Abstract
Macroautophagy (hereafter autophagy) is a highly evolutionarily conserved process essential for sustaining cellular integrity, homeostasis, and survival. Most eukaryotic cells constitutively undergo autophagy at a low basal level. However, various stimuli, including starvation, organelle deterioration, stress, and pathogen infection, potently upregulate autophagy. The hallmark morphological feature of autophagy is the formation of the double-membrane vesicle known as the autophagosome. In yeast, flux through the pathway culminates in autophagosome-vacuole fusion, and the subsequent degradation of the resulting autophagic bodies and cargo by vacuolar hydrolases, followed by efflux of the breakdown products. Importantly, aberrant autophagy is associated with diverse human pathologies. Thus, there is a need for ongoing work in this area to further understand the cellular factors regulating this process. The field of autophagy research has grown exponentially in recent years, and although numerous model organisms are being used to investigate autophagy, the baker’s yeast Saccharomyces cerevisiae remains highly relevant, as there are significant and unique benefits to working with this organism. In this review, we will focus on the current methods available to evaluate and monitor autophagy in S. cerevisiae, which in several cases have also been subsequently exploited in higher eukaryotes.
Keywords: Atg8, autophagosome, mitophagy, PAS, phagophore, vacuole
1. Introduction
1.1 An introduction to autophagy and its significance
Autophagy (or cellular “self-eating”) plays an integral role in various aspects of cell physiology, whereas defects in this process are associated with numerous pathological conditions. Autophagy is ubiquitous in eukaryotes and occurs constitutively at a basal level. However, various stress conditions, including starvation or changing nutrient conditions, organelle deterioration, and pathogen infection, result in an upregulation of this process. Importantly, aberrant autophagy is associated with diverse human pathologies, such as cancer, neurodegeneration, aging, cardiovascular, pulmonary, and infectious diseases, macular degeneration, diabetes and lysosomal storage disorders [1, 2]. Thus, there is a need for ongoing work in this area to further understand the cellular factors regulating this inherently complex process.
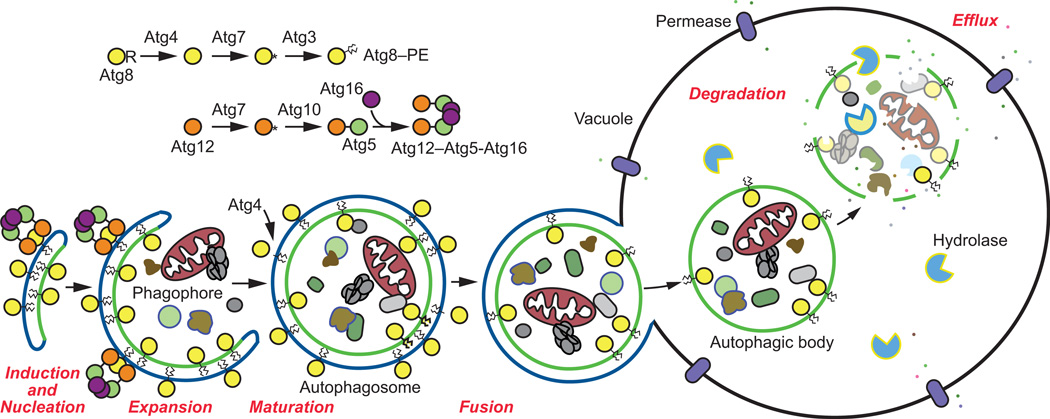
In brief, the most obvious morphological feature of autophagy is the double-membrane autophagosome that contains either bulk cytoplasm or select cargo, depending on the inducing condition. However, the autophagosome is essentially the end product of the sequestration process (although it will fuse with the vacuole). The dynamic membrane compartment involved in autophagic sequestration is the phagophore, the autophagosome precursor. The mechanism of phagophore formation is unique, and is quite distinct from other vesicle-mediated processes involved in cargo trafficking. For example, in contrast to the secretory pathway where the vesicles bud off from preexisting organelles already containing their cargo, the phagophore expands sequentially, providing autophagy with an extremely flexible capacity for cargo sequestration. In yeast, autophagosomes typically range from ∼300–900 nm in diameter [3, 4]. Nonetheless, efficient degradation of organelles such as peroxisomes and mitochondria involves their fission by the dynamin-related GTPase Dnm1 complex prior to, or during, engulfment by phagophores [5, 6].
Upon completion of expansion, the phagophore seals to form the autophagosome. Flux through the pathway culminates in autophagosome-vacuole fusion, which releases the inner autophagosome single-membrane vesicle into the vacuole lumen. In yeast, the resulting inner autophagosome vesicles, which transiently reside within the vacuole, are known as autophagic bodies. To date, these structures have only been identified in yeast and are subsequently lysed, allowing degradation of the cargo by vacuolar hydrolases. The resulting macromolecules are finally transported into the cytosol via membrane permeases.
1.2 Saccharomyces cerevisiae is a fundamental model organism for the study of autophagy
The phenomenon of autophagy was first observed in mammalian cells using electron microscopy (EM) in the 1950s, and was officially termed as such by Christian de Duve in 1963 at the CIBA Foundation Symposium on Lysosomes (reviewed in [7, 8]). However, autophagy was not described in yeasts until the 1980s [9]. S. cerevisiae and other fungi are fundamental model organisms for the study of autophagy. For example, much of our current understanding of autophagy is due to work conducted in S. cerevisiae, Pichia pastoris, and Hansenula polymorpha using genetic screens and biochemical methods. At present, 38 AuTophaGy-related (ATG) genes have thus far been identified in fungi [10]. Importantly, at least half of these genes are clearly conserved up to human, and analogous proteins are present for many others, reflecting a high degree of overall pathway conservation.
The field of autophagy research has grown exponentially in recent years, and although numerous model organisms exist to investigate autophagy, S. cerevisiae continues to be an important system for studying this process. In addition to the high degree of conservation, there are significant and unique benefits to working in this organism, including the capability to do in vivo genetics work quickly and relatively easily, and the myriad unique assays that are available to monitor different steps of autophagy. This review will focus on an overview of the current methods that can be used to evaluate and monitor the various stages of autophagy in S. cerevisiae (see the accompanying article by Guimaraes et al. for additional information and protocols).
1.3 Overview of autophagy in S. cerevisiae
There are two major forms of autophagy in yeast—macroautophagy and microautophagy (reviewed in [11]); this review focuses on macroautophagy. Briefly, microautophagy can be selective or nonselective, and is morphologically distinct from macroautophagy [11–13]. During the process that is strictly defined as microautophagy, tubules invaginate directly from the vacuolar membrane into the lumen [12]. Following scission, these tubules release single-membrane vesicles that may appear similar to autophagic bodies, although the mechanism of formation is completely distinct; microautophagy does not directly involve the Atg proteins, and its physiological function is not fully understood. In contrast to microautophagy, there are also microautophagy-like processes such as micropexophagy (the selective microautophagic degradation of peroxisomes), micromitophagy (which is used to eliminate mitochondria), and piecemeal microautphagy of the nucleus/PMN or micronucleophagy (to remove small portions of the nucleus). These types of sequestration involve direct uptake at the vacuolar membrane, and require the Atg proteins (for further reviews on microautophagy, see [11–14]).
There are four main stages of autophagic activity, enabling progression (otherwise known as flux or autophagic turnover) through the pathway (Figure 1). As a number of excellent reviews already exist detailing the complex molecular interactions that occur throughout the various stages of autophagy [8, 11, 13, 15–17], and as this review primarily focuses on methods, we will only concisely discuss each phase of autophagic activity before moving on to the most common assays that may be used to assess each.
Figure 1. An overview of the autophagy pathway in S. cerevisiae.
Shown is a schematic overview of each of the four key stages of autophagy: induction and nucleation of the phagophore, expansion of the phagophore and closure to form the autophagosome, autophagosome-vacuole fusion, and cargo degradation and efflux. Prior to the elongation of the phagophore membrane, Atg8 is converted to its lipidated form, Atg8–PE following a series of proteolytic events involving the Atg8 conjugation system (Atg3, Atg4, Atg7, and Atg8). The Atg12 ubiquitin-like conjugation system (Atg5, Atg7, Atg10, Atg12, and Atg16) is also required for membrane elongation.
2. Induction and nucleation of the phagophore
2.1 Background
During the induction and nucleation phase, a physiological stimulus such as nutrient deprivation, or a change in nutrient conditions, results in a shift from basal, constitutive autophagy to induced autophagy (Figure 2). Stimulation of autophagy can also be achieved through genetic or pharmacological means such as occurs following treatment with rapamycin, which inhibits the activity of TOR (target of rapamycin), a serine/threonine kinase that is a major regulator or cellular metabolism, and a negative regulator of autophagy [18, 19]. In yeast, the intracellular location of autophagosome formation is the phagophore assembly site (PAS), which localizes adjacent to the vacuole [15]. The “core” Atg proteins assemble at the PAS to initialize phagophore nucleation [17]. The initial protein complex recruited to the PAS is comprised of Atg1, Atg13 and the Atg17-Atg31-Atg29 ternary subcomplex [13, 20, 21]. Atg1 is another serine/threonine kinase, and its activity is regulated by Atg13 [17, 22–24] and Atg17-Atg31-Atg29 [8, 20, 25, 26]. Following recruitment of the Atg1 complex, Atg9 (and interacting proteins including Atg2 and Atg18) localize to the PAS [8]. Thus far, Atg9 is the only integral membrane protein that is absolutely essential for autophagosome formation, although the exact role of Atg9 has not yet been determined [27]. The Pho23-Rpd3 complex regulates ATG9 transcription, and the expression level of Atg9 controls the frequency of autophagosome formation [28, 29], which fits with a general model whereby Atg9 directs the delivery of membranes to allow formation of the phagophore. Furthermore, various SNARE proteins that control the localization of Atg9 have been implicated in autophagosome biogenesis [30, 31].
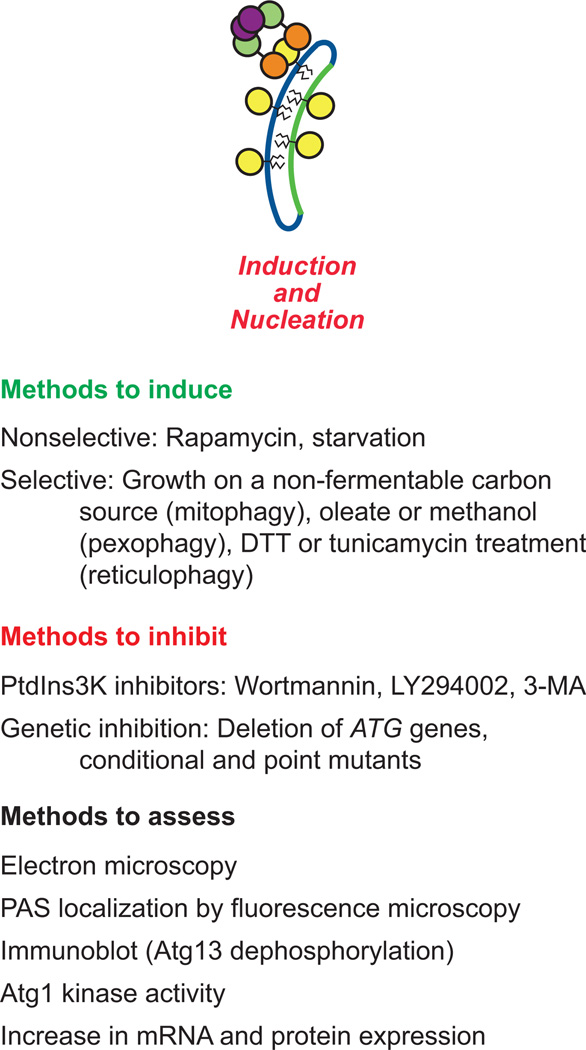
Figure 2. Methods to assess induction and nucleation.
Shown are methods to induce and inhibit nonselective or selective forms of autophagy. The most commonly used methods to assess autophagic induction are listed. See the text for details
Although the detailed mechanism is not understood, the PAS may be a nucleation site that is converted into a phagophore. In yeast and mammals, various intracellular compartments have been identified as the probable source(s) of the phagophore membrane, including the ERGIC (ER-Golgi intermediate compartment) [32, 33], the ER [34–36], the Golgi apparatus [37], the mitochondrial-associated membrane (MAM) at ER-mitochondria contact sites [38], the mitochondria [39], the plasma membrane [40, 41] and recycling endosomes [42]. However, it is possible that the source varies according to the type of autophagy that the cell is undergoing (nonselective versus selective and what form of selective), the organism, and the signaling pathway(s) involved.
2.2 Methods to induce autophagy
There are various methods available to initiate autophagy (Figure 2, Table 1). As mentioned above, rapamycin can be used to induce TOR-dependent autophagy [18, 43]; however, in higher eukaryotes TOR-independent autophagy pathways have also been identified [44–48]. Furthermore, rapamycin does not appear to induce autophagy as strongly as when nitrogen starvation is used as a stimulus [43]. Culturing cells in nitrogen starvation medium (SD-N; synthetic medium with dextrose, minus nitrogen: 0.17% yeast nitrogen base without ammonium sulfate or amino acids, containing 2% glucose) for 2–4 h also initiates nonselective autophagy.
Table 1.
Methods for the analysis of autophagy progression in yeast and mammalian cells.
| Yeast | Mammals | |
|---|---|---|
| 1. Induction and nucleation | ||
| A. Induction | ||
| Nutrient deprivation | X | X |
| Rapamycin | X | X |
| B. Inhibition | ||
| Pharmacological | Xa | X |
| Genetic | X | Xb |
| C. Assessment | ||
| Electron microscopy | X | X |
| PAS localization | X | |
| Atg13/ATG13 dephosphorylation | X | X |
| Atg1/ULK1 kinase activity | X | X |
| Upregulation of ATG gene expression | X | X |
| 2. Expansion and maturation | ||
| A. Assessment | ||
| GFP-Atg8/LC3 | X | X |
| Protease protection/sequestration assays | X | X |
| TAKA assay | X | |
| Atg4/ATG4 protease activity | X | X |
| 3. Fusion | ||
| A. Inhibition | ||
| Pharmacological | Xa | X |
| Genetic | X | Xb |
| B. Assessment | ||
| Electron microscopy | X | X |
| Fluorescence microscopy | X | X |
| Immunoblot (GFP processing) | X | Xc |
| Pho8Δ | X | |
| 4. Degradation and efflux | ||
| A. Inhibition | ||
| Pharmacological | Xa | X |
| Genetic | X | Xb |
| B. Assessment | ||
| Electron microscopy: autophagic body accumulation | X | |
| Fluorescence microscopy | X | X |
| Radioactive efflux | X | X |
| Assay for new protein synthesis | X | X |
| Long-lived protein degradation | X | X |
Although autophagy can be inhibited pharmacologically in yeast, it is more typical to use genetic methods such as null or conditional mutants.
Traditionally this has been done with RNAi, but now can also be achieved with Cas9 CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology.
The ability to detect GFP-LC3 processing is is cell type-dependent.
Alterations in the type of culture medium may be used to induce various forms of selective autophagy. Ribophagy, a selective type of autophagy targeting ribosomes, may also be induced by nitrogen starvation as described above [49]. Growing cells in media containing a non-fermentable carbon source such as lactate (2%), glycerol (3%), glycerol-lactate (2%-2%), or ethanol (3%) for 12–16 h to mid-log phase induces mitochondrial biogenesis; a subsequent shift to rapamycin or nitrogen starvation medium can initiate mitophagy, a selective form of autophagy targeting mitochondria [50–54] (see the accompanying review by Guimaraes et al. for a detailed protocol to assay this type of mitophagy). In general, this type of selective autophagy allows the cell to adapt to changing nutrient conditions by subcellular remodeling, where organelles that are now superfluous or in excess can be removed to lower the energetic requirements for their functional maintenance. Culturing to post-log phase for several days in media containing a non-fermentable carbon source can also stimulate mitophagy [55, 56] (see the accompanying review by Guimaraes et al. for a detailed protocol to assay this type of mitophagy). This form of mitophagy is also known as “stationary phase mitophagy,” and may serve as a quality control mechanism to rid the cell of accumulated mitochondria during quiescence [57]. During stationary phase, mitophagy may be induced due to the lower energy requirement of the cells; the elimination of excess mitochondria also reduces the production of reactive oxygen species that can otherwise be deleterious to cell physiology.
Similarly, pexophagy can be stimulated by culturing S. cerevisiae cells in oleate medium (YTO: 0.67% yeast nitrogen base without amino acids, 0.1% Tween-40, 0.1% oleic acid, pH 5.5; or YPO: 0.25% yeast extract, 0.5% peptone, 1% oleate, 5% Tween-40, 5 mM phosphate buffer, pH 5.5), and shifting to SD-N for at least 2 h [6, 58, 59] (see the accompanying review by Guimaraes et al. for a detailed protocol to measure pexophagy). Growth in medium containing oleic acid (or methanol in the case of methylotrophic fungi such as P. pastoris and H. polymorpha) as the sole carbon source causes peroxisome proliferation (in both size and number); the subsequent shift to glucose (or ethanol) medium with or without nitrogen starvation induces pexophagy to degrade the superfluous peroxisomes [6, 59–62]. Another form of selective autophagy, reticulophagy, which targets the endoplasmic reticulum (ER) can be stimulated by ER stress resulting from the overproduction of proteins that are prone to misfolding, or treatment with compounds such as DTT or tunicamycin that also interfere with protein folding [63]. It should be noted that the mechanism of reticulophagy in yeast is currently unresolved [64].
2.3 Methods to inhibit autophagy
As a corollary to induction, various methods can be utilized to inhibit the initiation phase of autophagy (Figure 2, Table 1). As a brief note – in mammals, the typical methods to prevent autophagy induction involve pharmacological approaches that target phagophore initiation including the use of wortmannin, LY294002, and 3-methyladenine (3-MA). These three chemicals are inhibitors of both the class I phosphoinositide 3-kinase and the class III phosphatidylinositol 3-kinase (PtdIns3K), and accordingly are not specific to autophagy [65–69]. Even though they block the class I enzyme, which is inhibitory for autophagy, the net result of these chemicals is to block induction because the downstream PtdIns3K enzyme (PIK3C3 in mammals, Vps34 in yeast) is essential for autophagy [70]. The product of this kinase, phosphatidylinositol-3-phosphate (PtdIns3P), is required for phagophore formation [65, 66].
Although these chemicals are commonly used in mammalian cells, they are not typically used in yeast. Substantially higher concentrations of drugs are often needed to inhibit enzymes in yeast compared to mammals, due to the presence of a cell wall, differences in plasma membrane lipid composition, and/or the presence of efficient efflux pumps. For example, the concentrations of wortmannin and LY294002 routinely used in mammalian cells have little effect on yeast Vps34 [71]. As an alternative to pharmacological treatment, genetic approaches are used to inhibit both nonselective and selective autophagy in yeast; the deletion of key ATG genes may block induction of the entire pathway [23, 67, 72, 73], and conditional (temperature sensitive) mutants have also been generated for some of these genes [11, 74, 75] as well as others (i.e., non-ATG genes) involved in autophagy initiation [30, 76]. One advantage of using molecular genetics is specificity. For example, rather than using wortmannin and other nonspecific chemicals, it is possible to use conditional mutants of Vps34 (which is the only PtdIns 3-kinase in yeast) to block PtdIns3P production [77]. Vps34, however, is part of two PtdIns3K complexes, and only complex I is specific to autophagy; a deletion of ATG14 inactivates complex I, but has no affect on complex II, which is involved in the vacuolar protein sorting (Vps) pathway. In addition to the VPS34 or ATG14 genes, induction can be blocked through the use of alleles with mutations in genes encoding regulatory components. For example, the hyperactive Ras2G19V mutant [78], the use of a mutant expressing constitutively active Gtr1 (a GTPase that activates TOR complex 1 [TORC1]) [79, 80], or a mutant lacking expression of the protein kinase A (PKA) regulatory subunit Bcy1 [81] will prevent the induction of autophagy.
If loss of only a selective form of autophagy is desired (or to serve as a negative control in a specific assay), genetic deletion of the cargo recognition receptor required for that particular form of autophagy (but not for another) might be performed. For example, deletion of ATG32, the selective receptor for the targeting of mitochondria, significantly impairs only mitophagy [82, 83]. Similarly, in S. cerevisiae, Atg36 functions as the pexophagy receptor, and its loss inhibits the selective degradation of peroxisomes [84, 85]. In the cytoplasm-to-vacuole targeting (Cvt) pathway, a biosynthetic pathway that utilizes the autophagy machinery to deliver resident hydrolases to the vacuole under nutrient-rich conditions [86], Atg19 selects the cargo proteins prApe1 (precursor aminopeptidase I), Ams1 (α-mannosidase), and Ape4 (aspartyl aminopeptidase) for targeting to the vacuole [87–90]. During induced autophagy, Ams1 is selected for vacuolar targeting by Atg34 [91]. (For further explanation, see section 3.2 “Methods to assess phagophore expansion and closure” below.) In contrast to these specific receptors, Atg11 is a scaffold protein that is required for most types of selective autophagy [55, 85, 92].
2.4 Methods to assess the induction of autophagy
In addition to either directly stimulating or inhibiting autophagy, there are various methods that are commonly used to measure nucleation of the phagophore, including microscopy and biochemical assays (Figure 2, Table 1). In yeast, identification of the PAS by transmission electron microscopy (TEM) is not currently practical through morphological analysis alone [67]. In contrast, immunogold labeling can be applied to aid in the detection of phagophores by EM [93]. In contrast to autophagosomes, several components of the Atg core machinery are associated with phagophores including Atg8–phosphatidylethanolamine (PE) and Atg12–Atg5-Atg16. Phagophores can also be conveniently visualized during the Cvt pathway by monitoring the Cvt complex (composed primarily of oligomeric prApe1 bound to Atg19), because the large oligomeric Ape1 complex is electron dense and relatively easy to identify even without immunogold labeling [94, 95].
Alternatively, phagophore initiation can also be assessed by fluorescence microscopy utilizing the green fluorescent protein (GFP; or another fluorophore) conjugated to almost any Atg protein because most of these components are transiently associated with the PAS [67], although most also exist as a diffuse cytosolic pool. GFP-Atg8 is particularly useful to monitor phagophore nucleation and expansion because it is relatively abundant, is recruited to the PAS as the last Atg protein, and its fluorescence intensity increases as the phagophore expands, followed by disappearance of the distinct punctate signal as the completed autophagosome fuses with the vacuole [96]. Although it is preferable to use genome-tagged proteins to follow these processes, the plasmid-based overexpression of fluorescently-tagged Atg proteins does not seem to have a major effect on autophagy activity, but may induce slightly elevated overall levels of autophagy and augment the cytoplasmic pool of Atg proteins rather than the population associated with the PAS [29]. Furthermore, it is also possible to calculate the stoichiometry of assembling Atg proteins at the PAS utilizing fluorescence microscopy, but in this case expression should be from endogenous promoters [97].
The PAS appears as a single distinct perivacuolar punctum when visualized by tagging most of the Atg proteins; however, when monitoring only the PAS it is best to avoid proteins such as Atg9, Atg23, and Atg27 that may be found at other intracellular locations (in addition to the PAS) [98]. As mentioned above with regard to both TEM and fluorescence microscopy, a good combination of protein markers that can be used to distinguish the PAS from other autophagic compartments is the heterotrimeric complex of Atg12–Atg5-Atg16, as this complex dissociates upon phagophore closure [67, 68, 99]. Fluorescently-tagged prApe1 is also a marker of the PAS, particularly under growing conditions. It is often helpful to delineate the vacuole (rim or lumen) to assist in the detection of the peri-vacuolar PAS. This can be achieved through the use of dyes such as FM 4–64 (vacuole membrane marker), CellTracker Blue CMAC (vacuole lumen marker), or the expression of fluorescently-tagged vacuolar proteins such as GFP-Pho8 [100–102].
Aside from microscopy, other approaches can be used to follow autophagy induction. For example, Atg13 is hyperphosphorylated by TORC1 and PKA under basal conditions [8, 22, 24, 103]; however, under conditions that stimulate autophagy, Atg13 is largely dephosphorylated [103, 104]. The change in phosphorylation status can be readily monitored by western blot [67]. A complete protocol for assessing the phosphorylation status of Atg13 has been published [103]. Additionally, Atg1 kinase activity can be monitored to measure the initiation of autophagy [67]. Finally, autophagic induction can be further monitored by increased Atg8 expression levels or by following the change in ATG8 mRNA [43, 93, 105, 106]. It should be noted that changes in Atg8 levels or the corresponding transcript are not by themselves adequate evidence of autophagy induction. For example, ATG8 (or the mammalian homolog LC3) can be induced, and Atg8/LC3 conjugation to PE can proceed, in the absence of a functional PtdIns3K complex, even though autophagy will not take place [70, 107, 108]. Thus, it is critical to monitor more than one parameter to ensure an accurate interpretation. Other transcripts that are upregulated upon autophagy induction are those of ATG14 and ATG9, which are under the control of the Gln3 and Pho23 transcription factors, respectively [29, 109].
3. Phagophore expansion and closure
3.1 Background
In the second stage of autophagy (also known as the expansion or elongation step), extension of the phagophore and maturation into the completed autophagosome occurs through two separate, conserved ubiquitin-like conjugation systems (Figure 1). In brief, the first system requires the Atg12 conjugation system consisting of the heterotrimeric complex Atg12–Atg5-Atg16, which is localized to the phagophore and functions in part as an E3-like enzyme for the Atg8 conjugation system [110]. Atg7, an E1-like enzyme that activates Atg12, and Atg10, an E2-like enzyme, promote the conjugation of Atg12 to Atg5 at the phagophore [111, 112].
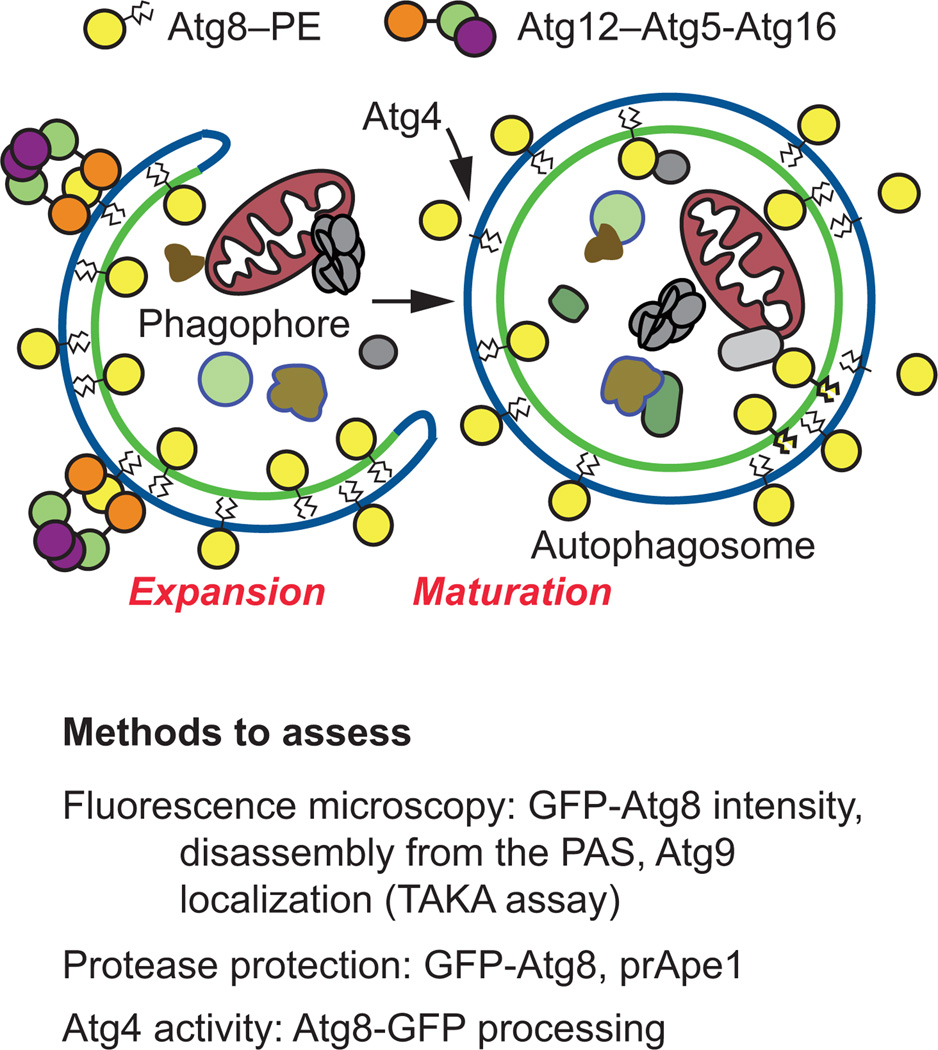
The second ubiquitin-like conjugation system involves Atg8 [or microtubule-associated protein 1 light chain 3 (LC3) in mammalian cells], and is required for membrane expansion and closure. The Atg8-conjugation system is comprised of Atg8 and the additional factors Atg7, Atg4, and Atg3 [8]. Atg8 is converted to its lipidated form, Atg8–PE, following Atg4-mediated proteolytic processing of its C terminus, Atg7-dependent activation, and Atg3-facilitated conjugation at a conserved C-terminal glycine exposed by the Atg4 cleavage [111, 113]. Atg8–PE lines both sides of the phagophore; upon formation and maturation of the autophagosome the Atg8 on the outer surface is removed from PE by deconjugation, a second Atg4-mediated cleavage event, which is required for efficient autophagosome biogenesis [110, 111, 113, 114]. The activity of Atg4 can be monitored directly, by expressing Atg8-GFP where the fluorescent tag is added at the C terminus [115].
In yeast, the closed phagophore—now referred to as the autophagosome—is approximately 300–900 nm in diameter [3, 4]. Cvt vesicles, the sequestering compartments that form during the Cvt pathway, are much smaller (140–160 nm) and exclude bulk cytoplasm [27, 116]. As noted above, the autophagosome is a double-membrane structure containing either bulk cytoplasm (in the case of nonselective autophagy) or selective cargo that is typically destined for degradation and recycling (in the case of the Cvt pathway, the cargos are resident vacuolar enzymes that are delivered to their site of function as described above).
3.2 Methods to assess phagophore expansion and closure
There are several assays used in yeast to evaluate autophagosome maturation (Figure 3), and most of these can also be utilized in mammalian cells (Table 1). For visualizing and determining whether membrane closure of the phagophore has occurred, microscopy techniques are problematic. Unless three-dimensional tomography is performed, it can be difficult to fully visualize vesicle elongation and phagophore closure by EM [94, 117]. For further detail on the use of EM in investigating autophagy, see section 4.2.3 “Electron microscopy” below. Accumulation of GFP-Atg8 puncta can be visualized by fluorescence microscopy, but it may be difficult to distinguish whether this pool of Atg8 is localized distinctly to the completed autophagosome or the PAS, unless additional markers are used. For further discussion on the use of GFP-Atg8 to assay autophagy by fluorescence microscopy, see section 4.2.1 “Fluorescence-based microscopy assays”. Finally, it is possible to monitor Atg protein disassembly by fluorescence microscopy to assess autophagosome closure [114, 118]. This latter assay relies on the observation that most of the Atg proteins, with the notable exception of Atg8, are released from the completed autophagosome; mutants that are defective in phagophore closure accumulate Atg proteins.
Figure 3. Methods to assess phagophore expansion and closure.
See the text for details.
As mentioned above, Atg9 exists in multiple puncta in wild-type cells, including the PAS, and peripheral sites that may correspond to membrane donors for phagophore formation [13, 119, 120]. In cells defective for delivery of Atg9 to the PAS (anterograde trafficking), this protein is seen in multiple puncta, but not at the PAS (e.g., it does not colocalize with prApe1) [121]. In contrast, in mutants such as atg1Δ, Atg9 accumulates almost entirely at the PAS. This phenotype is the basis for the transport of Atg9 after knocking out ATG1 (TAKA) assay [67, 68, 122], which provides an epistasis analysis for the effect of a mutation combined with atg1Δ. Thus, Atg9 localization can be monitored to provide an assessment of the state of the cycling machinery involved in Atg9 trafficking.
The GFP-Atg8 protease protection assay may also be used as a method to monitor autophagosome maturation. This technique may complement microscopy studies to determine whether a completed autophagosome has formed. The protease protection assay relies on determining whether or not a particular cargo, in this case GFP-Atg8, is sensitive to degradation by the addition of exogenous protease, or whether the cargo is protected within the completed autophagosome structure, following the osmotic lysis of spheroplasts under conditions that retain the integrity of intracellular compartments such as the vacuole and autophagosomes. GFP-Atg8 is an ideal cargo for use in this analysis if the goal is to examine nonselective autophagy. A detailed protocol on how to perform this technique and analysis has been published [117].
A comparable assay can be performed to investigate the biosynthetic Cvt pathway for selective autophagy. In this case the primary cargo, prApe1, is a zymogen that is synthesized with an amino-terminal propeptide that is normally cleaved upon vacuolar delivery; the propeptide is quite sensitive to proteolytic removal, whereas the remainder of the protein is extremely resistant to degradation, similar to other vacuolar hydrolases. A protease protection assay can therefore be used to determine whether the Cvt vesicle is complete, in which case the propeptide will not be accessible to degradation [123]. Nutritional conditions dictate whether prApe1 will be sequestered within an autophagosome or a Cvt vesicle. Under nutrient rich conditions, the Cvt pathway biosynthetically delivers prApe1 to the vacuole, whereas during autophagy the Cvt complex is selectively incorporated into an autophagosome. In either case, the proteolytic processing step can be monitored by western blot. Comprehensive summaries on performing the prApe1 protease protection assay have been published [117, 123].
4. Fusion of the autophagosome with the vacuole
4.1 Background
The third major step of the autophagic pathway occurs when there is fusion between the outer membrane of the autophagosome and the vacuole (Figure 1). The vesicle formed by the remaining inner autophagosome membrane is released into the vacuole lumen, where it is now termed an autophagic body. Although normally broken down rapidly, autophagic bodies can be induced to accumulate within the vacuole either through genetic or pharmacological means [4, 124].
4.2 Methods to assess autophagosome-vacuole fusion
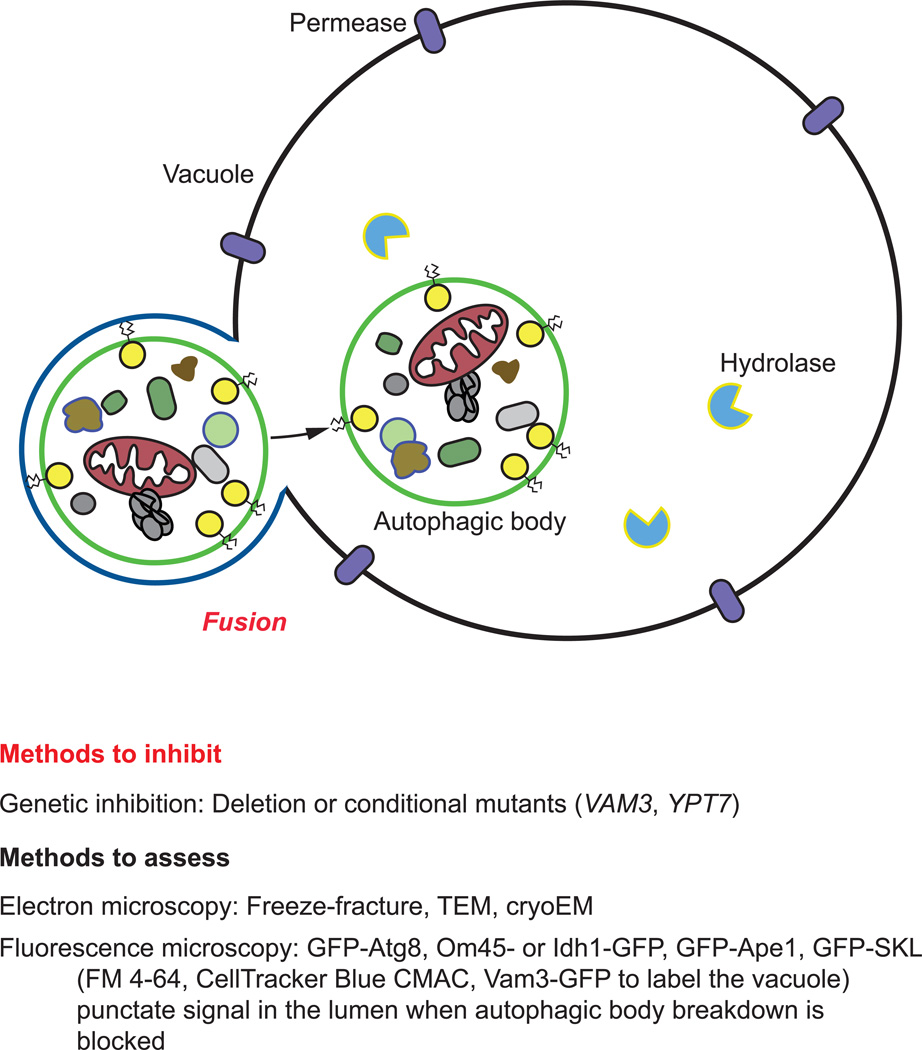
Autophagosome-vacuole fusion can be monitored for both nonselective and selective forms of autophagy utilizing a variety of microscopy-based and biochemical assays (Figure 4, Table 1).
Figure 4. Techniques to evaluate autophagosome maturation and autophagosome-vacuole fusion.
See the text for details
4.2.1 Fluorescence-based microscopy assays
As described earlier, Atg8 is a ubiquitin-like protein essential for autophagy, and a component of the core autophagy machinery that is often used as a marker protein of the PAS, phagophores, and autophagosomes [96, 125]. Cells that have not yet undergone autophagosome-vacuole fusion (but are actively undergoing autophagy) will display the presence of a single GFP-Atg8 punctum outside the vacuolar compartment (corresponding to the PAS, or a completed autophagosome prior to fusion with the vacuole). If fusion of autophagosomes with the vacuole is blocked, multiple GFP-Atg8 puncta will accumulate in the cytosol; this type of block can be achieved genetically (e.g., by deleting VAM3 or YPT7), or pharmacologically (e.g., by treating with the vacuolar-type H+-ATPase inhibitor bafilomycin A1). As mentioned above, it is usually helpful to mark the vacuole (e.g., with FM 4–64), although it can also be observed through light microscopy using differential interference contrast (DIC, or Nomarski optics). Note that GFP-Atg8 puncta do not correspond only to autophagosomes; therefore, it is critical to use other methods as described here (such as protease protection and TEM) to verify their nature.
4.2.2 Biochemical assays
Fusion can be monitored indirectly by assays that reflect subsequent steps of autophagy, in particular autophagic body lysis and cargo degradation. These assays are discussed below in section 5.2.2
4.2.3 Electron microscopy
TEM and its variations (freeze-fracture, cryoEM) remain essential techniques in autophagy research to visualize morphological changes in autophagosome structure and flux. In yeast, it can be difficult to precisely detail the early stages of autophagy due to issues with preservation and fixation techniques that allow for high-quality visualization of cytoplasmic structures, although it is possible to detect phagophores (for a review see [94, 124]). TEM is particularly useful to quantify the number and size of autophagic bodies [124] (see the accompanying review by Guimaraes et al. for a detailed protocol for the examination of autophagic bodies); the absence of these structures may indicate a fusion defect or a block at an earlier stage that prevents autophagosome formation.
As discussed above, in order to monitor autophagic bodies it is necessary to prevent their breakdown through genetic (e.g., pep4Δ) or pharmacological (e.g., PMSF) means. Additionally recommended, although not required, VPS4 may also be deleted in strains undergoing TEM analysis to eliminate the accumulation of multivesicular bodies within the vacuole. When performing TEM, it is also preferable that the studies be conducted at two different time points (with corresponding quantification of autophagic bodies at each time point) to allow for an analysis of flux [68]. TEM may be used as a way to determine whether nonselective or selective forms of autophagy are occurring. In nonselective autophagy, the contents of the sequestering body should be similar to cytoplasm. During selective forms of autophagy, the cargo should obviously be present in the autophagic body, and bulk cytoplasm should be excluded [53]. Comprehensive methodologies on conducting TEM studies in S. cerevisiae and subsequent analysis have been published [94, 124, 126].
Variations on TEM, particularly cryofixation and freeze-fracture studies, have been advantageous to gaining further structural understanding of the later stages of autophagy. In yeast, rapid high-pressure freezing cryofixation and low-temperature dehydration techniques are successful in further preserving the delicate intracellular membrane structure for TEM analysis and providing additional three-dimensional structural detail of the autophagosome [68, 127, 128]. Additionally, TEM performed using the freeze-fracture technique of platinum-carbon replicas allows for the visualization of autophagic structures along the hydrophobic plane of the membrane including integral membrane proteins [68, 127, 129]. More recently, novel electron microscopy techniques, such as the quick-freeze freeze-fracture replica labeling method (QF-FRL), allow for nanometer scale analysis of the distribution of membrane lipids in autophagic compartments [130, 131].
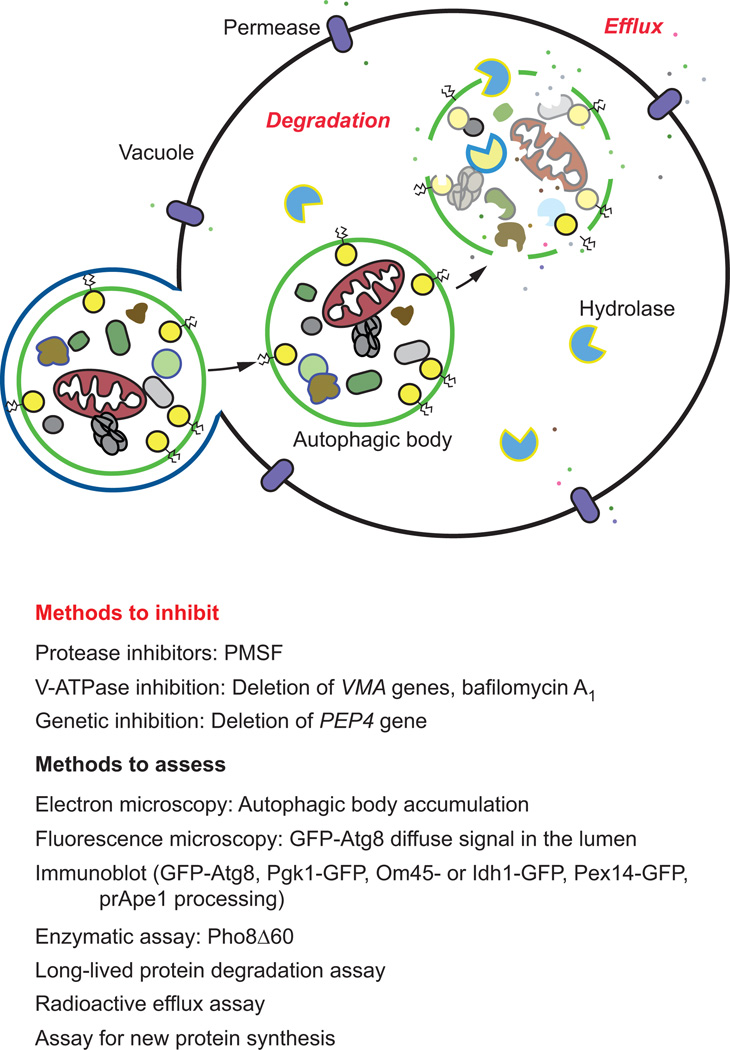
5. Cargo degradation and efflux
5.1 Background
The final steps in autophagy are cargo degradation followed by efflux of the resulting macromolecules (Figure 1). Although the vacuole has enzymes for breaking down each of the major types of macromolecules, efflux systems have only been identified for amino acids. Thus, the mechanism(s) by which nucleic acids, carbohydrates and/or lipids might be recycled for reuse in the cytosol are not known.
5.2 Methods to Inhibit Cargo Degradation
The majority of the methods available to monitor cargo degradation involve the use of pharmacological or genetic approaches to block this stage of autophagy (Figure 5, Table 1). In mammalian cells, for example, the inhibitor bafilomycin A1 is commonly used, and appears to function either at the level of autophagosome-lysosome fusion and/or sub-lysosomal vesicle/cargo breakdown (due to a block in vacuolar acidification), depending on the length of treatment [132, 133]. Thus, this agent may be used to assess autophagic flux, both at the level of fusion and autophagic body/cargo degradation within the vacuole. Bafilomycin A1 can be also be used to confine the autophagic cargo in a recognizable early state to determine the contents of the autophagic body and/or to inhibit degradation and turnover [68, 132, 134]. In yeast, autophagic body breakdown can also be blocked genetically through the deletion of VMA (vacuolar membrane ATPase) genes, encoding subunits of the vacuolar H+-ATPase [135].
Figure 5. Assays to assess cargo degradation and efflux.
Commonly used techniques to inhibit cargo degradation are listed, along with methods to measure degradation and efflux. See the text for details
Vacuolar proteolysis can be blocked through the combined use of pepstatin A (an aspartic protease inhibitor) and E-64D (a cysteine protease inhibitor) [68]. Alternatively, the threonine, cysteine, and serine protease inhibitor leupeptin may be used in combination with pepstatin A and/or E-64D. It is important to note that some of these inhibitors are not membrane permeable, but in some cases membrane permeable derivatives are available [68]. Additionally, the acidotropic agents chloroquine (a weak base amine) or NH4Cl may be used to prevent cargo degradation and turnover by neutralizing the vacuolar pH. As explained above in section 4.2.3 “Electron microscopy.” In yeast, the PEP4 gene can be deleted to inhibit degradation, or the inhibitor PMSF can be applied to block the activity of serine proteases such as Prb1, the major protease in the vacuole.
Efflux of the autophagic cargo by membrane permeases such as Atg22 enables the reuse of the resulting macromolecules in the cytosol [67, 136]. The putative lipase Atg15 is required for the intravacuolar lysis of Cvt and autophagic bodies, and defects in lysis can be monitored by the accumulation of these single-membrane vesicles in the vacuole lumen [137, 138]; a specific lipase assay for Atg15 function has not been described. Successful recycling of degraded components such as amino acids can be measured indirectly by assaying for protein synthesis of vacuolar hydrolases because these enzymes are induced under starvation conditions, and require amino acid efflux during starvation to supply needed amino acids [67, 136].
5.3 Methods to assess cargo degradation and efflux
In contrast to the lysosome, the yeast vacuole is not particularly acidic, but it does contain a range of hydrolases [139], allowing for lysis of the autophagic body, and breakdown of the autophagic cargo. Thus, most of the assays to monitor the final stages of autophagy rely on degradation or proteolysis of autophagic substrates.
5.3.1 Microscopy assays
As mentioned above in section 4.2.3 “Electron microscopy,” accumulation of autophagic bodies within the vacuole indicates a defect in degradation when examined ultrastructurally by EM [124]. Additionally, fluorescence microscopy techniques can be used to visualize cargo degradation. When GFP-Atg8 is delivered to the vacuole, the Atg8 moiety is rapidly degraded. In contrast, GFP is relatively stable in the vacuole and free GFP remains structurally intact (see the accompanying review by Guimaraes et al. for a detailed protocol for the GFP-Atg8 processing assay). This allows for GFP accumulation within the vacuolar compartment, which can be detected by microscopy (the level of vacuolar GFP derived from GFP-Atg8 during growing conditions is relatively low and is easily distinguished as background). Alternatively a nonselective autophagic cargo such as Pgk1-GFP may also be used to monitor autophagic body lysis and cargo degradation [98]. Another variation on this approach is to prevent the breakdown of autophagic bodies, as occurs for example in a pep4Δ mutant. In this case, GFP-Atg8 can be detected as discrete puncta within the vacuole lumen, rather than as a diffuse signal. More recently, the existence of an autophagy receptor, Cue5, that recognizes ubiquitinated proteins for autophagic degradation under nitrogen starvation conditions has been identified [140]. GFP-tagging of Cue5 or one of these cargo proteins [140] represents an alternative to Pgk1 for ubiquitinated substrates.
During selective types of autophagy, such as mitophagy, degradation of vacuolar contents can also be monitored by epifluorescence in much the same way as with GFP-Atg8. Mitochondrial proteins (such as Om45, an outer membrane protein, and Idh1, a matrix protein) can be C-terminally tagged with GFP, and monitored for GFP fluorescence in the vacuole [55] (see the accompanying review by Guimaraes et al. for a detailed protocol for the examination of mitophagy flux). Similarly, Pex14-GFP or GFP-SKL can be used to tag peroxisomes [67, 68] (see the accompanying review by Guimaraes et al. for a detailed protocol for the examination of pexophagy flux).
5.3.2 Biochemical assays
Similar to monitoring the presence of GFP by fluorescence microscopy, degradation can also be assessed by the release of free GFP from GFP-Atg8 on an immunoblot as an indicator of nonselective autophagy. Atg8 is sensitive to degradation within the vacuole; however, as mentioned above, the free GFP signal is relatively resistant to vacuolar hydrolysis, remains intact, and can be detected by western blot [98]. Similarly, other markers such as Pgk1-GFP may also be used to assess nonselective autophagy by blot [98]. It should be kept in mind that GFP-Atg8 is membrane associated (at least initially) and therefore reflects the surface area of the autophagic body (and hence indirectly the autophagosome), whereas Pgk1-GFP levels correspond to the volume. In much the same way, a comparable analysis may be conducted by western blotting for the detection of free GFP during mitophagy or pexophagy using Om45-GFP or Idh1-GFP [51, 55, 58], or Pex14-GFP [43], respectively.
Fusion of Cvt vesicles with the vacuole and lysis of the resulting Cvt body (analogous to an autophagic body) in the Cvt pathway can also be examined by immunoblot to assay for maturation of prApe1 (see the accompanying review by Guimaraes et al. for a detailed protocol for the analysis of Cvt pathway progression by western blot). As discussed above, delivery of prApe1 to the vacuole results in the proteolytic removal of the N-terminal propeptide [43]. This cleavage event can be detected by immunoblot; prApe1 is visualized as an approximately 61-kDa band, whereas mature Ape1 is 50 kDa. Alternatively, radiolabeled prApe1 can also be used in a pulse-chase experiment to assess the kinetics of prApe1 delivery to the vacuole [43].
A very useful assay to monitor autophagy in yeast relies on the measurement of alkaline phosphatase activity [141–144]. In S. cerevisiae, PHO8 encodes vacuolar alkaline phosphatase that transits to the vacuole through a part of the secretory pathway [142]. A truncated form of Pho8, Pho8Δ60, which lacks the N-terminal transmembrane domain that normally functions as an internal uncleaved signal sequence, accumulates in the cytosol and is only delivered to the vacuole through nonselective autophagy [141, 143, 144]. The magnitude of autophagy can therefore be monitored through an enzymatic assay in strains where the endogenous PHO8 gene has been replaced with pho8Δ60 (see the accompanying review by Guimaraes et al. for a detailed protocol for the Pho8Δ60 assay). Furthermore, the Pho8Δ60 assay can be modified to evaluate changes in selective forms of autophagy. For example, to assay for mitophagy, Pho8Δ60 can be fused to a mitochondrial localization sequence [145].
Finally, the proteasome is considered to be the primary means for the degradation of short-lived proteins, whereas autophagy is used to degrade long-lived proteins. Accordingly, autophagic degradation can be assessed by monitoring the turnover of radiolabelled long-lived proteins [67, 68]. Although this technique has fallen out of favor, likely due to the need to use radioisotopes, it is a useful method that can be used to follow autophagic flux.
Although relatively little attention has been paid to the final step of autophagy, efflux of the macromolecules resulting from the breakdown process, this step is critical as part of the stress response [136]. Efflux can be monitored non-radioactively by examining the synthesis of nascent proteins. Under starvation conditions, the translation of most proteins is inhibited. In contrast, a few proteins, such as vacuolar hydrolases are upregulated, and this synthesis depends on the release of amino acids from the vacuole as a result of autophagy. Thus, the increased synthesis of these enzymes can be followed by western blot. Direct measurement of the release of radioactive amino acids from labeled proteins can also be used to monitor efflux [67].
6. Conclusions
We present here an overview of the most commonly used methods to assess autophagy at each of the four main stages of the pathway. Due to the inhererent complexity and dynamic nature of autophagy, it is always recommended that multiple and complementing approaches be conducted to assay any phase. S. cerevisiae and other yeast species provide unique models for the study of autophagy due to the array of methods that are exclusively available for use in these organisms.
There remain many questions to be answered in this ever-expanding field. We hope that the survey of assays provided here are helpful to investigators of all disciplines who are interested in exploring autophagy in their research.
Acknowledgements
The authors apologize to those whose work was not included here due to space limitations. D.J.K. is supported by funding from the National Institutes of Health (GM053396). F.R. is supported by ALW Open Program (821.02.017 and 822.02.014), DFG-NWO cooperation (DN82-303), SNSF Sinergia (CRSII3_154421) and ZonMW VICI (016.130.606) grants.
Abbreviations
- 3-MA
3-methyladenine
- Atg
autophagy related
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- GFP
green fluorescent protein
- PAS
phagophore assembly site
- PKA
protein kinase A
- PMN
piecemeal microautphagy of the nucleus
- TEM
transmission electron microscopy
- TOR
target of rapamycin
- TORC1
TOR complex 1
- VMA
vacuolar membrane ATPase
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(7):651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 2.Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24(1):69–79. doi: 10.1038/cr.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge L, et al. The protein-vesicle network of autophagy. Curr Opin Cell Biol. 2014;29C:18–24. doi: 10.1016/j.ceb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Takeshige K, et al. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119(2):301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao K, et al. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26(1):9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao K, et al. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy. 2014;10(4):652–661. doi: 10.4161/auto.27852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12(9):814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y, et al. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veenhuis M, et al. Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch Microbiol. 1983;134(3):193–203. doi: 10.1007/BF00407757. [DOI] [PubMed] [Google Scholar]
- 10.Klionsky DJ. Citing recent declines in the discovery of new ATG genes, some scientists now suggest that the end of autophagy research may be within sight. Autophagy. 2014;10(5):715–716. doi: 10.4161/auto.28813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism machinery and regulation. Genetics. 2013;194(2):341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7(7):673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 13.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20(3):460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69(7):1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue Y, Klionsky DJ. Regulation of macroautophagy in Saccharomyces cerevisiae. Semin Cell Dev Biol. 2010;21(7):664–670. doi: 10.1016/j.semcdb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 18.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273(7):3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 19.Chang YY, et al. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem Soc Trans. 2009;37(Pt 1):232–236. doi: 10.1042/BST0370232. [DOI] [PubMed] [Google Scholar]
- 20.Mao K, et al. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci U S A. 2013;110(31):E2875–E2884. doi: 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao K, et al. The role of Atg29 phosphorylation in PAS assembly. Autophagy. 2013;9(12):2178–2179. doi: 10.4161/auto.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuura A, et al. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192(2):245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 24.Kamada Y, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30(4):1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151(7):1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawamata T, et al. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19(5):2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noda T, et al. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148(3):465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin M, Klionsky DJ. Transcriptional regulation of ATG9 by the Pho23-Rpd3 complex modulates the frequency of autophagosome formation. Autophagy. 2014;10(9) doi: 10.4161/auto.29641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin M, et al. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol. 2014;24(12):1314–1322. doi: 10.1016/j.cub.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146(2):290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nair U, Klionsky DJ. Autophagosome biogenesis requires SNAREs. Autophagy. 2011;7(12):1570–1572. doi: 10.4161/auto.7.12.18001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge L, et al. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge L, Schekman R. The ER-Golgi intermediate compartment feeds the phagophore membrane. Autophagy. 2014;10(1):170–172. doi: 10.4161/auto.26787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi-Nishino M, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11(12):1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 35.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182(4):685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yla-Anttila P, et al. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5(8):1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi Y, et al. Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy. Autophagy. 2011;7(1):61–73. doi: 10.4161/auto.7.1.14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495(7441):389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 39.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravikumar B, Moreau K, Rubinsztein DC. Plasma membrane helps autophagosomes grow. Autophagy. 2010;6(8):1184–1186. doi: 10.4161/auto.6.8.13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravikumar B, et al. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12(8):747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puri C, et al. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154(6):1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- 44.Kanazawa T, et al. Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes. J Biol Chem. 2004;279(9):8452–8459. doi: 10.1074/jbc.M306337200. [DOI] [PubMed] [Google Scholar]
- 45.Stephan JS, et al. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106(40):17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scarlatti F, et al. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem. 2004;279(18):18384–18391. doi: 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- 47.Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng X, Kinsella TJ. Mammalian target of rapamycin and S6 kinase 1 positively regulate 6-thioguanine-induced autophagy. Cancer Res. 2008;68(7):2384–2390. doi: 10.1158/0008-5472.CAN-07-6163. [DOI] [PubMed] [Google Scholar]
- 49.Kraft C, et al. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10(5):602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 50.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 51.Kanki T, Kang D, Klionsky DJ. Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy. 2009;5(8):1186–1189. doi: 10.4161/auto.5.8.9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanki T, Klionsky DJ. The molecular mechanism of mitochondria autophagy in yeast. Mol Microbiol. 2010;75(4):795–800. doi: 10.1111/j.1365-2958.2009.07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kissova I, et al. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3(4):329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- 54.Kissova I, et al. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279(37):39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 55.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283(47):32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tal R, et al. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem. 2007;282(8):5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- 57.Abeliovich H. Stationary-phase mitophagy in respiring Saccharomyces cerevisiae. Antioxid Redox Signal. 2011;14(10):2003–2011. doi: 10.1089/ars.2010.3807. [DOI] [PubMed] [Google Scholar]
- 58.Mao K, et al. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol. 2011;193(4):755–767. doi: 10.1083/jcb.201102092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;112(Pt 22):4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- 60.Farre JC, et al. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14(3):365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin M, Liu X, Klionsky DJ. SnapShot: Selective autophagy. Cell. 2013;152(1–2):368–368. doi: 10.1016/j.cell.2013.01.004. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunn WA, Jr., et al. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1(2):75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 63.Yorimitsu T, et al. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281(40):30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuck S, Gallagher CM, Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci. 2014 doi: 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petiot A, et al. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275(2):992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 66.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8(11):931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 67.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3(3):181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 68.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blommaart EF, et al. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243(1–2):240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 70.Kihara A, et al. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152(3):519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stack JH, Emr SD. Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J Biol Chem. 1994;269(50):31552–31562. [PubMed] [Google Scholar]
- 72.Thumm M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349(2):275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 73.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1–2):169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 74.Suzuki K, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20(21):5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.George MD, et al. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol Biol Cell. 2000;11(3):969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yen WL, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188(1):101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stack JH, et al. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129(2):321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Budovskaya YV, et al. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279(20):20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Binda M, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35(5):563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 80.Kira S, et al. Reciprocal conversion of Gtr1 and Gtr2 nucleotide-binding states by Npr2-Npr3 inactivates TORC1 and induces autophagy. Autophagy. 2014;10(9):1565–1578. doi: 10.4161/auto.29397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yorimitsu T, et al. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18(10):4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanki T, et al. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17(1):98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17(1):87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 84.Motley AM, Nuttall JM, Hettema EH. Atg36: the Saccharomyces cerevisiae receptor for pexophagy. Autophagy. 2012;8(11):1680–1681. doi: 10.4161/auto.21485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012;31(13):2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584(7):1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scott SV, et al. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7(6):1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shintani T, et al. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3(6):825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem. 2001;276(23):20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuga M, et al. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J Biol Chem. 2011;286(15):13704–13713. doi: 10.1074/jbc.M110.173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Suzuki K, et al. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J Biol Chem. 2010;285(39):30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16(4):1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kirisako T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147(2):435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eskelinen EL, et al. Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy. 2011;7(9):935–956. doi: 10.4161/auto.7.9.15760. [DOI] [PubMed] [Google Scholar]
- 95.Baba M, et al. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139(7):1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19(8):3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Geng J, Klionsky DJ. Determining Atg protein stoichiometry at the phagophore assembly site by fluorescence microscopy. Autophagy. 2010;6(1):144–147. doi: 10.4161/auto.6.1.10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Welter E, Thumm M, Krick R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy. 2010;6(6):794–797. doi: 10.4161/auto.6.6.12348. [DOI] [PubMed] [Google Scholar]
- 99.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152(4):657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128(5):779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Devenish RJ, et al. Monitoring organelle turnover in yeast using fluorescent protein tags. Methods Enzymol. 2008;451:109–131. doi: 10.1016/S0076-6879(08)03209-6. [DOI] [PubMed] [Google Scholar]
- 102.Urbanowski JL, Piper RC. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J Biol Chem. 1999;274(53):38061–38070. doi: 10.1074/jbc.274.53.38061. [DOI] [PubMed] [Google Scholar]
- 103.Miller-Fleming L, et al. Detection of Saccharomyces cerevisiae Atg13 by western blot. Autophagy. 2014;10(3):514–517. doi: 10.4161/auto.27707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fujioka Y, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21(6):513–521. doi: 10.1038/nsmb.2822. [DOI] [PubMed] [Google Scholar]
- 105.Bartholomew CR, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A. 2012;109(28):11206–11210. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang WP, et al. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275(8):5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 107.Itakura E, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fogel AI, et al. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol. 2013;33(18):3675–3688. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chan TF, et al. Regulation of APG14 expression by the GATA-type transcription factor Gln3p. J Biol Chem. 2001;276(9):6463–6467. doi: 10.1074/jbc.M008162200. [DOI] [PubMed] [Google Scholar]
- 110.Yu ZQ, et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8(6):883–892. doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Klionsky DJ, Codogno P. The mechanism and physiological function of macroautophagy. J Innate Immun. 2013;5(5):427–433. doi: 10.1159/000351979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nakatogawa H, et al. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 113.Kirisako T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151(2):263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nair U, et al. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy. 2012;8(5):780–793. doi: 10.4161/auto.19385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Klionsky DJ. For the last time, it is GFP-Atg8, not Atg8-GFP (and the same goes for LC3) Autophagy. 2011;7(10):1093–1094. doi: 10.4161/auto.7.10.15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jin M, Klionsky DJ. Regulation of autophagy: modulation of the size and number of autophagosomes. FEBS Lett. 2014;588(15):2457–2463. doi: 10.1016/j.febslet.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nair U, et al. GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy. 2011;7(12):1546–1550. doi: 10.4161/auto.7.12.18424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cebollero E, et al. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol. 2012;22(17):1545–1553. doi: 10.1016/j.cub.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reggiori F, et al. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1(2):101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reggiori F, et al. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6(1):79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 121.He C, et al. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell. 2008;19(12):5506–5516. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cheong H, et al. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16(7):3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yen WL, Klionsky DJ. Proteinase protection of prApe1 as a tool to monitor Cvt vesicle/autophagosome biogenesis. Autophagy. 2012;8(8):1245–1249. doi: 10.4161/auto.20916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Backues SK, et al. Estimating the size and number of autophagic bodies by electron microscopy. Autophagy. 2014;10(1):155–164. doi: 10.4161/auto.26856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lang T, et al. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17(13):3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Griffith J, et al. A cryosectioning procedure for the ultrastructural analysis and the immunogold labelling of yeast Saccharomyces cerevisiae. Traffic. 2008;9(7):1060–1072. doi: 10.1111/j.1600-0854.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- 127.Baba M. Electron microscopy in yeast. Methods Enzymol. 2008;451:133–149. doi: 10.1016/S0076-6879(08)03210-2. [DOI] [PubMed] [Google Scholar]
- 128.Baba M, et al. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124(6):903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20(6):465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- 130.Fujimoto T, Yamamoto H, Ohsumi Y. Different phosphatidylinositol 3-phosphate asymmetries in yeast and mammalian autophagosomes revealed by a new electron microscopy technique. Autophagy. 2014;10(5):933–935. doi: 10.4161/auto.28489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cheng J, et al. Yeast and mammalian autophagosomes exhibit distinct phosphatidylinositol 3-phosphate asymmetries. Nat Commun. 2014;5:3207. doi: 10.1038/ncomms4207. [DOI] [PubMed] [Google Scholar]
- 132.Klionsky DJ, et al. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4(7):849–850. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 133.Yamamoto A, et al. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23(1):33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 134.Zhu J, Dagda RK, Chu CT. Monitoring mitophagy in neuronal cell cultures. Methods in molecular biology. 2011;793:325–339. doi: 10.1007/978-1-61779-328-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Graham LA, Powell B, Stevens TH. Composition and assembly of the yeast vacuolar H(+)-ATPase complex. J Exp Biol. 2000;203(Pt 1):61–70. doi: 10.1242/jeb.203.1.61. [DOI] [PubMed] [Google Scholar]
- 136.Yang Z, et al. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17(12):5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Epple UD, et al. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J Bacteriol. 2001;183(20):5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Teter SA, et al. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J Biol Chem. 2001;276(3):2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54(3):266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158(3):549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 141.Klionsky DJ. Monitoring autophagy in yeast: the Pho8Delta60 assay. Methods Mol Biol. 2007;390:363–371. doi: 10.1007/978-1-59745-466-7_24. [DOI] [PubMed] [Google Scholar]
- 142.Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8(8):2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Noda T, Klionsky DJ. The quantitative Pho8Δ60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 144.Noda T, et al. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210(1):126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 145.Campbell CL, Thorsness PE. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J Cell Sci. 1998;111(Pt 16):2455–2464. doi: 10.1242/jcs.111.16.2455. [DOI] [PubMed] [Google Scholar]