Summary
Drugs that inhibit the MAPK pathway have therapeutic benefit in melanoma, but responses vary between patients, for reasons that are still largely unknown. Here we aim at explaining this variability using pre- and post-MEK inhibition transcriptional profiles in a panel of melanoma cell-lines. We found that most targets are context-specific – under the influence of the pathway in only a subset of cell-lines. We developed a computational method to identify context-specific targets, and found differences in the activity levels of the interferon pathway, driven by a deletion of the interferon locus. We also discovered that IFNα/β treatment strongly enhances the cytotoxic effect of MEK inhibition, but only in cell lines with low activity of interferon pathway. Taken together, our results suggest that the interferon pathway plays an important role, and predicts, the response to MAPK inhibition in melanoma. Our analysis demonstrates the value of system-wide perturbation data in predicting drug response.
Introduction
Advances in the identification and understanding of oncogenic pathways, as well as the development of highly specific drugs, allow clinicians to tailor treatments based on tumor genomics. However, drug response is variable in both experimental systems and in the clinic, even when all tumors harbor mutations that activate the pathways targeted by the drugs (Flaherty et al., 2010; Joseph et al., 2010; Pratilas et al., 2009; Slamon et al., 2001).
Here, we focus on the variability in response to ERK-MAPK pathway inhibition in melanoma. At least 70% of melanoma tumors harbor an oncogenic mutation in the ERK-MAPK pathway (Hodis et al., 2012), and drugs targeting this pathway have been approved with observed clinical success (Sosman et al., 2012). However, phenotypic responses to MAPK pathway inhibitors, both in patients and in vitro, vary dramatically (Flaherty et al., 2010; Joseph et al., 2010).
Several molecular mechanisms have been proposed to explain response heterogeneity. Feedback reactivation of the pathway attenuates the inhibitory effects of the drugs (Lito et al., 2012; Poulikakos et al., 2010). Other studies found PTEN and MITF status correlated to response heterogeneity (Johannessen et al., 2013; Paraiso et al., 2011; Xing et al., 2012), but these explain only part of the observed variability. While these factors may contribute to the heterogeneous response, they are limited by characterizing the overall phenotypic response, without distinguishing cytotoxic from cytostatic phenotypes.
We aim to explain the phenotypic variability in response to MAPK inhibition by studying the transcriptional response to this inhibition. While most studies use correlation between genetic and genomic features and phenotypic outcome to identify predictive features (Barretina et al., 2012; Garnett et al., 2012), we take a different approach. We use pre- and post- MEK inhibition expression data in a panel genetically diverse cell lines to get a better understanding of the targets and pathways regulated by ERK-MAPK, and use these regulation patterns, and how they differ between tumors, to explain the variability in response to treatment. In this study, genes with changes in their mRNA levels following MEK inhibition are defined as targets of the MAPK pathway.
We found extensive heterogeneity in the transcriptional response to MEK inhibition between cell lines. Although all cell lines harbor a MAPK pathway activating mutation (either NRAS or BRAF), a vast majority of MAPK targets are context-specific – under the control of the pathway in only a subset of cell lines (hereafter, a context refers to any subset of the cell lines, with or without a known, shared and unique genetic feature). As these differences could reveal the molecular mechanisms underlying phenotypic variance, we developed a computational tool, COSPER (COntext SPEcific Regulation), to identify context-specific targets using pre- and post-perturbation gene expression data.
Analysis with COSPER revealed that the IFN-Type I pathway presents context-specific behavior. While studying this pathway, we found that Type-I Interferon (IFNα/β) strongly enhances the cytotoxic response of MEK inhibition. We show that cell lines with high basal activity of the interferon pathways are resistant to MEK inhibition alone or its combination with IFNα/β. We identified that a deletion of the interferon locus is correlated with that differential basal activity level of the interferon pathway and predicts the cytotoxic response of MEK inhibition.
Our results demonstrate that inhibition of a key oncogenic pathway leads to substantially different transcriptional programs in different cell lines. We show that a better understanding of the interactions and activity state of different pathways would enable clinicians to tailor new and unexpected drug combinations to individual patients, which may lead to better clinical responses.
Results
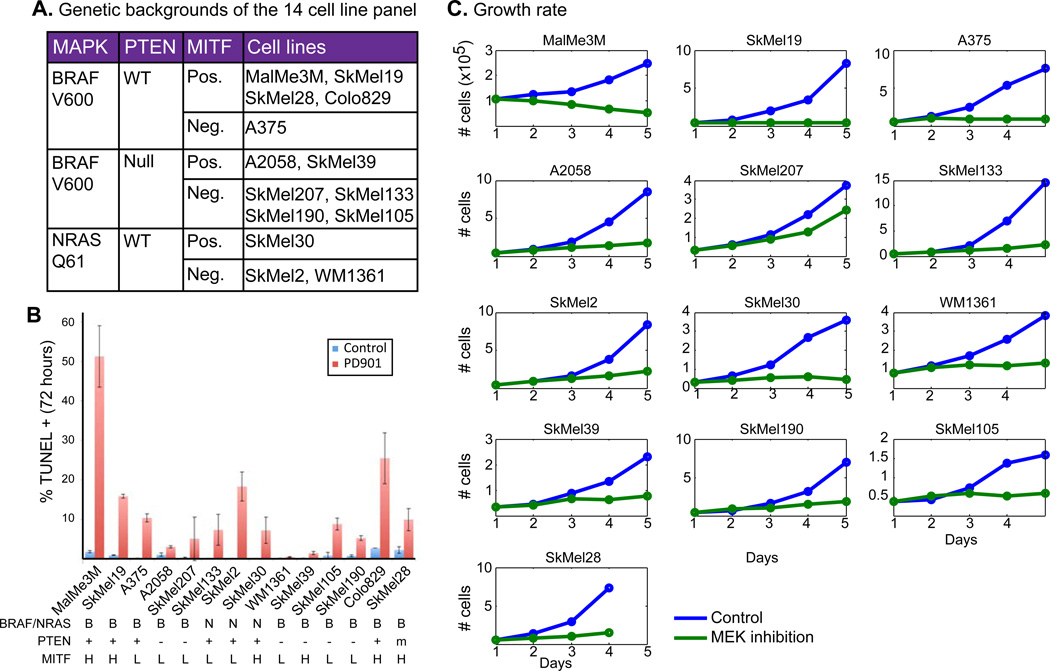
Cell lines harboring MAPK-activating mutations vary in their response to inhibition of the pathway, both in rate of proliferation and death (Xing et al., 2012). To characterize the targets and crosstalk of the ERK-MAPK pathway, we chose a panel of 14 genetically diverse melanoma cell lines. This panel represents the spectrum of common genetic aberrations in melanoma – MAPK mutations, MITF amplification and PTEN deletion (figure 1A).
Figure 1.
Phenotypic heterogeneity in response to MEK inhibition in melanoma. A. BRAF, NRAS, PTEN and MITF status show the genetic diversity of our panel of 14 cell line panel. We used 50nM of PD325901 that fully inhibits the pathway in both NRAS and BRAF mutant cell lines (figure S1A). B. Mean percentage ± SD of TUNEL+ cells after 72 hours of treatment with DMSO (control) or PD901 (50nM). MAPK mutation, PTEN status and MITF status are listed at the bottom. C. Growth curves of untreated (blue) and MEK-inhibited (green) cells showing dramatically different responses.
We compared the transcriptional and phenotypic response to MAPK pathway inhibition of both NRAS-mut and BRAF-mut cell lines using a MEK inhibitor (PD325901, 50nM) that fully inhibits the pathway in all cell lines at 8 hours (figure S1A), and not the clinically used BRAF inhibitor, which works on BRAF-mut cells only. A comparison of the MEK inhibitor with a BRAF inhibitor (PLX4720 (Tsai et al., 2008)) in a BRAF-V600E cell line shows almost identical transcriptional response, both in the genes affected and the extent of transcriptional change (see supplementary information and figure S1B for more information).
We first characterized the cell lines’ phenotypic responses to MEK inhibition. The cell lines display a wide range of cytotoxic responses, as well as differences in proliferation under MEK inhibition (figure 1B,C). Notably, and contrary to previously published results (Barretina et al., 2012; Xing et al., 2012), we found that key genetic aberrations common in melanoma, including MITF and PTEN status, and MAPK mutation type, fail to fully explain the response heterogeneity (figure 1B, S1C–D).
Heterogeneity in transcriptional response to MAPK inhibition
To identify MAPK transcriptional targets, and how these differ across cell lines, we characterized the transcriptional response before and after MEK inhibition. We measured gene expression 8 hours following MEK inhibition to capture the peak of the transcriptional changes following inhibition (Pratilas et al., 2009).
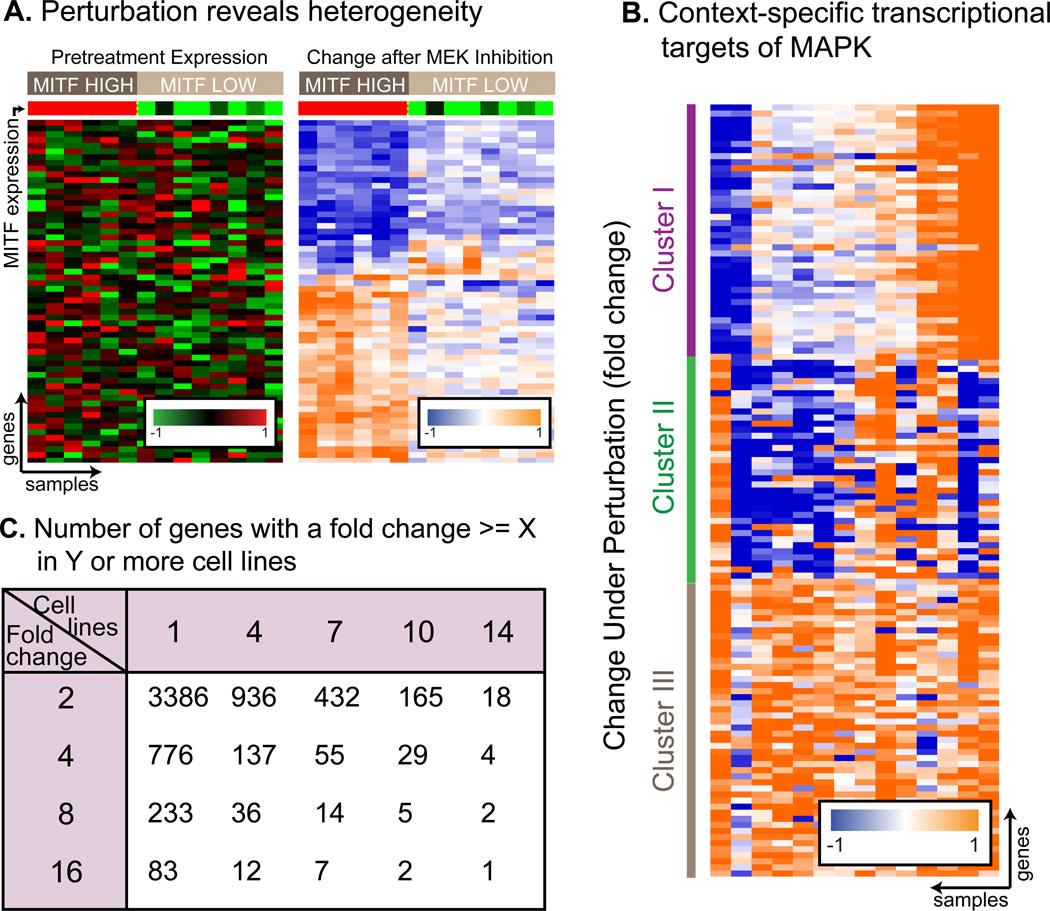
Our data show that MEK inhibition reveals transcriptional patterns that are not observable in steady state expression - genes that display no correlation in their gene expression levels before pathway inhibition, become strongly correlated following MEK inhibition. In figure 2A (list of genes in Table S1) we see a set of genes that is only correlated with MITF levels in its transcriptional response to MEK inhibition. These same genes show no correlation between themselves and with MITF levels in their baseline, pre-inhibition levels. Their association with MITF can therefore only be revealed when measuring their post-perturbation response. We found this phenomenon widespread with almost 4000 genes that behave in concert only after perturbation (full details in Supplementary Information).
Figure 2.
Transcriptional heterogeneity in response to MEK inhibition in melanoma. A. MEK inhibition reveals transcriptional targets of MEK (right), undetectable in steady-state conditions (left). These genes are only regulated by MEK in high MITF cell lines. In this and other heat - map figures, columns are samples and rows are genes. Red-Black-Green plots represent pre-treatment levels comparing between cell lines, and Orange-White-Blue plots show expression fold change 8 hours after treatment (both in log2 scale). The same genes (in the same order) are shown in both heatmaps, table S1 lists the genes in the figure. B. 3 gene clusters demonstrating the extent of context-specificity of MAPK targets. In each of the 3 clusters, cell lines show different response to MEK inhibition. Moreover, each cell line is unique and responses for each cell lines are different in each cluster. C. Number of differentially expressed genes as a function of fold change and number of cell lines. Arbitrarily choosing the cutoff is likely to mislabel hundreds of genes. BRAF and PTEN status are not correlated with transcriptional response to MEK inhibition (figure S2).
The most striking phenomenon observed in post inhibition data is the heterogeneity in response to MEK inhibition across different cell lines. Although all cell lines harbor a MAPK activating mutation, most genes are regulated by the MAPK pathway in only a subset of the cell lines, and no two cell lines behave similarly (figure 2B). For example, only 18 genes change by >2 fold in all 14 cell lines, but 936 genes pass this threshold in 4 or more cell lines (figure 2C). We term those genes context-specific targets – under the control of the MAPK pathway in only a subset of cell lines. The term “context” is used to represent a known or unknown genetic or genomic background that is shared by a subset of cell lines, but not by the others. Notably, we didn’t find a significant enrichment of genes regulated by MAPK only in BRAF-mut or NRAS-mut cell lines (figure S2A–B).
Our data show that MEK inhibition leads to different phenotypic responses in different cell lines, and that MAPK regulates different genes, and presumably different pathways, in different cell lines. We hypothesized that that differential regulation of pathways and genes underlies the phenotypic variability, and identifying context-specific targets might explain it. Therefore, we investigated the patterns of context-specific regulation.
Context-specific regulation
The first step in the analysis was to identify targets of the MAPK pathway using post-inhibition changes in expression levels. However, figure 2C show that choosing an arbitrary fold-change threshold and number of tumors to classify genes as targets and not-targets can lead to misclassification. We therefore developed a method that specifically searches for contextspecific MAPK regulated genes using both pre- and post-perturbation data.
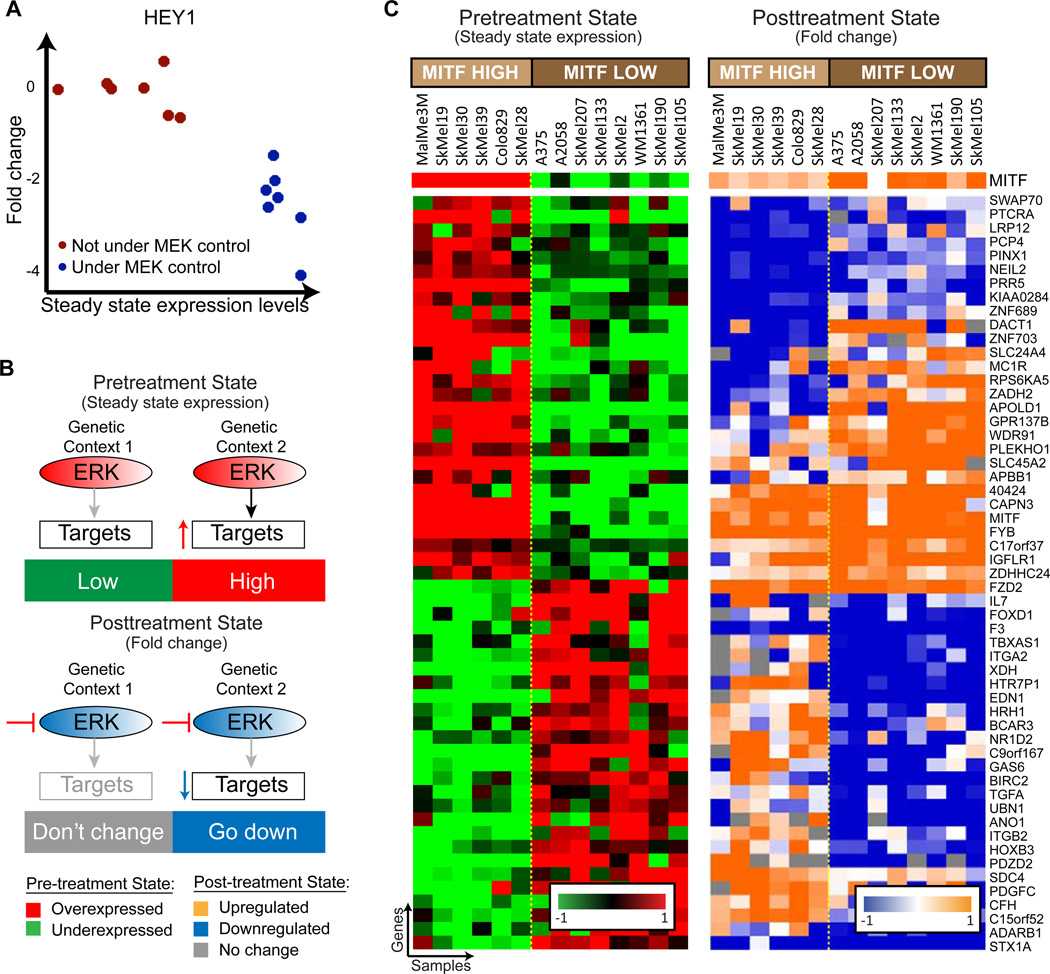
At the core of our method is the observation that some genes show distinct patterns of context-specific regulation both before and after MAPK inhibition. HEY1 is used as an example of a context-specific regulated gene (figure 3A). HEY1 has two states, or contexts, that are detectable both in pre- and post- inhibition expression levels. In one context (i.e. one set of cell lines) it is not under the control of MAPK, and shows low basal expression levels when MAPK is active, and its expression doesn’t change after inhibition of the pathway (figure 3A, red dots). In the second group of cell lines (blue dots), HEY1 is up-regulated by MAPK and therefore shows high basal expression levels before pathway inhibition, and its expression drops following MEK inhibition.
Figure 3.
COSPER identifies COntext-SPEcific Regulation – genes are under the control of MAPK in only a subset of cell lines, both before and after inhibition. A. HEY1 is an example for a context-specific target - regulated by MAPK in only a subset of cell lines (blue dots). MEK inhibition doesn’t affect its expression in the other group of cells (red), and its basal expression is lower in these cell lines. B. A cartoon of context-specific regulation exhibited by HEY1. ERK up-regulates a set of targets only in genetic context 2, while it has no effect in the context 1 (upper panel). Therefore, the genes are down-regulated following MEK inhibition only in genetic context 2 (lower panel). C. COSPER identifies gene clusters with context-specific regulation. The cluster contains genes controlled by MAPK only in cell lines with high MITF mRNA expression. MITF expression, which is not part of this cluster, is in the top row. Several patterns of regulation (up- and down-regulations) are shown.
As genes are often co-regulated, we expect clusters of context-specific co-regulated genes (figure 3B). Using clusters of genes to identify contexts and context-specific targets enables us to computationally reduce the experimental noise, and increase the probability that the association between a context and a gene is a product of an underlying biological phenomenon rather than a spurious association.
We developed a computational method - COSPER (COntext-SPEcific Regulation) - that uses pre- and post- inhibition transcriptional data to identify context-specific co-regulated clusters of genes.
COSPER identifies context-specific MAPK regulated genes
COSPER can be viewed as a bi-clustering algorithm – designed to identify gene clusters that show context-specific regulation patterns (figure 3B). In each cluster, the cell lines are divided into two groups, or contexts, and the genes have a distinct but different behavior in each context, both before and after pathway inhibition. Combining data from both pre- and post- pathway inhibition focuses the search to genes that are likely to be regulated by the MAPK pathway. By identifying the genes regulated by the pathway in only a subset of cell lines, e.g. sensitive versus resistant, COSPER helps focus the analysis on genes and pathways that are likely to contribute to the phenotypic response to pathway inhibition.
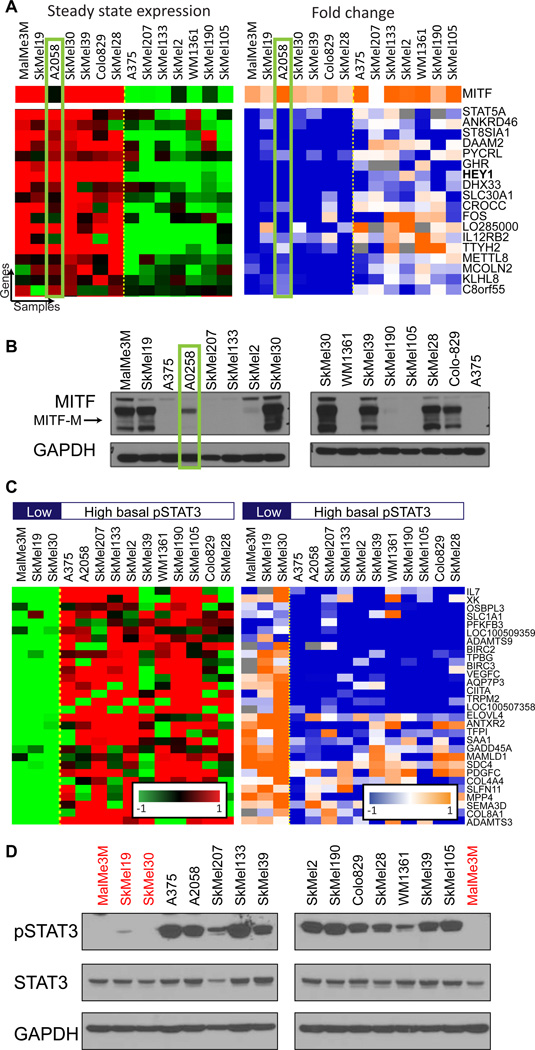
COSPER is not restricted to the patterns depicted in figure 3A, and can identify any context-specific pattern of regulation (figure 3C). Overall, COSPER identified 70 context-specific clusters with 5 genes or more, and assigned a total of 1024 genes to clusters (genes are allowed to belong to more than one cluster, list of all clusters appears in Table S2, for full algorithmic detail see supplementary information). Fifteen clusters associate with MITF, containing 401 genes in total. These clusters either have a perfect correlation with MITF expression, such as the cluster in figure 3C, or have 1–2 cell lines that “switch sides” - they behave similarly to cell lines with the opposite MITF status (figures 4A and S3A, which include HEY1).
Figure 4.
Analysis of the clusters’ genes allows facilitates the identification of pathways that exhibit context-specific interactions with the MAPK pathway. A. A cluster associated with MITF-M protein levels identified by COSPER. Its genes are overexpressed in high-MITF-M cell lines, and are down-regulated only in these cells after MEK inhibition. MITF expression is in the top row. The cluster is almost perfectly correlated with MITF-expression, except for one cell line highlighted in green. The binding site of MITF is overrepresented in the promoters of the cluster genes (pvalue=10−3). Only part of this cluster’s genes is shown (full cluster appears in figure S3A). B. MITF protein levels in all 14 cell lines. A2058 (green rectangle) is the only low mRNA-MITF cell line that expresses the MITF-M isoform. C. Additional cluster identified by COSPER. The cluster’s genes are enriched for STAT3-related GO annotations (full cluster appears in figure S3D). A bar indicating pSTAT3 levels appears in top row. D. As predicted by COSPER, pSTAT3-Y705 levels are correlated with the cluster. Cell lines with low-pSTAT3 are marked in red, matching the first 3 pSTAT3-low cell lines shown in C.
Notably, none of the clusters correlate with the oncogenic activation of MAPK (BRAF or NRAS), or with the cells’ PTEN status. Moreover, we also explicitly tested for genes correlated with these aberrations, but no gene’s expression of was found to be significantly associated with these mutations (see supplementary information and figure S2).
Inferring pathway activity using COSPER
COSPER identifies clusters of genes downstream of MAPK that show context-specific behavior. Using standard gene set enrichment analysis methods, we can postulate the pathways that govern the differential expression of those genes, and the activity of the clusters’ regulators.
For example, the clusters in figures 3C and 4A demonstrate the different roles of MITF isoforms. While the cluster in figure 3C correlates with MITF mRNA expression, the cluster in 4A correlates with the abundance of the MITF-M protein isoform (figure 4B). MITF itself is also regulated by MAPK, both at the mRNA and protein levels (figure S3B,C)..
The different functional annotations of the genes in the two clusters suggest that different MITF isoforms regulate different processes. The promoters for genes in the MITF-M cluster are highly enriched for the MITF binding site (CACATG)(Levy et al., 2006) (pvalue=10−3 compared with 0.7 for genes in MITF-expression cluster, see supplementary information for details). However, the MITF-expression cluster, but not the MITF-M cluster, is enriched for the GO annotation “melanocyte differentiation” (qvalue=10−4), suggesting that another isoform of MITF is responsible for cellular differentiation.
An additional cluster COSPER identified is correlated with STAT3 activity (figures 4C, S3D). Gene ontology enrichment analysis found that the genes in the cluster are enriched for cytokine-cytokine receptor pathway (qvalue<10−3), and with miR-19 and miR-17 (qvalue<10−3), two miRs known to be regulated by pSTAT3 (Dai et al., 2011; Zhang et al., 2012), which led us to suspect that this cluster is associated with STAT3 regulation. We confirmed these predictions by measuring STAT3 activity in the cell lines. Levels of pSTAT3-Y705, an indicator for STAT3 activity, but not of pSTAT3-S727, match the cluster’s contexts (figure 4D, S3E).
Using the MITF and STAT3 examples, we showed that by combining information from both steady state and post-perturbation data, COSPER infers both network state and interactions between pathways. However, when running COSPER on each data set alone, the resulting clusters are much larger, less specific, and therefore less informative than the clusters resulting from using both conditions (for full comparison analysis see supplementary information). Steady state data give a point of reference and provides information on basal pathway activation state, while post-inhibition data enables the identification of genes directly regulated by the MAPK pathway. Therefore, only by using both data sets COSPER identifies the state and interconnectivity of pathways.
Interferon-STAT1 pathway is differentially regulated in cell lines
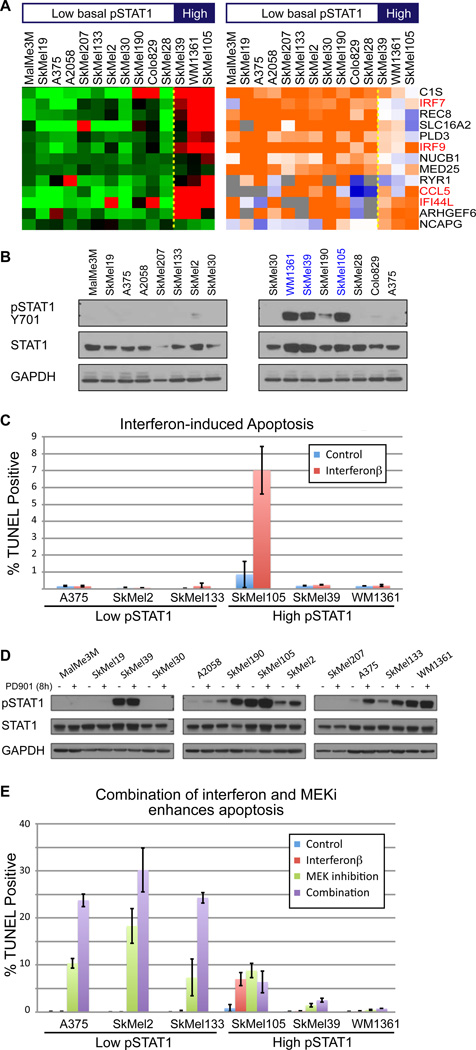
COSPER also identified a cluster that contains several interferon targets, IRF7, IRF9, CCL5 and IFI44L (figure 5A), which reflect the activity of the Type I interferon pathway (Hecker et al., 2013). Since Type I interferon (IFNα/β) is one of the few approved drugs for metastatic melanoma, we decided to focus on this cluster.
Figure 5.
IFNβ enhances cytotoxic response of MEK inhibition in low-pSTAT1 cell lines. A. COSPER identified a cluster containing several known interferon targets (marked in red). Three cell lines have high target expression, and MEK inhibition upregulates the pathway in the other 11 cell lines. A bar indicating pSTAT1 levels at the top and these are different than the high pSTAT3 cell lines of figure 4. B. pSTAT1-Y701, a marker for interferon-STAT1 pathway activity, is correlated with the gene expression and shows high basal activation level in the 3 high cell lines (blue). C. High interferon pathway activity is necessary, but not sufficient, for IFN-induced death. We used TUNEL staining as a marker for apoptosis 72 hours after IFNβ treatment (mean levels ± SD). Only one out of 3 high-pSTAT1 cell lines respond to IFNβ (red) and none of the low-PSTAT1 lines respond to IFNβ. We used IFNβ, and not IFNα, due to its higher efficacy (see figure S4A). D. MEK inhibition leads to up-regulation of pSTAT1 in all cell lines. E. MEK inhibition induces death in low-pSTAT1 cell lines only (green). IFNβ dramatically enhances the cytotoxic effect of MEK inhibition in low pSTAT1 cell lines (purple). High pSTAT1 cell lines show only mild response to the MEK inhibitor and its combination with IFBβ (right). IFNβ alone (red) has almost no cytotoxic response.
The cluster splits the cell lines into two groups; the first contains 3 cell lines with an upregulation of interferon response genes, and cell lines in the second context express these genes at lower levels. Levels of pSTAT1-Y701, an indicator of the interferon-STAT1 activity levels (Platanias, 2005), confirmed that the high basal expression levels of the pathway targets correspond with high signaling activity of the pathway (figure 5B). Notably, the cell lines with upregulation of the STAT1-interferon response genes are not the same 3 cell lines with low activity of STAT3.
High basal activity of the STAT1-interferon pathway has been previously shown to be necessary, but not sufficient, for IFNα/β-induced apoptosis (Jackson et al., 2003). To test this claim, 3 low- and 3 high-pSTAT1 cell lines were treated IFNβ and apoptosis levels were assessed by TUNEL. All low-pSTAT1 and 2 high-pSTAT1 cell lines were resistant to the cytotoxic effects of IFNβ, and one high-pSTAT1 cell line was marginally sensitive (figure 5C). Both IFNα and IFNβ were tested, and as previously shown (Leaman et al., 2003), IFNβ led to a stronger apoptotic response than IFNα (figure S4A); thus, IFNβ was chosen for further analysis. Our results confirmed the previous findings that STAT1 activity is necessary, but not sufficient, for IFNα/β sensitivity.
IFNβ enhances the cytotoxic response of MEK inhibition in low pSTAT1 cell lines
According to the expression data, MEK inhibition leads to an up-regulation of the IFNα/β pathway. Analysis of protein levels by Western blots indicates an increase in pSTAT1 levels after MEK inhibition, confirming a crosstalk between MAPK and STAT1 (figure 5D). Because interferon activity seems to be required for IFN-induced death, we hypothesized that IFN might synergize with MEK inhibition to increase apoptosis.
The cytotoxic effect of MEK inhibition on both high- and low-pSTAT1 cell lines was assessed. We found that high-pSTAT1 cell lines are mostly resistant to the cytotoxic effects of MEK inhibition, while low-pSTAT1 cells are sensitive (figure 5E). Notably, both groups contain NRAS and BRAF mutant cell lines, and cell lines with high and low MITF expression, although both MITF-low cell lines and NRAS mutant cell lines have been previously reported to be less sensitive to MAPK pathway inhibition(Barretina et al., 2012; Solit et al., 2006). Moreover, the results show that the cytotoxic response of MEK inhibition is independent of its cytostatic response. For example, SkMel133 continues to grow rapidly under MEK inhibition (figure 1C), but has relatively high apoptosis levels under MEK inhibition.
We then examined the cytotoxic effect of the combination of MEK inhibition and IFNβ. While IFNβ as a single agent has no cytotoxic effect on low-pSTAT1 cell lines, it notably enhances the cytotoxic response of MEK inhibition, increasing TUNEL-positive cells by almost two-fold (figure 5E, S4B). Moreover, while low-pSTAT1 cell lines show a strong sensitivity to the combination of MEK inhibition and IFNβ, high pSTAT1 cell lines seem to be resistant to the cytotoxic effects of both MEK inhibition alone and the dual treatment (figure 5E). To confirm that the effect of IFNβ on the cytotoxic response of MAPK pathway inhibition is not specific to MEK inhibition, we show a similar behavior, albeit slightly weaker, between a BRAF inhibitor (PLX4720) and IFNβ (figure S4C).
Transcriptional response to IFN is similar in all cell lines
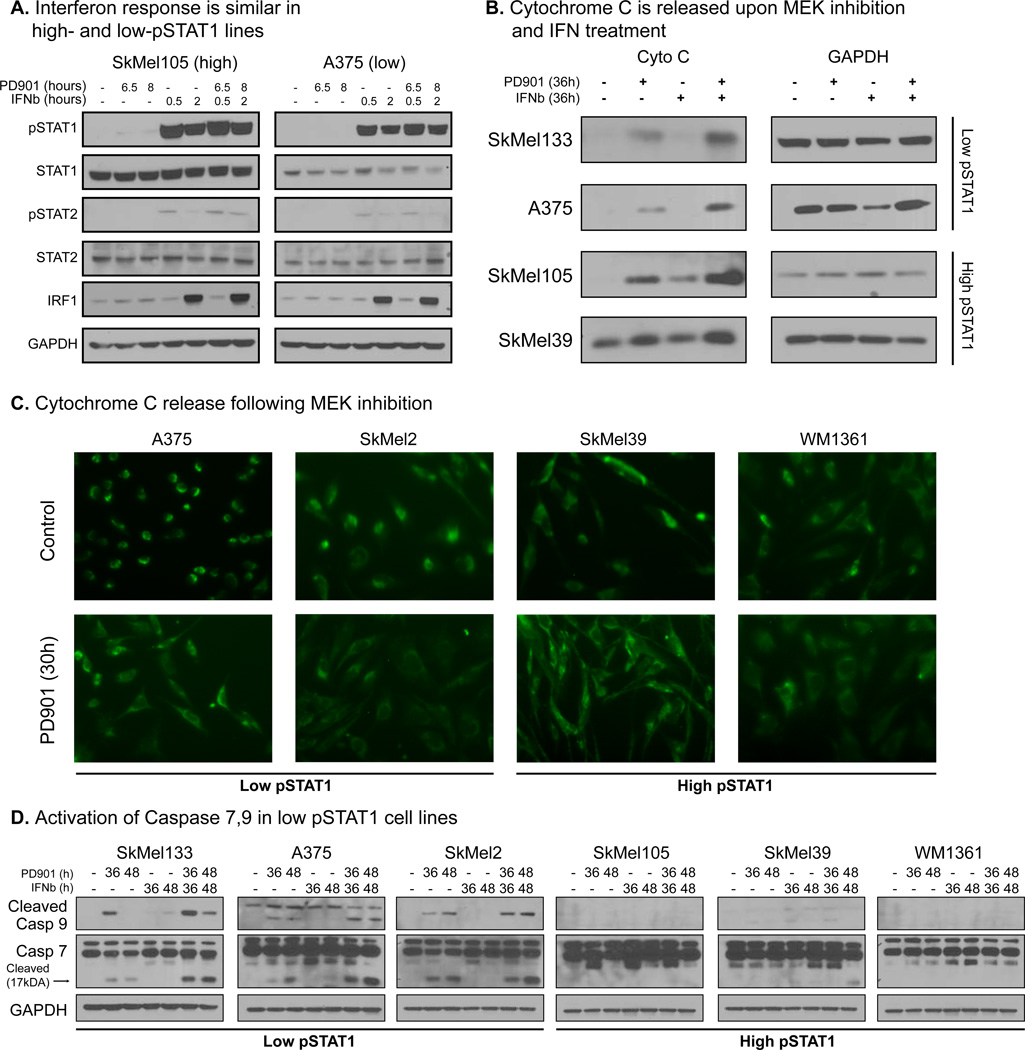
Our data demonstrated that basal activation level of the interferon pathway predicts the cytotoxic response to MEK inhibition. We hypothesized that these phenotypic differences are associated with changes in the interferon pathway and its response to IFNα/β treatment. We therefore characterized the signaling and transcriptional responses to IFNβ and MEK inhibition.
Western blots show that activation of STAT1 by IFNβ is identical, in both timing and extent, in a low-pSTAT1 and a high-pSTAT1 cell lines, and inhibition of MEK does not alter the this response (figure 6A, S5A). Interestingly, we found that the levels of pSTAT1 in the so-called “high-pSTAT1 cell lines” are substantially lower than pSTAT1’s levels following IFNβ treatment (figure 6A compared with figure 5B).
Figure 6.
Elucidating the cytotoxic response of IFNβ and MEKi. A. Response to IFNβ, as measured by pSTAT1 and IRF1 levels, is similar in both high- and low- pSTAT1 cell lines, and MEK inhibition doesn’t alter the response (for transcriptional response see figure S5B). Notably, basal activity level of the pathway in high-pSTAT1 cell lines is much lower than the induction in pathway activity after IFNβ treatment. B. MEKi activates the intrinsic apoptotic pathway by cytochrome C release from the mitochondria, approx. 36 hours after treatment. IFNβ enhances the response in all cell lines, including the high-pSTAT1 resistant cell lines. C. Fluorescent microscopy staining of CytoC shows similar patterns of CytoC release in both high- and low-pSTAT1 cell lines. CytoC is released from the mitochondria in all cells following MEK inhibition. See figure S5D for the images with nucleus staining. D. Caspase 7 and 9 are cleaved and activated following MEK inhibition in low pSTAT1 cell lines only. IFNβ enhances this effect, but fails to activate the pathway by itself. Both caspases are not cleaved in high-pSTAT1 cell lines. To reinforce the association between STAT1 levels and response to MEK inhibition we tested 4 more cell lines. Both high- and low-pSTAT1 levels respond with accordance to their STAT1 levels (figure S5E).
To search for more global regulatory differences in the interferon response, and to assess the effects of IFNβ more quantitatively, we measured gene expression levels 8 hours after treatment with PD325901, IFNβ or their combination in three low- and three high-pSTAT1 cell lines (figure S5B). No significant differences in the transcriptional response following the treatments are apparent after 8 hours of treatment. Additionally, MEK inhibition does not alter the IFNβ response, and no synergy between treatments is revealed (see supplementary information and figure S5C).
These data suggest that the differences in the phenotypic response are not due to the basal activation level of the interferon pathway.
The caspase pathway is only activated in low pSTAT1 cell lines
Since the transcriptional response to IFNβ fails to explain the differences in the cytotoxic response between the cell lines, we moved to characterize the apoptotic pathway directly.
The intrinsic apoptotic pathway is initiated by the release of cytochrome C (CytoC) from the mitochondria, which together with Apaf-1, cleaves and activates initiator and executioner caspases (Bratton and Salvesen, 2010). Surprisingly, we found that inhibition of MEK is sufficient to induce release of CytoC in all cell lines, and the release is enhanced by co-treatment with IFNβ (FIGURE 6B). We further confirmed this behavior is single cell fluorescent microscopy, which demonstrates that CytoC is released in all cells in both high- and low-pSTAT1 cell lines (figures 6C and S5D). However, although MEK inhibition initiates the intrinsic pathway in high-pSTAT1 cell lines, and this response is enhanced by IFN, these cell lines fail to undergo apoptosis.
CytoC release leads to apoptosis by activating the caspase pathway. We found that caspase 9, an initiator caspase, and caspases 7 and 3, executioner caspases, are cleaved following the release of CytoC by MEK inhibition in low-pSTAT1 cell lines only (figure 6D and S5E). Combinatorial treatment leads to a stronger and faster activation of these two caspases, but IFNβ treatment alone does not activate them (figures 6D, S5E). Importantly, caspases are not cleaved in high-pSTAT1 cell lines, although CytoC is released. To confirm the association between pSTAT1 levels and caspase activation, we extended our panel to 10 cell lines, adding 2 additional high- and 2 additional low- pSTAT1 cell lines. As with the original set of cell lines, caspases are cleaved only in low-pSTAT1 cell lines (figure S5E). The lack of caspase activation may explain the cytotoxic resistance to treatment.
Additional components, such as APAF-1, cIAP1–2 and XIAP, play part in the activation of the caspase pathway We therefore assessed the baseline expression levels of these proteins in our cell line panel, but found no correlation between their levels and the cytotoxic response to the treatments (figure S5F–G). Additionally, we confirmed that caspase 9, the upstream caspase of the caspase pathway (Riedl and Shi, 2004), is expressed in comparable levels in all cell lines (figure S5F).
Deletion of interferon locus correlates with cytotoxic response
Basal activity of the interferon pathway predicts the cytotoxic response to MEK inhibition and its combination with IFNα/β. Levels of pathway inhibitors from the SOCS and PIAS family are similar in all cell lines and fail to explain the differences in the basal activation of the pathway (figure S6A). We therefore sought to identify genetic lesions that could be responsible for the differential basal activation of this pathway.
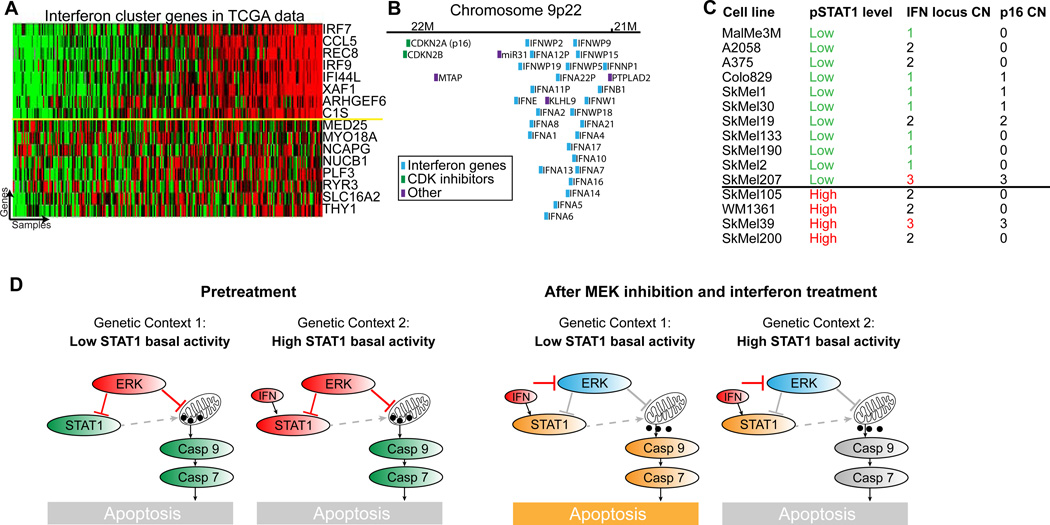
Using the large number of samples in The Cancer Genome Atlas (TCGA) melanoma dataset, we associated STAT1 pathway activity levels with genetic aberrations (see materials and methods). To infer pathway activity, we used the genes in the STAT1 cluster identified by COSPER. This gene cluster reflects pSTAT1 levels and is also highly correlated in the TCGA dataset (figure 7A).
Figure 7.
Deletion of interferon locus and IFN expression levels explains the two interferonpathway states and predicts drug response. A. The interferon gene cluster identified by COSPER is highly correlated in the TCGA melanoma expression data set. This allows us to infer pathway activity in the TCGA tumors and associate it with DNA aberrations. Genes above the yellow line were used for association with DNA copy number. B. The interferon locus contains 26 interferon genes, and is only 0.5Mb downstream of CDKN2A (p16), a known melanoma tumor suppressor. C. Interferon locus copy number is also correlated with pathway activity in our 14 cell line panel (pvalue=0.05). p16 however, only 0.5Mb upstream, is not, suggesting that interferon deletion and p16 deletion are two independent events. SkMel200, a high-pSTAT1 cell line, was added for purposes of CNV analysis. Copy number of the interferon locus is also correlated with expression levels of interferon genes (figure S6B), and conditioned media experiment shows that cytokines are released from high pSTAT1 cell lines (figure S6C). D. A cartoon depicting the two network states, before and after MEKi and IFN treatment. Inhibition of MEK leads to cytochrome C release in both cellular contexts, and IFN treatment enhances the response. However, caspase 9 is cleaved and activated only in low pSTAT1 cell lines.
The copy number alteration most significantly associated with the STAT1 gene signature is a deletion of the interferon locus (qvalue=10−4, FDR(Storey and Tibshirani, 2003)), located in chromosome 9p22. The locus contains a cluster of 26 interferon genes (figure 7B) and deletion of this locus corresponds to low basal activity of the interferon pathway. Our panel confirms this association - most cell lines with low pathway activity have 0 or 1 copies of the 9p22 locus, while all cell lines with high activity have 2 or 3 copies (figure 7C, pvalue=0.05, see materials and methods for copy number assessment).
Interestingly, the interferon gene cluster on locus 9p22 is only 0.5Mbs downstream of p16 (CDKN2A) (figure 7B), a known tumor suppressor gene deleted in roughly 60% of melanoma tumors (Reed et al., 1995). Deletion of both p16 and the interferon locus was previously reported (Naylor et al., 1997), but as research focused on the role p16 in cancer, deletion of the interferon locus was viewed as a passenger mutation. However, copy number data show that both events are independent, and copy number of the interferon locus and not p16 is associated with the cytotoxic response to MEK inhibition (figure 7C).
We confirmed that an autocrine loop is responsible for the lower levels of pSTAT1, using a conditioned media experiment. In these experiments media from high pSTAT1 cell lines lead to activation of STAT1 in low pSTAT1 cell lines (figure S6C), confirming that high pSTAT1 cell lines that harbor two copies of the interferon locus produce and release cytokines, presumably IFN, which leads to STAT1 activation.
To summarize, our results show that cell lines with fewer copies of the interferon locus and lower expression of the interferon genes are sensitive to the cytotoxic effects of MEK inhibition (figure 7D). Furthermore, IFNα/β enhances this cytotoxic response via an increase in CytoC release from the mitochondria. However, cell lines with high basal activity of the interferon pathway are resistant to the cytotoxic effects of the treatments, and although MEK inhibition leads to CytoC release in these cell lines, it seems that an impairment of the caspase activation mechanism leads to apoptosis aversion. Taken together, we postulate that constitutive exposure to IFN is adverse to cancer cells, and they overcome it by either deactivation of the interferon pathway, or by an impairment of the apoptotic pathway.
Discussion
Contemporary cancer drug development focuses on targeting recurring oncogenic events, such as gene amplification and overexpression (HER2) or activation (BRAF). This approach is based on the principle of oncogene addiction. The underlying assumption is that the downstream targets of the oncogenes are the same in all tumors. Drug combinations are also suggested based on the principle of similar network structure and pathway dependencies in tumors harboring specific oncogenic mutations.
However, our analysis of MAPK targets in MAPK-activated melanomas reveals tremendous differences in underlying network structure between tumors. Although we analyzed the transcriptional output of MEK inhibition only in MAPK-activated melanoma cell lines, each cell line had a unique transcriptional response. Moreover, a vast majority of downstream targets of the MAPK pathway are context-specific – under the control of the pathway in only a subset of cell lines.
To detect context-specific targets using pre- and post-inhibition expression data, we developed COSPER, a bi-clustering algorithm that identifies co-expressed genes that are under the control of the MAPK pathway in only a subset of cell lines. There are several benefits to identifying clusters of context-specific, co-regulated genes. First, we can apply enrichment analysis to the co-expressed genes and identify the pathway that likely regulates their expression. Second, by using post-inhibition data to narrow the gene set to only those that respond to perturbation, we specifically search for pathways regulated by the MAPK pathway. Third, the context – partitioning the cell lines into two groups, can assist in the identification of genetic aberrations that are more frequent in one group versus the other, thus also associating a genetic lesion with pathway activation. Fourth, the subgrouping of cell lines can also be associated with a phenotype, such as growth rate, response to treatment, “stem cell-ness” and others. Together, context-specific co-regulated clusters link genetic lesions to a MAPK-regulated pathway and a phenotype, and can assist in the understanding of response heterogeneity.
Using COSPER, we identified a possible interaction between MEK inhibition and IFNα/β, two approved treatments for melanoma. An experimental validation uncovered two key findings: first, IFNα/β enhances the cytotoxic response of MEK inhibition; second, cell lines with high basal activity of the interferon pathway exhibit much lower cytotoxicity under MEK inhibition. We found that a deletion of the interferon locus is correlated, and explains, the basal activity level of the interferon pathway, and therefore predicts the cytotoxic response to MEK inhibition. However, our results indicate that the basal activity level is not the mechanism for the sensitivity and resistance to IFNα/β and MEK inhibition. Instead, we found an impairment of the caspase activation mechanism that may explain the cytotoxic resistance.
Although MEK inhibition leads to, and IFNβ increases, the release of CytoC from the mitochondria in all cell lines, regardless of their interferon-pathway basal activity level, caspases 9, 7 and 3 are activated only in cell lines with low interferon pathway activity. We failed, however, to identify the lesion that prevents caspase activation cell lines with high interferon pathway activity. Understanding the mechanism of resistance can support the development of new drugs and treatments.
Taken together, these results suggest that constitutive exposure to IFN is adverse to cancer cells, and they overcome it by either deactivation of the interferon pathway, or by an impairment of the apoptotic pathway. Weichselbaum et al. have previously linked interferon to drug response (Weichselbaum et al., 2008), showing that interferon pathway activity predicts survival of breast cancer patients following chemotherapy and radiation. Our analysis of the TCGA data show that a lower basal activity of the interferon pathway in breast cancer is associated with a deletion of IRF1, Interferon Response Factor 1, a necessary protein for interferon-induced death (data not shown) (Sanceau et al., 2000)..
The interferon pathway might have important clinical implications in melanoma and other cancers. Since interferon pathway activity predicts the cytotoxic response to MEK inhibition in vitro, it is possible that its signaling activity, interferon expression levels and/or interferon locus copy number can be used as a biomarker for treatment by MAPK pathway inhibitors. The clinical implications of IFNα/β treatment, however, are less straightforward, and further studies are necessary to check whether in the therapeutic window of IFN, it has tumor-specific effect.
To summarize, our work demonstrates that tumor networks are more complex and varied than previously appreciated. Although only MAPK-activated melanoma cell lines were examined, these were found to be heterogeneous and immensely varied. Moreover, while all BRAF-mutant tumors are grouped together and treated similarly in the clinic, the targets and pathways regulated by BRAF in different cell lines are vastly different. Even with a small sample size of only 14 cell lines, pre- and post- perturbation expression data empower the discovery of dependencies and interactions between pathways. We believe that a similar analysis of a larger datasets of pre- and post-inhibition expression data can help identify additional context-specific interactions.
Post-perturbation data significantly enhance the ability to identify downstream targets (Niepel et al., 2013; Sachs et al., 2005). Perturbations break correlated patterns, resolve cause and effect, and reveal regulation patterns that are not observed in steady state expression levels. It was previously shown that response to perturbation varies significantly, even in cancer subtypes that share similar oncogenic mutations (Duncan et al., 2012; Niepel et al., 2014). However, analysis of post perturbation protein levels typically focus only on post-perturbation changes. When an important pathway such as MAPK is inhibited, many of the differentially expressed genes involve response to stress, rather than genes that were regulated by the pathway prior to the perturbation. Typical methods would consider these MAPK targets (and indeed these respond to MAPK inhibition), however these are not regulated by MAPK in physiological conditions, prior to MAPK inhibition. COSPER can distinguish these using expression patterns prior to perturbation. Moreover, COSPER takes context into account. This allows us to identify gene clusters that only change in subsets of cell lines, which would likely be dismissed by other methods. By comparing both the pre- and post- perturbation gene expression, and taking context into account, we can better identify pathways that are regulated by MAPK in each cancer cell line. Therefore, by combining information from both pre- and post- perturbation levels we reveal the network structure governed by MAPK, and the differences in this structure in difference cell lines.
The full scale of these differences is only revealed when examining a perturbed network, which highlights the importance of post-inhibition data, compared with steady-state data only. We believe that our research has only scratched the surface, and future studies with larger cohort size should be conducted, as our data demonstrate the value of system-wide perturbation analysis of tumors in the era of personalized medicine.
Materials and Methods
Cell Culture and drug treatment
Cell lines were obtained from A. Houghton (Memorial Sloan-Kettering Cancer Center), except for Colo829 and A2058 that were purchased from ATCC. All cell lines were maintained in RPMI 1640 (Invitrogen 21870-092), supplemented with 2 mM glutamine, 50 units/mL penicillin, 50 units/mL streptomycin, and 10% FBS (Omega Scientific), and incubated at 37 °C in 5% CO2. Samples for protein and gene expression analysis were plated at 60–80% confluency and incubated for 20–24h. Then treated with PD325901 (50nM), Interferon alpha (20000U/mL, R&D 11100) or Interferon beta (1000U/mL, R&D 11415). Control samples were collected untreated at time of treatment.
Gene expression and microarrays
Agilent’s 8×60 human gene expression were used, and samples were harvested 8h post treatment. Experimental procedures and data normalization are described in supplementary material.
We used Agilent’s 1M SurePrint CGH arrays to assess copy number. DNA was extracted using Qiagen’s DNeasy kit and labeled and hybridized according to Agilent’s protocol. All microarray data are available on GEO under accession number GSE51115.
TCGA data analysis
TCGA expression and CGH data were downloaded from the TCGA website. Genes for the STAT1 gene signature were a subset of COSPER’s STAT1 signature. All genes with a Pearson r2 > 0.5 with at least 3 additional genes were included. Association with copy number was performed using Pearson correlation between the mean of the gene signature and copy number levels of each gene. Pearson’s pvalues were corrected by FDR (Storey and Tibshirani, 2003).
Protein levels
Samples for protein analysis were lysed using RIPA buffer. Protein concentration was assessed using BCA staining. Samples were then normalized to a fixed concentration and mixed with a 5× glycerol/SDS/DTT loading buffer. Lysates were run on gradient (4–12%) Bis-Tris gels. Primary antibodies are listed in table S3. After incubation with horseradish peroxidase- conjugated secondary antibodies, proteins were detected using chemiluminescence.
Cytochrome C release was assessed on fresh unfrozen pellets using Sucrose/Mannitol buffer (Majewski et al., 2004). Full details in the supplementary material. Protocol for CytoC staining in fluorescent microscopy is detailed in supplementary material.
Growth curves and Apoptosis levels
For growth curve measurement, 50K cells were plated in 6-well plates with 2mL of growth media. Cells were counted every 24h following treatment using a cell counter (Coulter Z1), in triplicates. Apoptosis was assessed by TUNEL staining. Cells were plated in 6-well plates at 200K cells/well. 24h after plating cells were treated with PD325901, and both floating and adherent cells were collected 72h after treatment. TUNEL was performed using Invitrogen BrdU TUNEL kit.
Context-specific computational model
A full description of COSPER can be found in the supplementary information. In short, all genes are scored for all possible splits using both pre- and post-treatment expression using the NormalGamma function. Genes with a strong association with a split joins its cluster. Then, similar clusters are merged, leaving fewer clusters with more genes each.
Supplementary Material
Acknowledgments
The authors would like to thank Meehan Crist, Ramon Parsons, Jacob Levin, Ran Reshef, Sagi Shapira and Catherine Wu for valuable comments. This research was supported by Stand Up To Cancer Innovative Research Grant (IRG08), National Institutes of Health (R01CA164729) and National Centers for Biomedical Computing Grant 1U54CA121852-01A1. D.P. holds a Packard Fellowship for Science and Engineering. O.L. is supported by the HHMI pre-doc fellowship program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton SB, Salvesen GS. Regulation of the Apaf-1-caspase-9 apoptosome. Journal of cell science. 2010;123:3209–3214. doi: 10.1242/jcs.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, Brown RD, Della Pelle P, Dias-Santagata D, Hung KE, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B, Meng J, Peyton M, Girard L, Bornmann WG, Ji L, Minna JD, Fang B, Roth JA. STAT3 mediates resistance to MEK inhibitor through microRNA miR-17. Cancer research. 2011;71:3658–3668. doi: 10.1158/0008-5472.CAN-10-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Hartmann C, Kandulski O, Paap BK, Koczan D, Thiesen HJ, Zettl UK. Interferon-beta therapy in multiple sclerosis: the short-term and long-term effects on the patients' individual gene expression in peripheral blood. Molecular neurobiology. 2013;48:737–756. doi: 10.1007/s12035-013-8463-1. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DP, Watling D, Rogers NC, Banks RE, Kerr IM, Selby PJ, Patel PM. The JAK/STAT pathway is not sufficient to sustain the antiproliferative response in an interferon-resistant human melanoma cell line. Melanoma research. 2003;13:219–229. doi: 10.1097/00008390-200306000-00001. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, Narayan R, Flaherty KT, Wargo JA, Root DE, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, Halilovic E, Persaud Y, Xing F, Viale A, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman DW, Chawla-Sarkar M, Jacobs B, Vyas K, Sun Y, Ozdemir A, Yi T, Williams BR, Borden EC. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2003;23:745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends in molecular medicine. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, Huang A, Wong WL, Callahan MK, Merghoub T, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer cell. 2012;22:668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski N, Nogueira V, Bhaskar P, Coy PE, Skeen JE, Gottlob K, Chandel NS, Thompson CB, Robey RB, Hay N. Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Molecular cell. 2004;16:819–830. doi: 10.1016/j.molcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Naylor MF, Brown S, Quinlan C, Pitha JV, Evertt MA. 9p21 deletions in primary melanoma. Dermatology online journal. 1997;3:1. [PubMed] [Google Scholar]
- Niepel M, Hafner M, Pace EA, Chung M, Chai DH, Zhou L, Muhlich JL, Schoeberl B, Sorger PK. Analysis of growth factor signaling in genetically diverse breast cancer lines. BMC biology. 2014;12:20. doi: 10.1186/1741-7007-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M, Hafner M, Pace EA, Chung M, Chai DH, Zhou L, Schoeberl B, Sorger PK. Profiles of Basal and stimulated receptor signaling networks predict drug response in breast cancer lines. Science signaling. 2013;6:ra84. doi: 10.1126/scisignal.2004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, Wood E, Fedorenko IV, Sondak VK, Anderson AR, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer research. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nature reviews Immunology. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JA, Loganzo F, Jr, Shea CR, Walker GJ, Flores JF, Glendening JM, Bogdany JK, Shiel MJ, Haluska FG, Fountain JW, et al. Loss of expression of the p16/cyclin-dependent kinase inhibitor 2 tumor suppressor gene in melanocytic lesions correlates with invasive stage of tumor progression. Cancer research. 1995;55:2713–2718. [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nature reviews Molecular cell biology. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- Sanceau J, Hiscott J, Delattre O, Wietzerbin J. IFN-beta induces serine phosphorylation of Stat-1 in Ewing's sarcoma cells and mediates apoptosis via induction of IRF-1 and activation of caspase-7. Oncogene. 2000;19:3372–3383. doi: 10.1038/sj.onc.1203670. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. The New England journal of medicine. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman JG, Gerald WL, Lazebnik YA, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. The New England journal of medicine. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, Bremer R, Gillette S, Kong J, Haass NK, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DS, Baker SW, Khodarev N, Su AW, Shaikh AY, Roach P, Kreike B, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing F, Persaud Y, Pratilas CA, Taylor BS, Janakiraman M, She QB, Gallardo H, Liu C, Merghoub T, Hefter B, et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene. 2012;31:446–457. doi: 10.1038/onc.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xiao Z, Lai D, Sun J, He C, Chu Z, Ye H, Chen S, Wang J. miR-21, miR-17 and miR-19a induced by phosphatase of regenerating liver-3 promote the proliferation and metastasis of colon cancer. British journal of cancer. 2012;107:352–359. doi: 10.1038/bjc.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.