease in African Americans (Exam 1 data)
| CAC, Agatston Score | Beta (SE) | p value |
|---|---|---|
| Model 1 | ||
| rs12938 | −15 (30) | 0.61 |
| rs3917768 | 16 (29) | 0.57 |
| rs4987361 | −16 (32) | 0.62 |
| Model 2 | ||
| rs12938 | −14 (30) | 0.64 |
| rs3917768 | 22 (29) | 0.45 |
| rs4987361 | −16 (33) | 0.63 |
| IMT, mm | ||
| Model 1 | ||
| rs12938 | −0.001 (0.01) | 0.88 |
| rs3917768 | 0.01 (0.01) | 0.38 |
| rs4987361 | −0.001 (0.01) | 0.89 |
| Model 2 | ||
| rs12938 | −0.001 (0.01) | 0.99 |
| rs3917768 | 0.01 (0.01) | 0.46 |
| rs4987361 | −0.002 (0.01) | 0.85 |
| Presence of Plaque | OR (95% CI) | p value |
| Model 1 | ||
| rs12938 | 1.19 (1.0 – 1.41) | 0.05 |
| rs3917768 | 0.99 (0.84 – 1.18) | 0.98 |
| rs4987361 | 1.01 (0.84 – 1.22) | 0.90 |
| Model 2 | ||
| rs12938 | 1.20 (1.0 – 1.43) | 0.05 |
| rs3917768 | 1.02 (0.86 – 1.21) | 0.81 |
| rs4987361 | 1.04 (0.86 – 1.26) | 0.69 |
| Coronary Heart Disease | HR (95% CI) | p value |
| Model 1 | ||
| rs12938 | 0.96 (0.69 – 1.34) | 0.82 |
| rs3917768 | 0.78 (0.56 – 1.10) | 0.16 |
| rs4987361 | 1.14 (0.81 – 1.59) | 0.46 |
| Model 2 | ||
| rs12938 | 0.96 (0.68 – 1.34) | 0.80 |
| rs3917768 | 0.79 (0.56 – 1.11) | 0.17 |
| rs4987361 | 1.15 (0.82 – 1.61) | 0.41 |
CAC coronary artery calcium, HDL high-density lipoprotein, IMT intima-media thickness, LDL low density lipoprotein, SE standard error
An additive multivariable genetic model was fit modeling 0, 1, or 2 copies of the minor allele (MAF) rs12938 (C), rs3917768 (C), rs4987361 (C)
Model 1 = age and sex
Model 2 = age, sex, Body mass index (BMI), smoking and alcohol use status, LDL and HDL cholesterol, triglycerides, and hypertension and diabetes status
Discussion
In this large, multi-ethnic population, we found that variants of the SELL gene are significantly associated with circulating levels of sL-selectin. In particular, we found that the C allele of SNP rs12938 is associated with lower levels of sL-selectin in all races/ethnicities. In African Americans two additional SNPs, rs4987361 and rs3917768, were identified using a penalized regression approach on the SELL/SELP/SELE region. Again, these variants are associated with lower levels of the circulating protein. However, none of these SNPs that account for a significant portion of the variance of sL-selectin levels was associated with subclinical or clinical atherosclerosis.
The SELL missense SNP rs1131498, previously reported to be associated with sL-selectin (Russell et al. 2005) was replicated in two of our racial subcohorts (Hispanic and non-Hispanic white Americans) but was not significant in the remaining two (African and Chinese Americans). Additionally, as Figure 1 demonstrates, there is strong LD (r2 > 0.6) present between rs1131498 and rs12938 in the same two subcohorts that have significant rs1131498 association findings, yet modest LD (r2 < 0.2) for the two that do not. The context of these findings suggest that rs12938 may be the underlying causal variant, and that rs1131498 is tagging rs12938 in populations where LD is present between the two SNPs.
SNP rs12938 is located in the 3′ untranslated region (UTR) of the SELL gene on Chromosome 1 and has been associated with sL-selectin levels in one previous study that included 278 lupus cases and 230 control siblings of European descent (Russell et al. 2005). In that study, three SNPs were identified as potentially relevant in determining sL-selectin levels; in particular, rs1131498 was suggested to be the putative causative SNP. Although Russell et al. surmised that an independent effect of rs12938 on sL-selectin may be driven by post-transcriptional regulation, they limited their analysis to in silico prediction of the rs12938 alternate allele on mRNA stability (Conne et al. 2000; Russell et al. 2005), concluding that there was little evidence of such a mechanism in place and that SNP’s effect was likely modest relative to the missense variant rs1131498. However, an alternative proposition for the functional relevance of rs12938 would be RNA silencing through modification of micro-RNA (miRNA) binding sites. We queried PolymiRTS v3.0 (Bhattacharya et al. 2014), an online database for SNP effects on RNA silencing-based post-transcriptional regulation, to evaluate the predicted impact of rs12938 on miRNA binding motifs. This analysis returned two putative miRNA binding sites created by the rs12938 C allele (Table 5), potentially inducing post-transcriptional down-regulation of SELL mRNA. Consistent with our results, previous expression quantitative trait loci (eQTL) studies reported rs12938 to be associated with SELL expression (Battle et al. 2014; Lappalainen et al. 2013; Xia et al. 2012). These previous eQTL association findings implicate this variant in the regulation of L-selectin mRNA levels, which in turn may correlate with protein expression on the leukocyte surface. The two additional SNPs identified in African Americans represent novel findings and have not been previously associated with sL-selectin levels. Additionally, rs2229569, reported by Wei et al. (Wei et al. 2011), was not significantly associated for any race/ethnicity (p > 0.05 for all analyses).
Table 5.
Predicted miRNA impacts of SELL rs12938 alternate allele from PolymiRTS
| SNP | Ref. Allele | Alt. Allele | miRBase ID | miRNA Motifa | Create/Disrupt | Experimental Validation? | Context Score Change |
|---|---|---|---|---|---|---|---|
| rs12938 | T | C | hsa-miR-197-5p | TCTACC[C]gac caa | Create | N | −0.255 |
| hsa-miR-3132 | TCTACC[C]gac caa | Create | N | −0.245 |
Binding motif in all capitalized letters; altered nucleotide bracketed and bolded.
SELL SNPs explained a significant portion of the variance of sL-selectin; however, there was no evidence of an association with subclinical atherosclerosis or CVD outcomes. These results are inconsistent with a previous study conducted within the Iranian population that found an association between the SNP rs1131498 on the SELL gene and CHD (Hajilooi et al. 2006). In fact, this SNP is in linkage disequilibrium with rs12938 among non-Hispanic whites in our study (r2 = 0.69). Several reasons could explain our results. First, the variance explained by the SNPs we identified ranged from 4% to 14% in the four race/ethnic groups. While statistically significant, the variation explained by SELL variants may be an insufficient surrogate of protein levels. Second, our ability to identify additional variants accounting for sL-selectin levels may be hindered by the inability of the circulating portion of sL-selectin to accurately reflect the cell-bound protein expression. There is some evidence suggesting that circulating L-selectin has a long half-life, and consequently does not correlate with the cell expression of the protein (Tu et al. 2002). Third, it is possible that, while the SNPs that we identified have some influence on the protein levels, other factors, genetic or not, may be more important and thus more closely related to the outcome. For example there may be non-synonymous SNPs that affect the protein structure and ultimately the function of L-selectin, but do not influence protein levels. Therefore, any SNPs that affect those characteristics may not be identified by looking for associations with circulating levels. Finally, these results could suggest a limited involvement of L-selectin in atherosclerosis. Other components of the selectin family may be more important in the initiation and progression on atherosclerosis compared to L-selectin.
Limitations and strengths
Some limitations need to be acknowledged. In this study we did not measure the cellular expression of L-selectin, as only frozen serum samples were available and these were not suitable for flow cytometry. In addition, while comprehensive genetic data have been used in our analysis, rare variants may not be captured. Finally, as the MESA population is a relatively young, the number of CVD events observed was lower than in other studies, which may have hindered our ability to fully explore the association between SELL genetic variants and clinical CVD. The main strength of our study is the large, multi-ethnic sample, coupled with the availability of comprehensive genetic data enriched for protein coding variation. Importantly, we demonstrate the utility of trans-ethnic analyses to provide additional insight into likely causal variants. Furthermore, MESA includes a large population with a reasonably long follow-up time for ascertainment of CVD events.
Conclusion
In conclusion, we identified variants in SELL accounting for a significant portion of the variance of circulating sL-selectin. Using trans-ethnic analyses, we show that rs12938, a variant previously associated with the quantitative regulation of mRNA and protein expression, was associated with sL-selectin in all four race/ethnic groups. Despite accounting for significant variance of protein level, this SNP, or other SNPs associated with the soluble protein levels, were not significantly related to clinical or subclinical CVD.
Supplementary Material
Acknowledgments
The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. MESA is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support is provided by grants and contracts N01 HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and RR-024156. Funding for adhesion protein levels was provided by NHLBI by grant R01HL98077. NIH/NIEHS P50 ES015915 “The Multi-Ethnic Study of Atherosclerosis and Air Pollution (MESA Air) is supported by the U.S. Environmental Protection Agency (EPA) under Science to Achieve Results (STAR) Program Grant # RD831697 and NIH/NIEHS P50 ES015915 award. Although the research described in this presentation has been funded wholly or in part by the United States Environmental Protection Agency through RD831697 to the University of Washington, it has not been subjected to the Agency’s required peer and policy review and therefore does not necessarily reflect the views of the Agency and no official endorsement should be inferred.” “The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center.”
Footnotes
Ethical standards The institutional review boards at each of the six field centers approved the study. Informed consent was obtained from all study subjects.
Conflict of interest The authors declare that they have no conflict of interest.
Contributor Information
Cecilia Berardi, Division of Epidemiology, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA. Montefiore Medical Center, NY 10467, USA.
Nicholas B. Larson, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Paul A. Decker, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Christina L. Wassel, Department of Epidemiology, University of Pittsburgh, PA 15261, USA
Phillip S. Kirsch, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
James S. Pankow, Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN 55454, USA
Michele M. Sale, Center for Public Health Genomics, University of Virginia, Charlottesville, VA 22908, USA
Mariza de Andrade, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA.
Hugues Sicotte, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA.
Weihong Tang, Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN 55454, USA.
Naomi Q. Hanson, Laboratory Medicine and Pathology, University of Minnesota, MN 55455, USA
Michael Y. Tsai, Laboratory Medicine and Pathology, University of Minnesota, MN 55455, USA
Yii-Der I da Chen, Institute for Translational Genomics and Population Sciences, Los Angeles BioMedical Research Institute and Department of Pediatrics at Harbor-UCLA, Torrance, CA, USA, 90502, USA.
Suzette J. Bielinski, Division of Epidemiology, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
References
- Battle A, Mostafavi S, Zhu X, Potash JB, Weissman MM, McCormick C, Haudenschild CD, Beckman KB, Shi J, Mei R, Urban AE, Montgomery SB, Levinson DF, Koller D. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardi C, Decker PA, Kirsch PS, de Andrade M, Tsai MY, Pankow JS, Sale MM, Sicotte H, Tang W, Hanson N, Polak JF, Bielinski SJ. Plasma and serum L-selectin and clinical and subclinical cardiovascular disease: the Multi-Ethnic Study of Atherosclerosis (MESA) Transl Res. 2014;163:585–592. doi: 10.1016/j.trsl.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinski SJ, Pankow JS, Li N, Hsu FC, Adar SD, Jenny NS, Bowden DW, Wasserman BA, Arnett D. ICAM1 and VCAM1 polymorphisms, coronary artery calcium, and circulating levels of soluble ICAM-1: the multi-ethnic study of atherosclerosis (MESA) Atherosclerosis. 2008;201:339–344. doi: 10.1016/j.atherosclerosis.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinski SJ, Reiner AP, Nickerson D, Carlson C, Bailey KR, Thyagarajan B, Lange LA, Boerwinkle EA, Jacobs DR, Jr, Gross MD. Polymorphisms in the ICAM1 gene predict circulating soluble intercellular adhesion molecule-1(sICAM-1) Atherosclerosis. 2011;216:390–394. doi: 10.1016/j.atherosclerosis.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr, Kronmal R, Liu K, Nelson JC, O’Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- Carr JJ, Nelson JC, Wong ND, McNitt-Gray M, Arad Y, Jacobs DR, Jr, Sidney S, Bild DE, Williams OD, Detrano RC. Calcified coronary artery plaque measurement with cardiac CT in population-based studies: standardized protocol of Multi-Ethnic Study of Atherosclerosis (MESA) and Coronary Artery Risk Development in Young Adults (CARDIA) study. Radiology. 2005;234:35–43. doi: 10.1148/radiol.2341040439. [DOI] [PubMed] [Google Scholar]
- Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr, Jones DW, Materson BJ, Oparil S, Wright JT, Jr, Roccella EJ. Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- Eriksson EE, Xie X, Werr J, Thoren P, Lindbom L. Importance of primary capture and L-selectin-dependent secondary capture in leukocyte accumulation in inflammation and atherosclerosis in vivo. J Exp Med. 2001;194:205–218. doi: 10.1084/jem.194.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom AR, Kronmal RA, Detrano RC, O’Leary DH, Bild DE, Bluemke DA, Budoff MJ, Liu K, Shea S, Szklo M, Tracy RP, Watson KE, Burke GL. Coronary artery calcification compared with carotid intima-media thickness in the prediction of cardiovascular disease incidence: the Multi-Ethnic Study of Atherosclerosis (MESA) Arch Intern Med. 2008;168:1333–1339. doi: 10.1001/archinte.168.12.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornage M, Boerwinkle E, Doris PA, Jacobs D, Liu K, Wong ND. Polymorphism of the soluble epoxide hydrolase is associated with coronary artery calcification in African-American subjects: the Coronary Artery Risk Development in Young Adults (CARDIA) study. Circulation. 2004;109:335–339. doi: 10.1161/01.CIR.0000109487.46725.02. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Galkina E, Kadl A, Sanders J, Varughese D, Sarembock IJ, Ley K. Lymphocyte recruitment into the aortic wall before and during development of atherosclerosis is partially L-selectin dependent. J Exp Med. 2006;203:1273–1282. doi: 10.1084/jem.20052205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Judd SE, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Mackey RH, Magid DJ, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, 3rd, Moy CS, Mussolino ME, Neumar RW, Nichol G, Pandey DK, Paynter NP, Reeves MJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajilooi M, Tajik N, Sanati A, Eftekhari H, Massoud A. Association of the Phe206Leu allele of the L-selectin gene with coronary artery disease. Cardiology. 2006;105:113–118. doi: 10.1159/000090212. [DOI] [PubMed] [Google Scholar]
- Humbria A, Diaz-Gonzalez F, Campanero MR, Arroyo AG, Laffon A, Gonzalez-Amaro R, Sanchez-Madrid F. Expression of L-selectin, CD43, and CD44 in synovial fluid neutrophils from patients with inflammatory joint diseases. Evidence for a soluble form of L-selectin in synovial fluid. Arthritis Rheum. 1994;37:342–348. doi: 10.1002/art.1780370307. [DOI] [PubMed] [Google Scholar]
- Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stancakova A, Stringham HM, Sim X, Yang L, Fuchsberger C, Cederberg H, Chines PS, Teslovich TM, Romm JM, Ling H, McMullen I, Ingersoll R, Pugh EW, Doheny KF, Neale BM, Daly MJ, Kuusisto J, Scott LJ, Kang HM, Collins FS, Abecasis GR, Watanabe RM, Boehnke M, Laakso M, Mohlke KL. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlof J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HP, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Montgomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Hasler R, Syvanen AC, van Ommen GJ, Brazma A, Meitinger T, Rosenstiel P, Guigo R, Gut IG, Estivill X, Dermitzakis ET. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Larson MG, Lunetta KL, Dupuis J, Rong J, Keaney JF, Lipinska I, Baldwin CT, Vasan RS, Benjamin EJ. Clinical and genetic correlates of soluble P-selectin in the community. J Thromb Haemost. 2008;6:20–31. doi: 10.1111/j.1538-7836.2007.02805.x. [DOI] [PubMed] [Google Scholar]
- Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak JF, Szklo M, Kronmal RA, Burke GL, Shea S, Zavodni AE, O’Leary DH. The value of carotid artery plaque and intima-media thickness for incident cardiovascular disease: the multi-ethnic study of atherosclerosis. J Am Heart Assoc. 2013;2:e000087. doi: 10.1161/JAHA.113.000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponthieux A, Herbeth B, Droesch S, Haddy N, Lambert D, Visvikis S. Biological determinants of serum ICAM-1, E-selectin, P-selectin and L-selectin levels in healthy subjects: the Stanislas study. Atherosclerosis. 2004;172:299–308. doi: 10.1016/j.atherosclerosis.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Morris N, Kang SJ, Li M, Tayo B, Lyon H, Hirschhorn J, Cooper RS, Zhu X. Interrogating local population structure for fine mapping in genome-wide association studies. Bioinformatics. 2010;26:2961–2968. doi: 10.1093/bioinformatics/btq560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Carlson CS, Thyagarajan B, Rieder MJ, Polak JF, Siscovick DS, Nickerson DA, Jacobs DR, Jr, Gross MD. Soluble P-selectin, SELP polymorphisms, and atherosclerotic risk in European-American and African-African young adults: the Coronary Artery Risk Development in Young Adults (CARDIA) Study. Arterioscler Thromb Vasc Biol. 2008;28:1549–1555. doi: 10.1161/ATVBAHA.108.169532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- Russell AI, Cunninghame Graham DS, Chadha S, Roberton C, Fernandez-Hart T, Griffiths B, D’Cruz D, Nitsch D, Whittaker JC, Vyse TJ. No association between E- and L-selectin genes and SLE: soluble L-selectin levels do correlate with genotype and a subset in SLE. Genes Immun. 2005;6:422–429. doi: 10.1038/sj.gene.6364222. [DOI] [PubMed] [Google Scholar]
- Tu L, Poe JC, Kadono T, Venturi GM, Bullard DC, Tedder TF, Steeber DA. A functional role for circulating mouse L-selectin in regulating leukocyte/endothelial cell interactions in vivo. J Immunol. 2002;169:2034–2043. doi: 10.4049/jimmunol.169.4.2034. [DOI] [PubMed] [Google Scholar]
- Venturi GM, Tu L, Kadono T, Khan AI, Fujimoto Y, Oshel P, Bock CB, Miller AS, Albrecht RM, Kubes P, Steeber DA, Tedder TF. Leukocyte migration is regulated by L-selectin endoproteolytic release. Immunity. 2003;19:713–724. doi: 10.1016/s1074-7613(03)00295-4. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, Burtt NP, Fuchsberger C, Li Y, Erdmann J, Frayling TM, Heid IM, Jackson AU, Johnson T, Kilpelainen TO, Lindgren CM, Morris AP, Prokopenko I, Randall JC, Saxena R, Soranzo N, Speliotes EK, Teslovich TM, Wheeler E, Maguire J, Parkin M, Potter S, Rayner NW, Robertson N, Stirrups K, Winckler W, Sanna S, Mulas A, Nagaraja R, Cucca F, Barroso I, Deloukas P, Loos RJ, Kathiresan S, Munroe PB, Newton-Cheh C, Pfeufer A, Samani NJ, Schunkert H, Hirschhorn JN, Altshuler D, McCarthy MI, Abecasis GR, Boehnke M. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedepohl S, Beceren-Braun F, Riese S, Buscher K, Enders S, Bernhard G, Kilian K, Blanchard V, Dernedde J, Tauber R. L-selectin--a dynamic regulator of leukocyte migration. Eur J Cell Biol. 2012;91:257–264. doi: 10.1016/j.ejcb.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Wei YS, Lan Y, Meng LQ, Nong LG. The association of L-selectin polymorphisms with L-selectin serum levels and risk of ischemic stroke. J Thromb Thrombolysis. 2011;32:110–115. doi: 10.1007/s11239-011-0587-4. [DOI] [PubMed] [Google Scholar]
- Williams KJ, Tabas I. Atherosclerosis and inflammation. Science. 2002;297:521–522. doi: 10.1126/science.297.5581.521. [DOI] [PubMed] [Google Scholar]
- Xia K, Shabalin AA, Huang S, Madar V, Zhou YH, Wang W, Zou F, Sun W, Sullivan PF, Wright FA. seeQTL: a searchable database for human eQTLs. Bioinformatics. 2012;28:451–452. doi: 10.1093/bioinformatics/btr678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.