Summary
Proper accumulation and function of miRNAs is essential for plant growth and development. While core components of the miRNA biogenesis pathway and miRNA-induced silencing complex have been well characterized, cellular regulators of miRNAs remain to be fully explored. Here we report that HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1) is a regulator of an important miRNA, mi168a/b, that targets the ARGONAUTE 1 (AGO1) gene in Arabidopsis. HOS1 functions to regulate plant cold stress responses as an ubiquitin E3 ligase, associate with the nuclear pores to regulate mRNA export and circadian clock and flowering time by binding to chromatin of the flowering regulator FLC. We isolated a loss-of-function Arabidopsis mutant of HOS1 in a genetic screen for enhancers of sic-1, which is defective in miRNA biogenesis. Like other hos1 mutant alleles, the new hos1-7 mutant allele flowered early and was smaller in stature than the wild type. Dysfunction in HOS1 reduced the abundance of miR168a/b but not of other miRNAs. In hos1 mutants, pri-MIR168b and pre-MIR168b levels were decreased, and RNA polymerase II occupancy was reduced at the promoter of MIR168b but not MIR168a. Chromatin immunoprecipitation assays revealed HOS1 protein is enriched at the chromatin of the MIR168b promoter. The reduced miR168a/b level in hos1 mutants results in an increase in the mRNA and protein levels of its target gene, AGO1. Our results reveal that HOS1 regulates miR168a/b and AGO1 levels in Arabidopsis by maintaining proper transcription of MIR168b.
Keywords: gene expression, transcriptional regulation, MicroRNA, MIR168b, high expression of osmotically responsive gene 1, Arabidopsis thaliana
Introduction
MicroRNAs (miRNAs) are 21-nucleotide (nt)-long small RNAs that induce post-transcriptional gene silencing through mRNA cleavage and/or translational repression. The functional importance of miRNAs in plant growth and development and stress responses has been extensively documented (Jones-Rhoades et al., 2004; Sunkar et al., 2007; Chen 2008; Zhu 2008; Chuck et al., 2009; Poethig 2009; Rubio-Somoza et al., 2009; Voinnet et al., 2009). In Arabidopsis, miRNA biogenesis and the core components involved in this process are well understood. First, RNA polymerase II transcribes the MIR genes, and then 5′ cap and 3′ poly A tails are added to produce pri-MIRNA transcripts. These pri-MIRNA transcripts fold into imperfect stem-loop secondary structures by base pairing within the transcripts. The stem-loop structure of pri-MIRNA is processed by DICER-LIKE1 (DCL1), an RNase III enzyme, to remove the 5′ and 3′ ends to produce pre-MIRNA, which is further processed by DCL1 into 21-nt-long miRNA/miRNA* duplexes.
Other components required for the proper functioning of DCL1 are HYPONNASTIC LEAVES1 (HYL1) (Dong et al., 2008), a dsRNA-binding protein, and SERRATE (SE), a C2H2 zine-finger protein (Dong et al., 2008). HUA ENHANCER1 (HEN1), a methyltransferase that catalyzes the 2′-O-methylation of the ribose sugar at the 3′ end of miRNA, which helps stabilize the miRNA (Yu et al., 2005). HASTY (HST), a homolog of mammalian EXPORTIN 5, guides the export of the methylated miRNA/miRNA* duplex from nucleus to cytosol (Park et al., 2005). The mature miRNAs exported to the cytosol are incorporated into the ARGONAUTE1 (AGO1) protein, which is a core component of the RNA-induced silencing complex (RISC). The RISC with a specific miRNA scans for complementary mRNA transcripts and directs the cleavage or translational repression at the target mRNAs (Jones-Rhoades et al., 2004; Baumberger and Baulcombe, 2005). Many other components play roles in the production of mature miRNAs, such as ABA HYPERSENSITIVE 1/CAP-BINDING PROTEIN 80 (ABH1/CBP80) and CBP20. Mutations in these genes cause dysfunction during the processing of pri-MIRNA transcripts into mature miRNA, which leads to reduced abundance of mature miRNAs (Laubinger et al., 2008). ABH1 may protect the capped miRNA from RNA decay and may function to bring pri-MIRNA to DCL1/HYL1/SE for processing of mature miRNA (Chen 2008). The hnRNP-like glycine-rich RNA-binding protein GRP7 showed its role in regulating pre-mRNA splicing (Köster et al., 2014). Recently, additional components involved in miRNA biogenesis have been identified. These include Erecta mRNA Under-expressed (EMU) (Furumizu et al., 2010), TOUGH (TGH) (Ren et al., 2012), STABILIZED1 (STA1) (Chaabane et al., 2013), SICKLE (SIC) (Zhan et al., 2012), and MODIFIER of SNC1 2 (MOS2) (Wu et al., 2013). However, the precise roles of these new components in miRNA biogenesis remain unclear.
Arabidopsis has 10 AGO proteins (Fagard et al., 2000; Carmell et al., 2002), among which AGO1 is the main protein that mediates miRNA-dependent silencing. Unlike its paralogs, the AGO1 transcript has a sequence complementary to miR168a/b, and AGO1 mRNA is cleaved at the site of miR168a/b complementarity (Vazquez et al., 2004). Furthermore, a decrease in mature miR168a/b in flowers of hen1-1 results in an increase in the AGO1 mRNA level (Vazquez et al., 2004).
The Arabidopsis HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1) functions as an ubiquitin E3 ligase (Dong et al., 2006). HOS1 is a negative regulator of cold-responsive genes like CBFs and of their downstream cold-regulated target genes such as RD29A and COR15A (Ishitani et al., 1998; Lee et al., 2001; Dong et al., 2006). HOS1 negatively regulates the cold response pathway at least in part by targeting the INDUCER OF CBF EXPRESSION1 (ICE1), which is a MYC transcription factor. ICE1 is important for induction of CBF genes under cold conditions (Chinnusamy et al., 2003; Lee et al., 2005), and it is marked by HOS1-mediated ubiquitination for protein degradation (Dong et al., 2006). HOS1 is also involved in regulating flowering time. Two different mechanisms by which HOS1 regulates the flowering pathway have been recently reported. First, HOS1 regulates the abundance of CONSTANS (CO), a photoperiod sensor (Jung et al., 2012; Lazaro et al., 2012). Previous report shows that CO is targeted by CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), a CUL4 E3 ligase, for degradation during dark photoperiods (Jang et al., 2008). Under cold stress conditions, CO is tagged by HOS1 for degradation (Jung et al., 2012). It has also been speculated that HOS1 may be the E3 ligase that targets CO for degradation during light photoperiods (Lazaro et al., 2013). With respect to the second mechanism, HOS1 regulates the transcription of FLOWERING LOCUS C (FLC) under cold stress by interacting with FVE and HDA6 (Jung et al., 2013). Transcriptional regulation of FLC by HOS1 does not involve the degradation of FVE or HDA6 (Jung et al., 2013). In addition, HOS1 associates with the nuclear pore, and is important for circadian clock that has a critical role in gating the cold response (MacGregor et al., 2013).
Here, we report the isolation of a new hos1 mutant allele, hos1-7, from an enhancer screen in the sic-1 mutant background. We discovered that HOS1 specifically regulates the level of miR168a/b. HOS1 modulates the level of miR168a/b by regulating the transcription of the MIR168b gene. We show that HOS1 is important for AGO1 mRNA and protein levels, and suggest that this helps explain the broad function of HOS1 in plant growth, development and stress tolerance.
Results
Identification of the hos1-7 mutant allele from a sic-1 enhancer screen
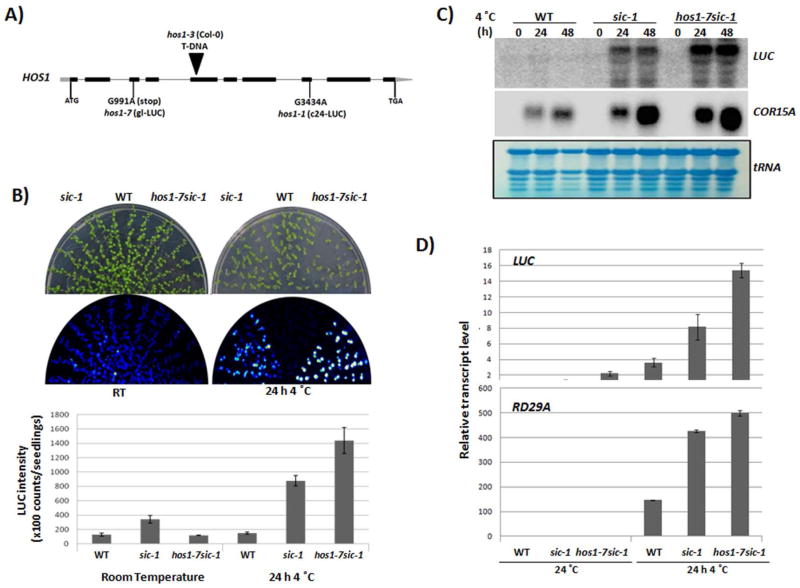
Previously, we found that a loss-of-function mutation in the SICKLE (SIC) gene resulted in an increase in proRD29A-LUC transgene expression under abiotic stresses such as cold, NaCl, and ABA (Zhan et al., 2012). SIC is a proline-rich protein involved in the biogenesis of some miRNAs as well as the degradation of some spliced introns (Zhan et al., 2012). To identify additional cellular factors that may regulate miRNA accumulation, we carried out a forward genetic screen in the sic-1 background. An EMS mutant population was generated in the sic-1 background for screening for putative enhancers of the sic-1 mutant. One such mutant was isolated based on its enhanced cold stress-induced LUC expression phenotype. Through map-based cloning, the enhancer mutation was found in the At2g39810/HOS1 gene (Figure 1A, and S1), which was previously identified as an ubiquitin E3 ligase that functions as a negative regulator of cold stress-responsive gene expression in Arabidopsis (Ishitani et al., 1998; Lee et al., 2001; Dong et al., 2006). The hos1-7sic-1 mutant displayed an enhanced LUC phenotype under cold conditions compared to sic-1 and the wild type (Col-0 ecotype with gl-1 mutation harboring proRD29A-LUC transgene) (Figure 1B). Based on Northern and quantitative real-time PCR (qRT-PCR) analyses, the hos1-7sic-1 had higher LUC transcript levels than sic-1 or the wild type under cold treatment (Figure 1C and 1D). Slight increases in endogenous RD29A (Figure 1D) and COR15A (Figure 1C) were also found in hos1-7sic-1 compared to sic-1 and the wild type under the same conditions. The hos1-7sic-1 mutant had smaller leaves and plant size than sic-1 or the wild type (Figure S2A). Furthermore, multiple siliques emerged from the same node in hos1-7sic-1 and in sic-1 (Figure S2B), and the mature plant was shorter for hos1-7sic-1 than for sic-1 or the wild type (Figure S2C and S2D). To confirm that these developmental phenotypes were due to mutations in the HOS1 gene, we isolated the single hos1-7 mutant from a backcross to the original wild type; we then compared the developmental phenotype of the hos1-7 single mutant with that of the previously identified hos1-3 mutant (SALK_069312, Col-0 background). Consistent with previous reports, hos1-7 displayed similar developmental phenotypes as hos1-3. The leaves were smaller for both mutants than for the wild type (Figure S2E). The hos1-7 also displayed an early flowering phenotype as reported in other hos1 mutants (Figure S2F, Ishitani et al., 1998; Lazaro et al., 2012). In addition, the mature hos1-7 mutant plant was shorter than the wild type (Figure S2G). All of the phenotypes were similar for the hos1-7 single and the hos1-7sic-1 double mutant except for the emergence of multiple siliques from one node in the hos1-7sic-1 double mutant, which may be caused by the sic-1 mutation. Our observations of the hos1-7 mutant are consistent with the notion that HOS1 is important for plant growth, development and cold stress responses (Ishitani et al., 1998).
Figure 1.
Identification and characterization of the hos1-7sic-1 mutant. (A) Diagram of the HOS1 gene and the hos1 mutant alleles used in this study. (B) Bioluminescence images of wild type (Col-0 ecotype (with gl-1 mutantion) harboring proRD29A-LUC transgene), sic-1, and hos1-7sic-1 under control and cold conditions, and the quantification of LUC intensity in wild type, sic-1, and hos1-7sic-1 DATA are average of 100 seedlings intensity, error bars represent s.d.’s (n = 100). (C) Northern analysis of transcript levels of transgene LUC and COR15A in gl-LUC, sic-1, and hos1-7sic-1. (D) qRT-PCR of the relative expression of LUC and RD29A in gl-LUC, sic-1, and hos1-7sic-1. Actin2 was used as internal control and error bars represent s.d.’s (n = 4).
HOS1 is required for proper accumulation of mature miR168a/b in Arabidopsis
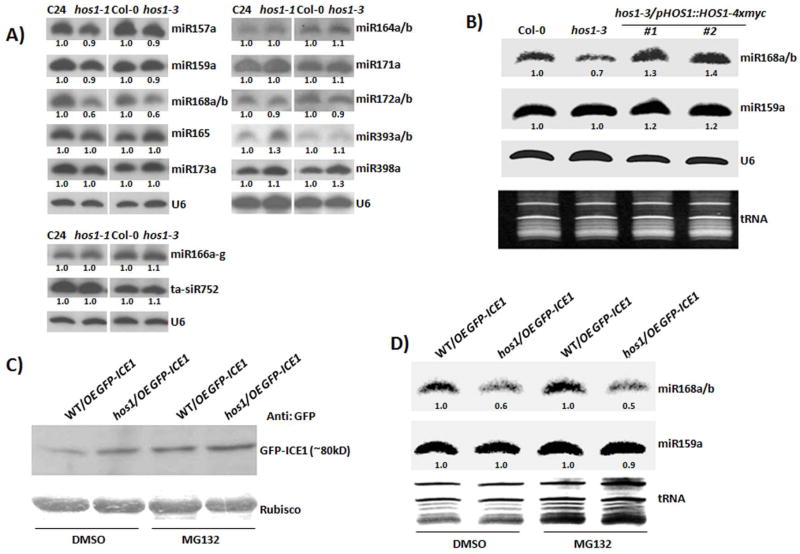
Because sic-1 is involved in miRNA biogenesis, we examined whether the mutation in HOS1, may also affect the accumulation of miRNA. Accumulation of miR159a and miR165 did not differ between hos1-7sic-1 and sic-1, but miR168a/b was less abundant in hos1-7sic-1 than in sic-1 (Figure S3A). We measured compared miRNA accumulation in hos1-1 (Ishitani et al., 1998) and hos1-3 (Figure 1A) and in their respective wild types. Accumulation of most of the examined miRNAs did not substantially differ between the wild types and hos1 single mutants except for miR168a/b (Figure 2A). Accumulation of mature miR168a/b was reduced in both the hos1 single mutants (Figure 2A). To further confirm the reduction of miR168a/b, we measured the accumulation of miR168a/b in the hos1-7 single mutant. Consistent with the results obtained with the other hos1 alleles, a clear reduction in the mature miR168a/b was evident in the hos1-7 single mutant (Figure S3B). Reduction in mature miR168a/b was compared between hos1-3 and mir168a-2 mutant, mature miR168a/b was significantly lower in mir168a-2 than in hos1-3 also when compared to WT (Figure S3C). Furthermore, increase in AGO1 transcript level was higher in mir168a-2 than hos1-3 (Figure S3D) consistent with reduction in mature miR168a/b.
Figure 2.
HOS1 is required for proper accumulation of mature miR168a/b. (A) miRNA accumulation in hos1 mutants. Two hos1 mutant alleles (hos1-1 and hos1-3) showed a reduction in mature miR168a/b, while other miRNAs were not affected by the hos1 mutations. hos1-1 in C24 ecotype harboring proRD29A-LUC transgene and hos1-3 is T-DNA insertion mutant (Salk_069312c). U6 indicates RNA loading control. Signal intensity was measured with ImageJ and normalized to loading control U6. Relative expression was normalized to wild type. (B) Restored mature miR168a/b in complemented transgenic lines. The genomic DNA sequence of HOS1 with its native promoter was transformed into the hos1-3 mutant. miR168a/b was restored to wild type levels in the complemented transgenic lines. U6 indicates RNA loading control. Signal intensity was measured with ImageJ and normalized to loading control U6. Relative expression was normalized to wild type. (C) GFP-ICE protein detection by Western blot analysis. Wild type and hos1 mutant harboring over-expression of GFP-ICE1 were treated with 50 μM MG132 for 24 h to inhibit the proteasome mediated protein degradation. DMSO was used as the treatment control. The increase in GFP-ICE1 protein level in the wild type under MG132 treatment compared to DMSO control treatment indicates the expected inhibition of the proteasome degradation pathway. Coomassie blue staining of Rubisco was used as loading control. (D) Accumulation of miR168a/b after MG132 treatment. MG132 treatment did not affect the accumulation of mature miR168a/b, indicating that the accumulation of miR168a/b was not affected by HOS1-mediated ubiquitination and degradation of ICE1. Signal intensity was measured with ImageJ and normalized to loading control tRNA. Relative expression normalized to wild type.
To determine whether a mutation in HOS1 is responsible for the reduction in miR168a/b, level, we used complementation lines harboring native proHOS1::HOS1-4xmyc in the hos1-3 mutant allele. The complemented lines had a restored hos1-3 developmental phenotype (Figure S2H). According to Northern analysis, accumulation of mature miR168a/b was similar in the complemented lines and the wild type but was reduced in hos1-3 (Figure 2B). Because HOS1 was previously determined to function as an ubiquitin E3 ligase, we next asked whether the reduction in miR168a/b might be due to a dysfunction of HOS1 as an ubiquitin E3 ligase. To help answer this question, we used previously reported transgenic lines, the wild type, and hos1 harboring over-expressed ICE1 tagged with GFP (WT/OE GFP-ICE1 and hos1/OE GFP-ICE1) (Dong et al., 2006). ICE1 was previously reported as a direct target of the ubiquitin E3 ligase activity of HOS1, i.e., HOS1 functions as an ubiquitin E3 ligase to target ICE1, which leads to ICE1 degradation through a proteasome degradation pathway (Dong et al., 2006). The wild type and hos1 transgenic lines with GFP-ICE1 were treated with 50 μM MG132 for 24 hours, which inhibits the proteasome degradation pathway. Treated samples were divided into two subsamples: miRNA levels were determined by Northern blot analysis in one subsample, and protein levels were determined by Western blot analysis in the other subsample. Consistent with Dong et al. (2006), the GFP-ICE1 protein level was higher in hos1 than in the wild type under the control DMSO treatment (Figure 2C), and GFP-ICE protein level was higher in both hos1 and the wild type under MG132 treatment than under the control DMSO treatment (Figure 2C), indicating the inhibition of proteasome degradation pathway by MG132 treatment. The reduction of miR168a/b was not recovered by MG132 treatment (Figure 2D), suggesting that the regulation of miR168a/b by HOS1 did not result from a loss of function for HOS1 to cause degradation of ICE1. These results indicated that HOS1 is needed for proper accumulation of mature miR168a/b and that the reduction in miR168a/b may be associated with a malfunction in an unidentified role of HOS1 that differs from its previously reported role as an E3 ligase.
HOS1 functions at the transcriptional level to regulate MIR168b
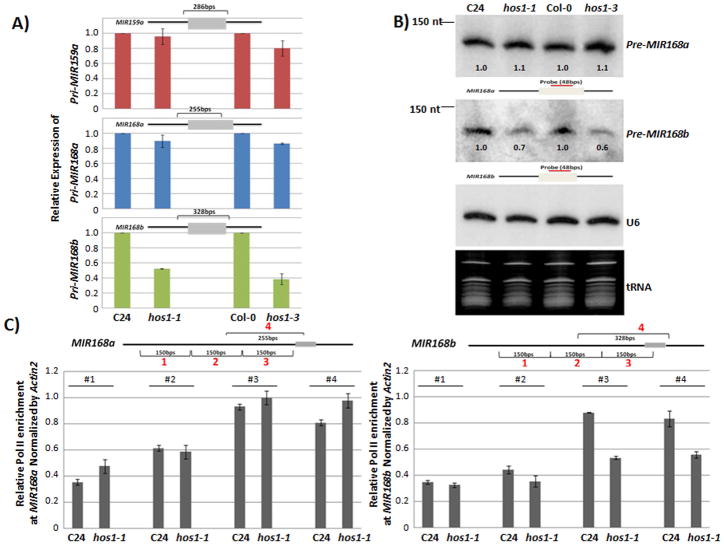
We detected a reduction in mature miR168a/b levels in hos1 mutant alleles (Figure 2A and S3). To further investigate whether the reduction is due to malfunction at the transcriptional or post-transcriptional level, we measured the levels of miR168a/b precursors in several experiments. Using qRT-PCR, we examined the expression level of pri-MIR159a, pri-MIR168a, and pri-MIR168b. The pri-MIR168b level was lower in hos1 mutants than in the respective wild types (Figure 3A), while pri-MIR168a and pri-MIR159a levels were similar between the hos1 mutants and their wild type controls (Figure 3A). To further confirm the qRT-PCR results, we used Northern blot analysis to measure the transcript levels of pre-MIR168a and pre-MIR168b. Consistent with the qRT-PCR results, the pre-MIR168b but not pre-MIR168a transcript level was lower in hos1 mutants than in the wild type controls (Figure 3B). The decrease in pri- and pre-MIR168b suggests in the hos1 mutants that HOS1 may affect the accumulation of mature miR168a/b at the transcriptional level and that HOS1 specifically affects MIR168b (Figure 3A and 3B).
Figure 3.
HOS1 regulates the transcription of MIR168b. (A) Reduction of pri-MIR168b in hos1 mutants. pri-MIR59a, pri-MIR168a, and pri-MIR168b levels were examined in hos1-1 and hos1-3 mutants. Actin2 was used as internal control and error bar represents s.d.’s (n = 4). (B) Reduction of pre-MIR168b in hos1 mutants as detected by Northern analysis. U6 was used as an internal control. Signal intensity was measured with ImageJ and normalized to loading control U6. Relative expression was normalized to wild type. (C) ChIP analysis showing the decrease in wild type and hos1-1 at MIR168a (left panel). The ChIP signal was normalized by actin2 and error bars represent s.d.’s (n = 4).
The qRT-PCR and Northern blot analysis results indicated that loss of HOS1 may reduce the transcription of MIR168b but not of MIR168a (Figure 3A and 3B). To further confirm these results, we performed chromatin immunoprecipitation (ChIP) using an RNA polymerase II-specific antibody to examine the enrichment of pol II at the MIR168a and MIR168b promoter regions. We detected a decrease in pol II occupancy at the promoter region of MIR168b in hos1-1 compared to the wild type (Figure 3C, right panel), while pol II occupancy was similar in hos1-1 and the wild type at the MIR168a region (Figure 3C, left panel). From these results, it is clear that loss-of-function hos1 mutants had a deficiency during the transcription of MIR168b but not MIR168a (Figure 3). This indicates that MIR168b is a target for transcriptional regulation by HOS1.
Enrichment of HOS1 protein at the promoter region of MIR168b
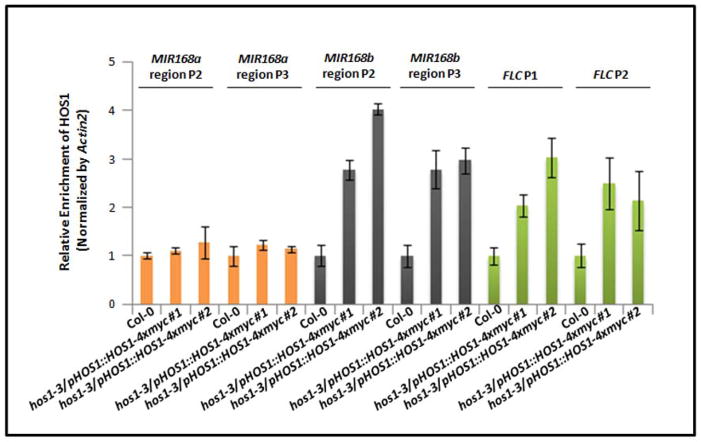
The specific regulation of MIR168b transcription by HOS1 indicates that HOS1 may bind to the MIR168b promoter region and regulate its transcription. To test this hypothesis, we conducted a ChIP assay using native proHOS1::HOS1-4xmyc transgenic lines to determine whether HOS1 enrichment can be found at the promoter region of MIR168b in vivo. HOS1 was previously reported to regulate the transcription of FLC (Jung et al., 2013). We used two regions near or at the FLC region as positive controls for the ChIP assay based on Jung et al. (2013). Consistent with the previous report, enrichment of HOS1 was detected at the FLC locus in our ChIP assay (Figure 4). Furthermore, we detected the enrichment of HOS1 protein at the MIR168b region but not at the MIR168a region or in the wild type without the myc-tagged HOS1 transgene (Figure 4). To test whether HOS1 protein may bind to the MIR168b promoter DNA directly, MBP-tagged HOS1 protein was expressed in E. coli, and DNA binding was examined in vitro by electrophoretic mobility shift assay (EMSA). Our repeated EMSA experiments failed to detect any HOS1 binding to the promoter region of MIR168b or MIR168a in vitro (Figure S4). Together, the EMSA and ChIP assay results suggested that HOS1 does not directly bind to the promoter DNA of MIR168b but associates with the chromatin of the MIR168b promoter to regulate transcription of MIR168b.
Figure 4.
HOS1 associates with the chromatin at the promoter region of MIR168b. ChIP assays using proHOS1::HOS1-4xmyc transgenic plants. ChIP assays were carried out using an anti-MYC antibody. FLC P1 and P2 were used as positive controls as reported previously (Jung et al., 2013). ChIP signal was normalized by actin2 and error bars represent s.d.’s (n = 4)
Mutation to HOS1 affects the proper balance of AGO1 due to reduced miR168a/b
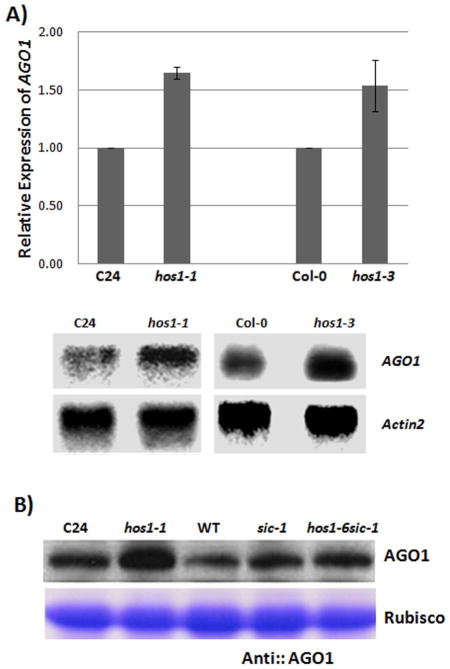
AGO1 is a core component of the RISC complex that represses miRNA targets (Vaucheret et al., 2006; Vaucheret 2009; Va’rallyay et al., 2010). Because the mutations in HOS1 results in a reduction of mature miR168a/b and because miR168a/b directs the cleavage of AGO1 mRNA to maintain the proper balance and production of AGO1 protein levels (Vaucheret et al., 2004), we suspected that the hos1 mutations may alter the proper balance and maintenance of AGO1 mRNA transcript level and AGO1 protein level. qRT-PCR and Northern blot analyses revealed an increase in AGO1 mRNA transcript level in the hos1 mutants compared to the wild type controls (Figure 5A). Similarly, the use of an AGO1 protein-specific antibody revealed an increase in AGO1 protein level in the hos1 mutants (Figure 5B). These results demonstrate that HOS1, by maintaining the proper transcript level of MIR168b, plays an important role in maintaining the proper balance of both the AGO1 transcript level and the AGO1 protein level.
Figure 5.
HOS1 is required for proper maintenance of AGO1. (A) Increase in AGO1 transcript in hos1 mutants. qRT-PCR (upper panel) and Northern analysis (lower panel) revealed an increase in AGO1 mRNA in hos1-1 and hos1-3 alleles compared to their respective wild types. Actin2 was used as internal control and error bars represents s.d.’s (n = 4) for qRT-PCR. Northern signal was measured with imageJ and normalized to loading control actin2. Relative expression was normalized to wild type. (B) Increase in AGO1 protein level in hos1 mutants. A Coomassie blue-stained band corresponding to the Rubisco largest subunit is shown as a loading control.
Discussion
HOS1 is an ubiquitin E3 ligase in Arabidopsis (Dong et al., 2006), and it regulates the cold response pathway by targeting ICE1, a transcription factor, which activates downstream cold-responsive genes (Lee et al., 2001; Dong et al., 2006). HOS1 can also regulate the flowering pathway by targeting CO for degradation (Jung et al., 2012; Lazaro et al., 2012). Mutants of miRNA biogenesis are known to accumulate reduced amounts of mature miRNAs, while the pri- and pre-MIRNA levels are increased compared to the wild type (Han et al., 2004; Vazquez et al., 2004; Lobbes et al., 2006; Yang et al., 2006; Dong et al., 2008; Kim et al., 2008; Laubinger et al., 2008; Zhan et al., 2012; and Chaabane et al., 2013). The hos1-7sic-1 double mutant showed decreases in miR168a/b compared to its background sic-1. Furthermore, other hos1 single mutant alleles also showed reduced accumulation of mature miR168a/b, while accumulation of other miRNAs tested was not affected by the hos1 mutations. We found that pri- and pre-MIR168b transcript levels were reduced in the hos1 mutants, while pri- and pre-MIR168a transcript levels were not affected. These results were further supported by ChIP results showing that the enrichment of RNA polymerase II was decreased at the promoter region of MIR168b but not of MIR168a. These results support the idea that HOS1 promotes proper transcription of MIR168b but not MIR168a. A similar case of regulation was previously reported in which a mutation of POWERDRESS (PWR) resulted in the reduction of MIR172a-c but not of MIR172d or MIR172e, in which transcription of MIR172a-c was affected by a mutation to PWR (Yumul et al., 2013). Also, the promoter region of MIR168a contains an ARBE motif where ABFs bind and positively regulate the transcription of MIR168a (Li et al., 2012). The overexpression of ABFs results in an increase in miR168a/b, and the increase is enhanced by treatment with abiotic stresses such as cold or ABA because of an increase in transcriptional activity (Li et al., 2012). Abiotic stress can also induce miR168a/b by increasing the transcription of MIR168a (Jia et al., 2009; Liu et al., 2008; Li et al., 2012). According to our ChIP results, HOS1 was enriched at the promoter region of MIR168b but not MIR168a, which provides further evidence that HOS1 regulates the transcription of MIR168b to maintain the proper accumulation of mature miR168a/b. The transcriptional regulatory role of HOS1 is also supported by a recent study where HOS1 was found to bind to FLC chromatin in the presence of FVE to prevent the binding of HDA6, which resulted in activation of FLC and a delay in flowering (Jung et al., 2013). The latter research also showed that enrichment of HOS1 at the FLC chromatin promoted the transcription of FLC (Jung et al., 2013). HOS1 interacts with FVE and HDA6, and the binding of HOS1 to FLC chromatin is dependent on FVE (Jung et al., 2013). At the present time, we do not know how HOS1 associates with the chromatin of the MIR168b promoter. Since HOS1 does not seem to bind directly to the promoter DNA, it is possible that HOS1 may associate with the promoter chromatin by interacting with other chromatin regulators.
The proper maintenance of AGO1 is essential for plant growth and development because excessive amounts of AGO1 can result in growth defects (Vaucheret et al., 2004). AGO1 homeostasis is mainly controlled by the presence of miR168a/b and the interaction between AGO1 and miR168a/b (Vaucheret et al., 2006; Vaucheret 2009; Va’rallyay et al., 2010). A mutated form of AGO1 mRNA in the miR168-binding region results in an increase in AGO1 mRNA due to the failure of miR168a/b to bind to the mutated form of AGO1 mRNA; this causes developmental defects, because a large excess of AGO1 protein interferes with RISC function (Vaucheret et al., 2004). The hos1 mutants showed reduced mature miR168a/b due to reduction in transcription of MIR168b. Consistent with reports in the literature, we detected an increase in AGO1 mRNA in the hos1 mutants, and this increase was translated into an increase in the AGO1 protein level. These results are also consistent with previous finding in which a mir168a-2 mutant resulted in a decrease in mature miR168a/b and an increase in AGO1 mRNA level (Vaucheret et al., 2009). Previously it was noted that MIR168a predominantly produces a 21-nt miR168 while MIR168b produces similar amounts of 21-nt and 22-nt miR168 (Rajagopalan et al., 2006). Some researchers have suggested that miR168b contributes less than miR168a, our result further support this idea, reduction of miR168a/b were lower in mir168a-2 than hos1-3 compared to WT. Accumulation of mature miR168a/b was significantly lower in mir168a-2 mutant compared to hos1-3, which indicates miR168a is represented more than the mir168b in mature miR168a/b. However, miR168b has been shown to rescue the developmental defects in 4m-AGO1 (mutated form of AGO1 mRNA) (Vaucheret et al., 2009). It has also been suggested that both 21-nt and 22-nt miR168a/b may be required for the proper maintenance of levels of AGO1 transcript and protein.
Mutation to the HOS1 gene results in developmental defects (Ishitani et al., 1998; Lazaro et al., 2012). Like other hos1 mutant alleles, the newly isolated hos1-7 allele displayed early flowering and a reduced plant size. Like the hos1 mutants, plants expressing miR168a/b-resistant forms of AGO1 mRNAs, 2m-AGO1 and 4m-AGO1, also over-accumulate AGO1 mRNAs and produce abnormally small leaves and small plants (Vaucheret et al., 2004). It is possible that the role of HOS1 in regulating miR168a/b and AGO1 levels contributes to the function of HOS1 in plant development. HOS1 also has an important function in cold stress tolerance (Ishitani et al., 1998). The function of HOS1 in plant cold tolerance could not be fully explained by its role as an ubiquitin E3 ligase to cause degradation of ICE1 (Dong et al., 2006). In hos1 mutant plants, the positive regulator of cold stress responsive genes, ICE1, is more stable; however, the hos1 mutant plants are more sensitive to freezing without cold acclimation (Ishitani et al., 1998; Dong et al., 2006). HOS1 regulates miR168a/b that in turn regulates AGO1 mRNA and protein levels. AGO1 is important for the function of all miRNAs and tasiRNAs (Jones-Rhoades et al., 2004). Therefore, by regulating miR168a/b and AGO1, HOS1 may affect the function of all miRNAs and tasiRNAs, some of which may be important for plant freezing tolerance in the absence of cold acclimation.
Experimental procedures
Plant materials and growth conditions
To screen for putative mutants affecting the accumulation of miRNAs, we introduced the miRNA biogenesis component sic-1 mutant (Zhan et al., 2012) to the EMS mutagen and then screened for putative sic-1 enhancer mutants, sic-1 was originally isolated in mutant screening in Col-0 ecotype (in gl-1 mutant) with proRD29A-LUC transgene refer to as wild type in this study (Zhan et al., 2012). One such mutant, hos1-7sic-1, showed an enhanced LUC phenotype under cold conditions and was identified as a sic-1 enhancer. See sic-1 map based cloning details in Supporting Methods S1. Two other Arabidopsis hos1 mutant alleles were used in this study: hos1-1 was isolated from the EMS pool from C24 ecotype harboring the same proRD29a-LUC transgene (Lee et al., 2001, Dong et al., 2006); and hos1-3 is a T-DNA insertion mutant allele in Col-0 background and was obtained from TAIR (SALK_069312c).
A 2314-bp sequence upstream of the HOS1 initiation start codon with HOS1 genomic sequence (without termination stop sequence and 3′ UTR sequence) was PCR amplified using Phusion high-fidelity DNA polymerase (M0530S, New England Biolab). The PCR product was then inserted into pENTR/D-Topo according to the manufacturer’s instructions (K2400-20, Invitrogen) and named pENTR/proHOS1::HOS1. Inserted sequences were confirmed by sequencing, and pENTR/proHOS1::HOS1 was used as a template for the LR Reaction using Gateway LR Clonase II (11791-100, Invitrogen) as instructed by the manual into a gateway destination vector pGWB-16 (-4xmyc). The construct carrying proHOS1::HOS1-4xmyc was transformed into the hos1-3 mutant via the traditional floral dip method (Clough and Bent, 1998).
Luciferase imaging
Twelve-day-old seedlings (wild type, sic-1, and hos1-7sic-1) were kept at 4 °C for 24 h to induce the proRD29A-LUC transgene. The LUC images were obtained using a low-light video imaging system (Princeton Instruments) with WinView software (Princeton Instruments; Chinnusamy et al., 2002).
RNA analysis
Accumulations of small RNAs were detected by Northern blot analysis. To obtain small RNAs, the total RNA from 12-day-old seedlings was extracted using Trizol (15596-026, Ambion) according to the manufacturer’s instructions. Low molecular weight RNAs were purified using PEG enrichment method, and small RNA Northern blot analysis was carried out as described previously (Zheng et al., 2007). The end labeling method was used to label the probes used in miRNA detection, and the random priming method was used to label the probe used to detect mRNA transcripts by Northern blot analysis. The probes used for Northern blot analysis are listed in Table S1. qRT-PCR was used to check the transcript levels. Total RNAs were extracted using Trizol (15596-026, Ambion) and were treated with Turbo DNase (AM1907, Ambion) to remove DNA contamination. DNase-treated RNAs were used to perform first-strand cDNA synthesis using the qScript Flex cDNA kit (95049-100, Quanta). qPR-PCR was performed as described previously (Zhan et al., 2012) with primers listed in Table S1.
Western blot analysis
Tissue from 12- to 14-day-old seedlings was ground in liquid N2 and then dissolved in protein extraction buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, pH 8.0, 10% glycerol (v/v), 1 mM DTT, 1 mM petablock, and protein inhibitor cocktail (11836153001, Roche)). The preparation was briefly mixed with a vortex apparatus and then centrifuged at 4 °C at 15000 g for 30 minutes. The supernatant containing total proteins was collected for protein normalization and western analysis. Anti-GFP tag antibody (11814460001, Roche) was used to determine the GFP-ICE protein level, and anti-AGO1 antibody (AS09 527, Agrisera) was used to determine the AGO1 protein level in hos1 mutants and the wild type.
Chromatin Immunoprecipitation (ChIP)
Twelve-day-old seedlings were collected for ChIP assay. Sample preparation and overall ChIP procedures were as previously described (Wierzbicki et al., 2008). Plants of the transgenic line harboring proHOS1::HOS1-4xmyc were first kept at 4 °C for 48 h to induce in vivo HOS1 protein activity so as to obtain a strong ChIP signal. The anti-myc antibody (05-724, Milipore) was used to check for the enrichment of HOS1 at MIR168a, MIR168b, and FLC. An anti-Pol II C-terminal domain (CTD) repeat antibody (ab817, Abcam) was used to check the enrichment of RNA polymerase II at MIR168a and MIR168b in the hos1-1 mutant and the wild type.
Supplementary Material
Figure S1. Identification of the hos1-7 mutant by map-based cloning.
Figure S2. Developmental phenotypes of hos1-7sic-1 and hos1-7.
Figure S3. Accumulation of miRNA in hos1-7sic-1 and hos1-7 mutants.
Figure S4. HOS1 does not directly bind to the MIR168a and MIR168b promoter.
Supporting material and methods.
Primer information (PDF).
Significance Statement.
Proper accumulation and function of miRNAs is essential for plant growth and development. Here we show a new role of previously identified HOS1 in regulating miR168a/b. We discovered HOS1 regulate MIR168b gene during transcription and it is required for proper expression of MIR168b. This role of HOS1 helps explain its broad function in plant growth, development and stress tolerance.
Acknowledgments
We thank Dr. Hervé Vaucheret and Dr. David P. Bartel for mir168a-2 seed. This work was supported by the National Institutes of Health Grants R01GM070795 and R01GM059138 (to J.-K. Z.).
Footnotes
The authors wish to state that there is no conflict of interest associated with any part of this manuscript.
References
- Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang M, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Chaabane SB, Liu R, Chinnusamy V, Kwon Y, Park JH, Kim SY, Zhu JK, Yang SW, Lee BH. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new play involved in miRNA biogenesis. Nucleic Acids Research. 2013;41:1982–1997. doi: 10.1093/nar/gks1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. A silencing safeguard: links between RNA silencing and mRNA processing in Arabidopsis. Dev Cell. 2008;14:811–812. doi: 10.1016/j.devcel.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Candela H, Hake S. Big impacts by small RNAs in plant development. Curr Opin Plant Biol. 2009;12:81–86. doi: 10.1016/j.pbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA. 2006;103:8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumizu C, Tsukaya H, Komeda Y. Characterization of EMU, the Arabidopsis homolog of the yeast THO complex member HPR1. RNA. 2010;16:1809–1817. doi: 10.1261/rna.2265710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell. 1998;10:1151–1161. doi: 10.1105/tpc.10.7.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008;27:1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia XY, Mendu V, Tang GL. An array platform for identification of stress-responsive microRNAs in plants. Methods Mol Biol. 2010;639:253–269. doi: 10.1007/978-1-60761-702-0_15. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M, Bartel D. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Jung JH, Park JH, Lee S, To TK, Kim JM, Seki M, Park CM. The Cold Signaling Attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 Activates FLOWERING LOCUS C Transcription via Chromatin Remodeling under Short-Term Cold Stress in Arabidopsis. Plant Cell. 2013;25:3478–4390. doi: 10.1105/tpc.113.118364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Seo PJ, Park CM. The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J Biol Chem. 2012;287:43277–43287. doi: 10.1074/jbc.M112.394338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell Physiol. 2008;49:1634–1644. doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster T, Meyer K, Weinholdt C, Smith L, Lummer M, Speth C, Grosse I, Weigel D, Staiger D. Regulationg of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic acid research. 2014;42(15):9925–9926. doi: 10.1093/nar/gku716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Ratsch G, Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Pineiro M, Jarillo JA. The Arabidopsis E3 Ubiquitin Ligase HOS1 Negatively Regulates CONSTANS Abundance in the Photoperiodic Control of Flowering. Plant Cell. 2012;24:982–999. doi: 10.1105/tpc.110.081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17:3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold regulated nucleo-cytoplasmic partitioning. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cui X, Meng Z, Huang X, Xie Q, Wu H, Jin H, Zhang D, Liang W. Transcriptional Regulation of Arabidopsis MIR168a and ARGONAUTE1 Homeostasis in Abscisic Acid and Abiotic Stress Responses. Plant Physiology. 2012;158:1279–1292. doi: 10.1104/pp.111.188789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14:836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor DR, Gould P, Foreman J, Griffiths J, Bird S, Page R, Stewart K, Steel G, Young J, Paszkiewicz K, Millar AJ, Halliday KJ, Hall AJ, Penfield S. HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 is required for circadian periodicity through the promotion of nucleo-cytoplasmic mRNA export in Arabidopsis. Plant Cell. 2013;25:4391–4404. doi: 10.1105/tpc.113.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci U S A. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. Small RNAs and developmental timing in plants. Curr Opin Genet Dev. 2009;19:374–378. doi: 10.1016/j.gde.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xie M, Dou Y, Zhang S, Zhang C, Yu B. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Nat Acad Sci USA. 2012;109:12817–12821. doi: 10.1073/pnas.1204915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Cuperus JT, Weigel D, Carrington JC. Regulation and functional specialization of small RNA-target nodes during plant development. Curr Opin Plant Biol. 2009;12:622–627. doi: 10.1016/j.pbi.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Várallyay E, Válóczi A, Agyi A, Burgyán J, Havelda Z. Plant virusmediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 2010;29:3507–3519. doi: 10.1038/emboj.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS ONE. 2009;4:e6442. doi: 10.1371/journal.pone.0006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathyway are crucial for plant development. Gene and Development. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajaqopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16 (1):69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Shi Y, Li J, Xu L, Fang Y, Li X, Qi Y. A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Research. 2013;23:645–657. doi: 10.1038/cr.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in. Arabidopsis Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhian S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumul RE, Kim YJ, Liu X, Wang R, Ding J, Xiao L, Chen X. POWERDRESS and Diversified of the MIR172 Gene Family Bolster the Floral Stem Cell Network. PLOS Genetics. 2013;9:e1003218. doi: 10.1371/journal.pgen.1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Wang B, Li H, Liu R, Kalia RK, Zhu JK, Chinnusamy V. Arabidopsis proline-rich protein important for development and abiotic stress tolerance in involved in microRNA biogenesis. Proc Natl Acad Sci USA. 2012;109:18198–18203. doi: 10.1073/pnas.1216199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26(6):1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Reconstituting plant miRNA biogenesis. Proc Natl Acad Sci USA. 2008;105:9851–9852. doi: 10.1073/pnas.0805207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Identification of the hos1-7 mutant by map-based cloning.
Figure S2. Developmental phenotypes of hos1-7sic-1 and hos1-7.
Figure S3. Accumulation of miRNA in hos1-7sic-1 and hos1-7 mutants.
Figure S4. HOS1 does not directly bind to the MIR168a and MIR168b promoter.
Supporting material and methods.
Primer information (PDF).