Abstract
Background
Two recent genome-wide association studies (GWAS) identified SNPs related to circulating 25-hydroxyvitamin D [25(OH)D] concentration in or near four genes. To examine the hypothesized inverse relationship between vitamin D status and breast cancer, we studied the associations between SNPs in these genes and breast cancer risk in a large pooled study of 9,456 cases and 10,816 controls from six cohorts.
Methods
SNP markers localized to each of four genes (GC, CYP24A1, CYP2R1, and DHCR7) previously associated with 25(OH)D were genotyped and examined both individually and as a 4-SNP polygenic score. Logistic regression was used to estimate the associations between the genetic variants and risk of breast cancer.
Results
We found no association between any of the four SNPs or their polygenic score and breast cancer risk.
Conclusions
Our findings do not support an association between vitamin D status, as reflected by 25(OH)D-related genotypes, and breast cancer risk.
Impact
These findings may contribute to future meta-analyses and scientific review articles, and provide new data about the association between vitamin D-related genes and breast cancer.
Introduction
A meta-analysis of prospective studies showed an inverse association between breast cancer and circulating 25(OH)D, an indicator of vitamin D status (1). Genome-wide association studies (GWAS) identified SNPs related to circulating 25(OH)D in or near four genes (2, 3): GC, encoding vitamin D binding protein, the major transporter of circulating vitamin D compounds; CYP24A1, encoding the cytochrome p450 24-hydroxylase that initiates intracellular catabolism of 25(OH)D and 1,25-dihydroxyvitamin D; CYP2R1, encoding the 25-hydroxylase which converts vitamin D to 25(OH)D in the liver; and DHCR7, encoding the enzyme that converts 7-dehydrocholesterol to cholesterol rather than vitamin D3 (2, 3). These four SNPs explain more variation in circulating 25(OH)D (5.2%) than a polygenic score including 9,000 SNPs (0.16%) (4).
The present study examines the four SNPs and their composite genetic score in relation to breast cancer in a large pooled analysis.
Materials and Methods
Details of the Breast and Prostate Cancer Cohort Consortium (BPC3) have been reported (5). Briefly, it is a consortium of breast cancer case-control sets nested in seven cohorts: the American Cancer Society Cancer Prevention Study II (CPS-II), the European Prospective Investigation into Cancer and Nutrition Cohort (EPIC), the Multiethnic Cohort (MEC), the Nurses’ Health Study I and II (NHS and NHSII), the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO), and the Women’s Health Study (WHS). This analysis was restricted to women of European ancestry (9,456 cases, 10,816 controls).
We chose SNPs identified in GWAS as associated with plasma/serum 25(OH)D concentrations (2, 3): rs2282679 (GC), rs10741657 (CYP2R1), rs12785878 (DHCR7), and rs6013897 (CYP24A1). Genotyping was conducted by Taqman or genome-wide scans as previously described (6).
Logistic regression was used to estimate odds ratios and 95% confidence intervals for breast cancer risk by individual SNPs and an unweighted polygenic score summing the number of lower 25(OH)D-associated alleles (i.e. “low vitamin D alleles”) (6). Models were adjusted for cohort and baseline age (continuous). Further adjustment for additional baseline factors did not alter the associations: body mass index (BMI), height, history of diabetes, smoking status, alcohol consumption, age at menarche, menopausal status, age at menopause, use of menopausal hormone therapy or oral contraceptives, age at first live birth, and family history of breast cancer. Details on collection and harmonization of these data have been published (5, 6). Analyses stratifying by estrogen (ER) and progesterone (PR) receptor status, cancer stage, menopausal status at diagnosis (postmenopausal at baseline or age 55+ at diagnosis vs. premenopausal or perimenopausal at baseline and age ≤51 at diagnosis), BMI, height, and cohort were conducted. SAS v9.3 was used.
Results
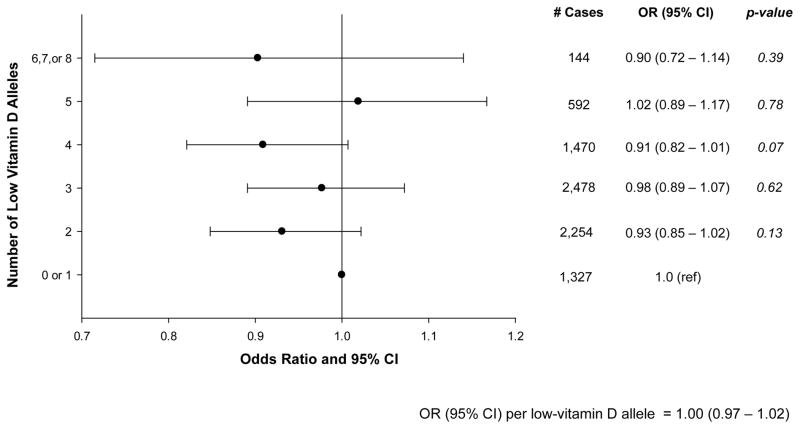
Characteristics of the cohorts have been published (5). Examination of the four SNPs individually revealed no association with breast cancer and no heterogeneity across cohorts for any SNP (Table). We observed no differences in the association for any SNP across subgroups of stage, menopausal status at diagnosis, or progesterone receptor (PR) status (data not shown), although women with two low vitamin D alleles for rs12785878 (DHCR7) appeared to be at lower risk of ER- breast cancer, consistent with a recessive genetic association (GG vs. TT: OR=0.56, 95% CI: 0.32–0.97; p=0.04). Consideration of multiple comparisons, however, rendered this finding not statistically significant (Bonferroni-corrected p-value threshold=0.00125). There was no statistically significant interaction with BMI or height after consideration of multiple comparisons. The polygenic score was also not associated with breast cancer risk (Figure), and this did not vary by height, BMI, ER or PR status, stage, or menopausal status at diagnosis (data not shown).
Table 1.
Individual SNP associations with risk of breast cancer
| SNP | Gene | Frequency of low vitamin D allele | # Low vitamin D alleles | # Cases | # Controls | OR (95% CI)* | p-value† | p for heterogeneity across cohorts§ |
|---|---|---|---|---|---|---|---|---|
| rs2282679 | GC | 0.28 | ||||||
| TT | 0 | 4,531 | 5,100 | 1.0 (ref) | ||||
| GT | 1 | 3,412 | 4,104 | 0.94 (0.88 – 1.00) | ||||
| GG | 2 | 713 | 781 | 1.03 (0.92 – 1.15) | 0.08 | |||
| Additive (per allele) | 0.98 (0.94 – 1.03) | 0.39 | 0.10 | |||||
|
| ||||||||
| rs6013897 | CYP24A1 | 0.21 | ||||||
| TT | 0 | 5,759 | 6,551 | 1.0 (ref) | ||||
| AT | 1 | 3,064 | 3,576 | 0.97 (0.91 – 1.03) | ||||
| AA | 2 | 447 | 491 | 1.03 (0.90 – 1.18) | 0.48 | |||
| Additive (per allele) | 0.99 (0.94 – 1.04) | 0.65 | 0.84 | |||||
|
| ||||||||
| rs10741657 | CYP2R1 | 0.61 | ||||||
| AA | 0 | 1,301 | 1,486 | 1.0 (ref) | ||||
| AG | 1 | 4,041 | 4,766 | 0.96 (0.88 – 1.05) | ||||
| GG | 2 | 3,276 | 3,708 | 1.00 (0.91 – 1.09) | 0.41 | |||
| Additive (per allele) | 1.01 (0.97 – 1.05) | 0.72 | 0.62 | |||||
|
| ||||||||
| rs12785878 | DHCR7 | 0.27 | ||||||
| TT | 0 | 4,935 | 5,674 | 1.0 (ref) | ||||
| GT | 1 | 3,620 | 4,052 | 1.03 (0.97 – 1.09) | ||||
| GG | 2 | 669 | 834 | 0.92 (0.83 – 1.03) | 0.16 | |||
| Additive (per allele) | 0.99 (0.95 – 1.03) | 0.60 | 0.13 | |||||
Adjusted for age at baseline (continuous) and study cohort.
For categorical analyses of genotype, the p-value is a 2 degree of freedom test comparing a model containing indicator variables for genotype and covariates with a model containing only the covariates. For the per allele analyses, the p-value is a 1 degree of freedom test comparing a model containing the number of alleles and covariates to a model containing only the covariates.
The p-value for heterogeneity across cohorts is based on the test for interaction between cohort and the additive.
Figure 1.
Association Between the 4-SNP Polygenic Score and Risk of Breast Cancer
Discussion
In this large, pooled analysis, we found no association between four SNPs related to vitamin D status (or the 4-SNP polygenic score) and breast cancer risk. Given that these SNPs explain ~5% of the 25(OH)D variation, if the OR of breast cancer per standard deviation increase of 25(OH)D were 0.83 or stronger we should have 80% power to detect it [6]. We observed no association with any of the breast cancer subtypes examined, with the possible exception of lower risk of ER- breast cancer among women with 2 low vitamin D alleles for rs12785878 near DHCR7, which encodes an enzyme that converts 7-dehydrocholesterol to cholesterol rather than to vitamin D3. As cholesterol is a precursor of sex steroids including estrogens and androgens, a breast cancer association with this gene, if substantiated, may have its basis in steroid hormone, not vitamin D, metabolism (7). The finding may, however, be due to chance.
Few studies have examined these SNPs in relation to breast cancer risk. One found no association for a polygenic score of the same four genes, but increased risk among women with two low vitamin D alleles for rs6013897 in CYP24A1 (8). Our findings do not support an association between SNPs associated with vitamin D status and breast cancer risk, but the possible association of DHCR7 with ER- disease should be examined further.
Acknowledgments
Financial Support: This work was supported by the U.S. NIH, National Cancer Institute cooperative agreements U01-CA98233-07 to D.J. Hunter, U01-CA98710-06 to M.J. Thun, U01-CA98216-06 to E. Riboli and R. Kaaks, and U01-CA98758-07 to B.E. Henderson and Intramural Research Program of NIH/National Cancer Institute, Division of Cancer Epidemiology and Genetics). I. M. Shui was supported by a National Research Service Award (T32 CA09001) from the National Cancer Institute, NIH. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Conflict of Interest: The authors have no conflicts to disclose.
References
- 1.Wang D, Velez de-la-Paz OI, Zhai JX, Liu DW. Serum 25-hydroxyvitamin D and breast cancer risk: a meta-analysis of prospective studies. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34:3509–17. doi: 10.1007/s13277-013-0929-2. [DOI] [PubMed] [Google Scholar]
- 2.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 19:2739–45. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 376:180–8. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiraki LT, Major JM, Chen C, Cornelis MC, Hunter DJ, Rimm EB, et al. Exploring the Genetic Architecture of Circulating 25-Hydroxyvitamin D. Genetic Epidemiology. 2012 doi: 10.1002/gepi.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter DJ, Riboli E, Haiman CA, Albanes D, Altshuler D, Chanock SJ, et al. A candidate gene approach to searching for low-penetrance breast and prostate cancer genes. Nat Rev Cancer. 2005;5:977–85. doi: 10.1038/nrc1754. [DOI] [PubMed] [Google Scholar]
- 6.Mondul AM, Shui IM, Yu K, Travis RC, Stevens VL, Campa D, et al. Genetic variation in the vitamin d pathway in relation to risk of prostate cancer--results from the breast and prostate cancer cohort consortium. Cancer Epidemiol Biomarkers Prev. 2013;22:688–96. doi: 10.1158/1055-9965.EPI-13-0007-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNamara KM, Sasano H. The intracrinology of breast cancer. J Steroid Biochem Mol Biol. 2014 doi: 10.1016/j.jsbmb.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Jorde R, Schirmer H, Wilsgaard T, Joakimsen RM, Mathiesen EB, Njolstad I, et al. Polymorphisms related to the serum 25-hydroxyvitamin d level and risk of myocardial infarction, diabetes, cancer and mortality. The tromso study. PLoS One. 2012;7:e37295. doi: 10.1371/journal.pone.0037295. [DOI] [PMC free article] [PubMed] [Google Scholar]