Abstract
Human reproduction is remarkably inefficient; nearly 70% of human conceptions do not survive to live birth. Spontaneous fetal aneuploidy is the most common cause for spontaneous loss, particularly in the first trimester of pregnancy. Although losses owing to de novo fetal aneuploidy occur at similar frequencies among women with sporadic and recurrent losses, some couples with recurrent pregnancy loss have additional associated genetic factors and some have nongenetic etiologies. Genetic testing of the products of conception from couples experiencing two or more losses may aid in defining the underlying etiology and in counseling patients about prognosis in a subsequent pregnancy. Parental karyotyping of couples who have experienced recurrent pregnancy loss (RPL) will detect some couples with an increased likelihood of recurrent fetal aneuploidy; this may direct interventions. The utility of preimplantation genetic analysis in couples with RPL is unproven, but new approaches to this testing show great promise.
Chromosomal abnormalities are observed in the majority of spontaneous pregnancy losses. Parental karyotyping and genetic testing of products of conception may aid in defining the underlying etiology and in counseling patients.
Spontaneous pregnancy loss is the most common complication of pregnancy. Approximately 70% of human conceptions fail to achieve viability, with almost 50% of all pregnancies ending in miscarriage before the clinical recognition of a missed period or the presence of embryonal heart activity (Edmonds et al. 1982; Wilcox et al. 1988). Recurrent pregnancy loss (RPL), or recurrent abortion, is less common, occurring in about one in 100 pregnant women (Alberman 1988).
RPL was previously defined as three or more consecutive clinically recognized spontaneous pregnancy losses before 20 wk of gestation. By this definition, one in 300 women experience RPL (Wilcox et al. 1988). Recent recommendations supporting clinical intervention after only two consecutive spontaneous abortions when other features of pregnancy loss are present define a higher prevalence of one in 100 women. These additional features include: detectible fetal heart activity preloss; normal fetal chromosomal content; advanced maternal age; or couple subfertility (Practice Committee of the American Society for Reproductive Medicine 2008a). Uterine anatomic abnormalities, endocrine abnormalities, infections, immunologic factors, environmental factors, metabolic or hormonal disorders, sperm quality, and maternal and paternal age have each been linked to RPL. The standard RPL evaluation presently incorporates testing for chromosomal translocations in each parent as well as maternal testing for endocrine (thyroid), autoimmune (lupus anticoagulant and antiphospholipid antibodies), anatomic (endometrial or uterine abnormalities), and, in some cases, single gene disorders (such as inherited thrombophilias) (Sierra and Stephenson 2006; Practice Committee of American Society for Reproductive Medicine 2012). Despite the number of proposed etiologies, parental chromosomal abnormalities and complications resulting from the antiphospholipid antibody syndrome continue to be the only undisputed causes of RPL. RPL remains unexplained in ∼45%–50% of patients (Stephenson 1996; Stephenson and Kutteh 2007). In most cases of RPL, the prognosis is far from bleak; researchers have shown that the overall probability of live birth subsequent to RPL is ∼70%–75%, even in women with advanced maternal age (Clifford et al. 1997; Brigham et al. 1999).
FETAL ANEUPLOIDY IN SPORADIC AND RECURRENT PREGNANCY LOSS
The remarkable inefficiency of human reproduction is largely the result of spontaneous fetal aneuploidy. Overall, 50%–70% of specimens from sporadic spontaneous losses show some type of cytogenetic abnormality, with the most common karyotypic defects being autosomal trisomies (60%), monosomy X (20%), and polyploidy (20%) (Silver and Branch 2007). Most result from random errors in germ cell development that, by definition, affect pregnancies in couples with and without a history of RPL equally. Typically, numerical aneuploidy results from meiotic nondisjunction in the germ cells of couples with normal parental karyotypes, and the recurrence of a particular abnormality in future pregnancies is rare in patients presenting with RPL and in the general population (Warren and Silver 2008; Suzumori and Sugiura-Ogasawara 2010). Supporting the concept that many losses among RPL patients are the result of random, nonrecurring events is the fact that the prognosis for subsequent pregnancies in RPL couples is better after an aneuploid miscarriage than after an euploid miscarriage (Warburton et al. 1987; Ogasawara et al. 2000; Carp et al. 2001).
The frequencies and specific types of chromosomal abnormalities found in tissues obtained from sporadic spontaneous pregnancy losses vary with the gestational age of the fetus at the time of demise and with maternal age. Losses occurring early in pregnancy appear to display a wide range of fairly unusual aneuploidies, whereas deaths that appear later in gestation show those aneuploidies more typically detected in live births, such as trisomies 21, 18, and 13 (Wapner and Lewis 2002). Fetal aneuploidy is present at a frequency of up to 90% in specimens obtained from losses aged 0–6 wk of gestation, ∼50% in sporadic losses occurring at 8–11 wk gestation, and 30% in tissues from losses at 16–19 wk gestation (Geraedts 1996). Six to 12% of miscarriage specimens obtained from demises that occur after 20 wk of gestational display chromosomal abnormalities (Wapner and Lewis 2002; Benkhalifa et al. 2005). Once a fetal heart rate is evident on ultrasound, the risk of aneuploidy is <5%.
Rates of sporadic pregnancy loss and of overall fetal chromosomal aberrations increase with maternal age (Angell 1994; Munné et al. 1995), although maternal age has a preferential effect on certain aneuploidies. There are no significant associations between advanced maternal age and rates of sex chromosome monosomy or polyploidy, but strong correlations can be seen with rates of autosomal trisomy (Hassold et al. 1984; Hassold and Chiu 1985; Eiben et al. 1990). The degree of the correlation differs significantly among specific types of trisomies. For example, the largest effect of advanced maternal age is seen in trisomies involving small chromosomes (8, 9, 10, 13, 14, 15, 18, 20, 21, and 22), whereas trisomy 16 is less closely correlated (Hassold et al. 1980).
There are several lines of evidence that suggest nonrandom genetic aberrations among couples with RPL. First, the frequency of parental karyotypic abnormalities, including balanced translocations, is higher among couples with a history of RPL (2%–5%) than in the general population (0.2%) (Royal College of Obstetricians and Gynaecologists 2011). Second, the prevalence of RPL among first degree relatives of women with RPL is increased approximately sixfold compared with controls (Christiansen et al. 1990). Third, preimplantation genetic screening (PGS) in age matched populations shows that embryos from women with RPL have a higher incidence of aneuploidy than those from women undergoing screening for reasons not related to pregnancy loss (Hassold 1980; Simón et al. 1998; Vidal et al. 1998). Although only a portion of aneuploid embryos are predicted to result in a clinically detectable loss, some preimplantation genetic studies of women with RPL have detected aneuploidy in all embryos at rates approaching 50% (Hassold 1980; Stern et al. 1996; Daniely et al. 1998; Ogasawara et al. 2000; Carp et al. 2001; Stephenson et al. 2002; Sullivan et al. 2004; Carp 2008). Both of the latter findings are frequently used to rationalize preimplantation genetic testing in RPL couples (discussed below). Although aneuploidy rates are higher in embryos from women with RPL than in controls, the frequency of cytogenetic abnormalities in miscarriage tissues obtained from women with RPL is lower than that in women experiencing sporadic loss, occurring in only 25%–50% of cases (Stephenson et al. 2002; Carp 2008). This suggests that noncytogenetic etiologies also occur more frequently in women experiencing RPL than in those with sporadic losses. In fact, a case control study comparing 420 karyotyped products of conception (POCs) specimens from 275 couples with RPL to two data sets from patients with sporadic losses detected more euploid specimens in women <36 yr with RPL than in women <36 yr with sporadic losses (Stephenson et al. 2002). No differences in the number of euploid specimens were seen between the two groups in women 36 yr or older, likely reflecting the fact that fetal chromosomal abnormalities are found in the POCs of >70% of loses among women older than 35 who experience RPL (Marquard et al. 2010). The detection of fewer chromosomal abnormalities in POCs from women with RPL strongly suggests that noncytogenetic etiologies also occur more frequently in women experiencing RPL than in those with sporadic losses and indicates a need for further evaluation in patients with two or more documented euploid losses.
The effects of paternal, as opposed to maternal, meiotic errors and paternal age on reproductive outcome are less clearly defined. Although errors of nondisjunction occur to a lesser extent in sperm than in oocytes, paternal errors are responsible for the majority of cases of the sex chromosome trisomies XXY and XYY (Chamley et al. 1993; Hawley et al. 1994). Sperm from couples with a history of recurrent miscarriage display an increase in sex chromosome disomy (an extra chromosome in the haploid state of the gamete) compared with control groups. Oligoasthenoteratozoospermic patients (patients with abnormalities in sperm number [low], motility [low], and morphology [too few normal forms]) have the highest rates of sex chromosome disomy as well as high rates of disomy of chromosomes 18 and 21 when compared with normozoospermic patients (Rubio et al. 2001). A 2003 case control study that examined the presence of sperm chromosomal aneuploidy and apoptosis in couples with unexplained RPL also showed a significantly increased mean aneuploidy rate in this group compared with the control groups (Carrell et al. 2003).
Several studies have linked increasing paternal age to decreased fecundity, increased sperm DNA damage, and rates of adverse reproductive outcomes, but the exact mechanisms have not been delineated (Ford et al. 2000; Vagnini et al. 2007). Only recently have studies begun to show possible connections between increased paternal age and both sporadic and recurrent pregnancy loss (Slama et al. 2005; de La Rochebrochard et al. 2006; Kleinhaus et al. 2006; Puscheck and Jeyendran 2007). Data addressing paternal age and fetal aneuploidy, however, is extremely varied and conflicting. A review in 2004 concluded that the literature on the effects of paternal age on autosomal aneuploidy is inconclusive, but an association between increased paternal age and the frequency of sex chromosome aneuploidies in live births has been more consistently shown (Sloter et al. 2004). To further investigate a potential link between paternal age and fetal aneuploidy, a 2010 study examined the karyotypes of the tissues collected from 50 sporadic first trimester pregnancy losses while controlling for maternal and gestational age, but found no association between fetal aneuploidy rates and paternal age (Kushnir et al. 2010).
GENETIC TESTING AS A DIAGNOSTIC TOOL IN RPL
When assessing a new patient with two or more spontaneous losses, it is important to obtain POC karyotype results from past losses, if available, or to obtain the tissues from those losses for subsequent genetic analysis. This allows the practitioner to determine more precisely whether a diagnostic work-up for RPL is indicated. Prior aneuploid losses likely reflect the high baseline rate of aneuploidy noted in all spontaneous miscarriages. If the fetal karyotype(s) of one or more of the reported losses is unknown, assuming euploidy may result in an unnecessary set of diagnostic tests in a patient who may not have experienced repeated unexplained losses and would be unlikely to benefit from further diagnostic testing (Stephenson and Kutteh 2007). Genetic analysis of POCs from new losses helps to detect baseline spontaneous aneuploidy that will occur in an RPL patient at the same rate as in an age-matched patient who has not been diagnosed with RPL (Stephenson et al. 2002). Post demise, genetic analysis of POCs for spontaneous karyotypic abnormalities offers a fiscally and emotionally responsible approach to the management of couples presenting with repeated losses. The detection of fetal aneuploidy in the index loss can be immensely reassuring to a woman experiencing RPL, particularly when therapeutic interventions have been instituted before or during the index pregnancy and appear to have failed. This information also aids the caregiver in future discussions with the patient and her partner about prognosis and/or additional testing.
Routine cytogenetic testing of POCs in couples experiencing RPL has been shown to be cost-effective (Wolf and Horger 1995) and to direct management decisions (Hogge et al. 2003), even at a time when analytical methods were based on karyotyping of cultured trophoblast cells and hindered by the occurrence of culture failure and maternal cell contamination. Genetic testing of POCs using nonculture-based techniques such as array comparative genomic hybridization with or without reflex microsatellite single nucleotide polymorphism (SNP) analysis (Rajcan-Separovic et al. 2010; Viaggi et al. 2013; Mathur et al. 2014) (discussed below) is more precise, more detailed, and more reliable than culture-based methods and should add to cost-efficiency and overall utility. Nonculture-based techniques have been used successfully in fresh and in preserved tissues (Kudesia et al. 2014; Mathur et al. 2014).
Fetal Aneuploidy Testing
Molecular Diagnostic Testing on POCs
Historically, the cytogenetic analysis of POCs in RPL most often used cell culture, Giemsa staining of cells arrested in metaphase, and analysis of banding patterns for both numeric and structural errors. This continues to be the method used for peripheral blood karyotyping of couples experiencing RPL. Despite its widespread utilization, the method has many inherent limitations, the most problematic of which is culture failure. In the analysis of POCs, further limitations include maternal cell contamination, difficulties in timely collection of viable placental cells, and an inability to use tissue samples stored in formalin (Bell et al. 1999; Mathur et al. 2014). Lack of adequate washing and separation of maternal decidua from miscarriage tissue, robust decidual cell overgrowth in cell culture, and absence of miscarriage tissue are the most common causes of maternal contamination (Bell et al. 1999; Lathi and Milki 2002). Recent improvements in testing methodologies have circumvented some of these problems. To assess whether maternal cell contamination is responsible for a 46,XX karyotype result from miscarriage tissues, some facilities now provide reflex microsatellite testing to compare miscarriage DNA to maternal DNA at several highly polymorphic DNA loci. Microsatellites are small sequences of dinucleotide, trinucleotide, or tetranucleotide repeats that are widely distributed throughout the human genome. Because of their high rates of polymorphism, fragment analysis of microsatellite sequences can be used to compare POC specimens with maternal blood samples. If a high rate of identical loci is detected, maternal contamination is confirmed (Jarrett et al. 2001).
An increasingly popular alternative method for cytogenetic analysis is comparative genomic hybridization (CGH). In CGH, reference control DNA and DNA extracted from miscarriage tissues are labeled with fluorescent molecules (fluorophores) of different colors and then hybridized in a 1:1 ratio to a set of normal metaphase chromosomes. A computer analysis of microscopic images of the hybridization compares relative fluorescence intensities of the sample and control across the metaphase spread to identify areas of imbalance. These areas correspond to chromosomal copy number variants such as trisomies and monosomies in the patient sample. Original studies of CGH revealed that the technique can be used when conventional cytogenetic analysis fails (Daniely et al. 1998; Barrett et al. 2001; Hu et al. 2006), cell culture fails, prominent maternal contamination is present (Bell et al. 1999; Lomax et al. 2000), or when miscarriage tissue has been preserved in formalin or embedded in paraffin (Bell et al. 2001). When compared with cytogenetic analysis by conventional Giemsa banding, CGH was found to have a higher overall success rate, improved accuracy, and fewer problems with maternal contamination (Lomax et al. 2000). CGH is not without limitations. Balanced structural chromosome rearrangements cannot be identified with this technique and flow cytometry is required to detect polyploidy.
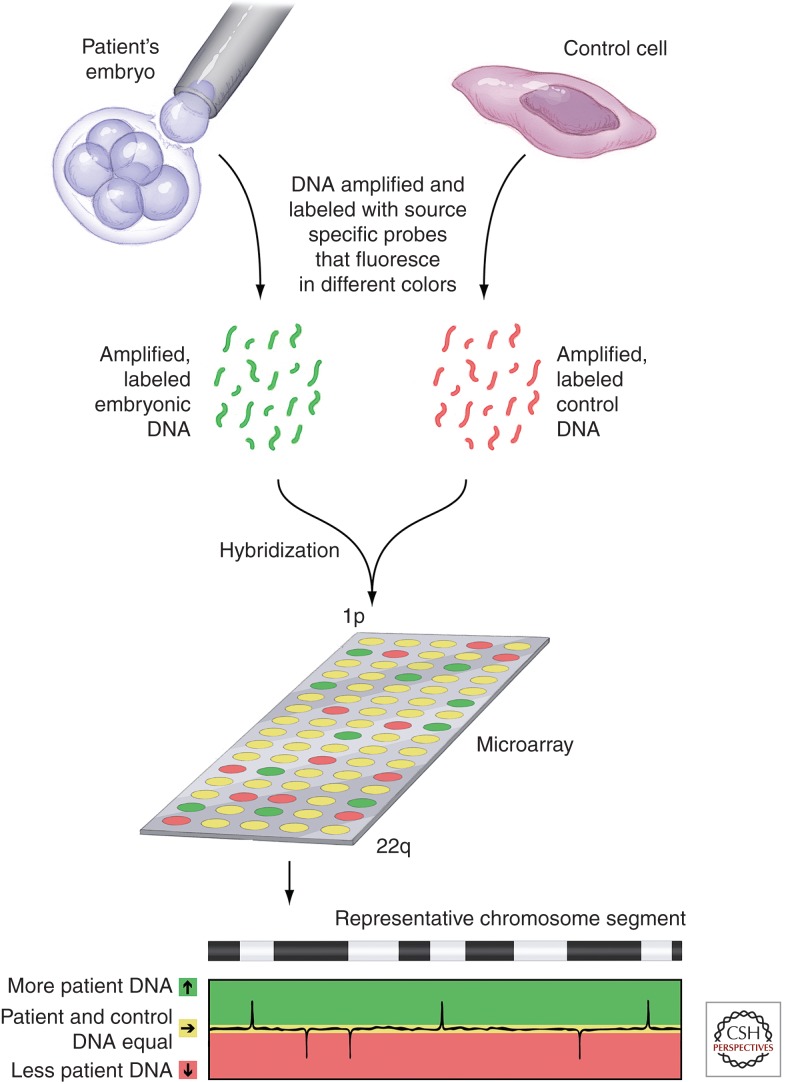
Advances in CGH analyses combined the technique with rapidly emerging microarray technologies. Array CGH (aCGH) testing (Thiesen 2008) of POCs mimics the standard metaphase spread CGH, except that the mixed and labeled reference and subject DNA samples are applied to a microarray (Fig. 1). The individual samples on the microarray determine the coverage and resolution of the test. For aCGH, the entire genome is represented on the array and resolution depends on the size of the specific DNA fragments that have been immobilized on the array chip. One great advantage of array-based approaches over those that use a metaphase spread is that there is no need for live cells, cell culture, or cell division. Array-based techniques can be performed much more rapidly than standard metaphase spread-based techniques. Perhaps the greatest power of array-based technology is its flexibility. Arrays can be produced that hone in on potential abnormalities in specific regions of specific chromosomes with very high resolution. Arrays based on SNP genotypes have recently been leveraged in POC molecular testing. In addition to what can be accomplished in standard whole genome microarrays, SNP chromosomal microarray analysis can detect the overall distribution of microdeletions and microduplications across a genome. In one study, the use of SNP chromosomal microarray analysis enabled the simultaneous detection of maternal cell contamination, triploidy, and uniparental disomy in miscarriage specimens (Levy et al. 2014). SNP arrays can also be used with CGH to detect aneuploidies and single gene disorders through linkage analysis (Brezina et al. 2011).
Figure 1.
Analysis of blastomere DNA using array comparative genomic hybridization (aCGH). DNA microarrays allow comparison of the amounts of specific segments of DNA within a test sample to that from a control sample. The portion of the genome tested by a given microarray depends on the specific pieces of DNA contained in the spots on the array. For complete genomic hybridization, these spots represent the entire genome, from the beginning of the short arm of chromosome 1 (1p) to the end of the long arm of chromosome 22 plus the X and Y chromosomes. In aCGH, DNAs from a single test cell (e.g., a blastomere) and from a control cell are amplified and labeled with colored fluorescent probes (each DNA source with its own color). Equal amounts of DNA from each source are hybridized to the array chip displaying the specific segments of DNA being tested. A computer then reads the chip and plots out the colors seen in each dot of the array. When one sample is labeled red and the other green, spots at which the DNA from each source is present in equal amounts will fluoresce yellow. Imbalances in the amounts of DNA will be seen as red or green. For example, in this diagram, red fluorescence indicates a less patient DNA than in the control. Areas of balance and imbalance can be mapped across the complete genome.
Parental Genetic Testing
Peripheral Blood Karyotyping
Parental numeric and structural cytogenetic abnormalities are perhaps the most thoroughly investigated genetic causes of RPL. Robertsonian translocations and balanced reciprocal translocations are detected in 2%–5% of couples with RPL. This should be followed by genetic counseling (Regan et al. 2011). Both the specific chromosome(s) affected and the types of rearrangement influence the probability of a future live birth. Genetic counselors can discuss with the affected couple their chances for transmission of the identified abnormality, their chances of future pregnancy loss, and the possibility of delivering a live affected child should they choose to continue to attempt pregnancy without intervention. Known carriers of structural chromosome abnormalities and some with single gene defects may also choose to proceed with assisted reproduction in combination with genetic analysis of any resulting embryos via preimplantation genetic diagnosis (PGD) to a select against affected embryos for transfer to the uterus.
Parental karyotypic abnormalities appear to be transmitted to the abortus at a frequency less than that predicted by simple Mendelian genetics. In one cohort study, known parental karyotype aberrations were detected in only 10% of abortus specimens, whereas 43.5% of abortuses were euploid (Carp et al. 2006). Some of this discrepancy is likely a reflection of preclinical loss of some affected embryos; still, the data support the concept that positive findings on parental karyotyping might not be directly predictive of subsequent embryonic karyotype abnormalities. The vast majority of fetuses with karyotypic abnormalities will not survive pregnancy. Parents with balanced translocations are less likely to deliver a live–born affected child than would be predicted by transmission rates.
Single Gene Defects
Single gene defects have been significantly less studied than karyotypic causes of sporadic miscarriage and RPL. Major groups of single gene defects that have been associated with pregnancy loss encompass musculoskeletal gene mutations including trinucleotide repeat disorders, genes involved in regulation of the immune system and implantation, thrombophilic gene mutations, and mutations in specific enzymes, including angiotensin-converting enzyme, ubiquitin-specific protease, and human alkaline phosphatase (Yang et al. 2012a; Asadpor et al. 2013; Wang et al. 2013; Vatin et al. 2014).
Musculoskeletal gene defects
Myotonic dystrophy, thanatoporic dysplasia, and type II osteogenesis imperfecta are among the single gene musculoskeletal disorders associated with RPL (Brook et al. 1992; Byrne and Ward 1994; Suthers 1996). From a genetic standpoint, myotonic dystrophy is particularly instructive as it is an example of a group of fairly unusual disorders called trinucleotide repeat diseases. These disorders are caused by DNA mutations in which a trinucleotide sequence (CTG for myotonic dystrophy) is repeated multiple times in a row. If the number of these repeats in a given segment of DNA is abnormal (i.e., exceeds a specific threshold), that segment of DNA is more prone to errors during mitosis. Although these errors may cause an increase or a decrease in the number of repeats present in the daughter cells, when increases occur, this is called a trinucleotide expansion. As expansions occur, more severe defects are seen in daughter cells and offspring. The principle of anticipation in myotonic dystrophy and related diseases describes the worsening of symptom severity and earlier age of onset as the disease is passed to subsequent generations. For myotonic dystrophy, analysis of tissues from stillborn babies has shown the highest number of CTG repeats when compared with living carriers (Kotzot 1999).
Immunologic gene defects
Because of the substantial immunologic mechanisms responsible for successful reproduction, studies on the role of single gene defects in sporadic and RPL have included the role of genes involved in immune regulation. The gene encoding the human leukocyte antigen-G (HLA-G), an important component of alloimmune recognition at the maternal–fetal interface, has been extensively studied. The presence of a null allele for the most common HLA-G isoform as well as distinct polymorphisms in the HLA-G promoter region, have been associated with recurrent miscarriage, suggesting that a functional protein is necessary for reproduction (Aldrich et al. 2001; Pfeiffer et al. 2001). Polymorphisms in genes including p53, p72, leukemia inhibiting factor (LIF), FAS-L, and the vascular endothelial growth factor (VEGF) gene have also been linked to increased rates of implantation failure and are undergoing investigation to determine their potential roles in women with RPL (Dumont et al. 2003; Steck et al. 2004; Brooks et al. 2007; Hu et al. 2007; Goodman et al. 2008; Banzato et al. 2013; Fraga et al. 2014). Despite evidence linking various polymorphisms in immune-related genes to pregnancy loss, these tests are not presently recommended as routine screening tests for couples with RPL owing to their low prevalence. Rather, their present utility lies in the insights they provide into particular mechanisms of the disease.
Thrombophilic gene defects
The genetic defects involved in the ever-expanding group of inherited thrombophilias are perhaps the best studied single gene mutations with reference to RPL. Among these, the majority of reports have addressed factor V Leiden, prothrombin gene promoter mutations, activated protein C resistance, and mutations in methylenetetrahydrofolate reductase, plasminogen activator inhibitor, thrombomodulin, and annexin A5 genes. Unlike trinucleotide expansion disorders and immune–related single gene mutations, the gene mutations causing several of the inherited thrombophilias are seen in relatively high prevalence in select populations. The data linking these defects to RPL, however, are conflicting. Mutations in factor V Leiden, the most common genetic cause of thrombosis, have a twofold higher prevalence in women experiencing repeated miscarriages compared with controls (Dizon-Townson et al. 1997; Grandone et al. 1997; Ridker et al. 1998; Ren and Wang 2006; Dawood et al. 2007; Mohamed et al. 2010). Mutations in the gene encoding Annexin A5, a protein that acts as an anticoagulant in placenta villi, have also been associated with a twofold increase in RPL risk (Bogdanova et al. 2007). Other cohort studies, however, have failed to confirm an association between RPL and inherited thrombophilias such as factor V Leiden and prothrombin promoter gene mutations (Dizon-Townson et al. 2005; Silver et al. 2010) and carriers and noncarriers of annexin A5 mutations have similar live birth rates, limiting the clinical significance of this particular mutation (Hayashi et al. 2013). In fact, there are data showing that when stratified for gestational age at the time of fetal demise, maternal carriage of the FVL mutation protects against pregnancy loss occurring before 10 wk of gestation (Roque et al. 2004). This finding aligns with earlier data showing increased implantation rates in carriers of FVL who become pregnant through in vitro fertilization (Gopel et al. 2001). Together these studies support the concept that FVL increases implantation of compromised embryos that would not typically implant and are ultimately lost.
Despite the growing evidence that thrombophilias caused by single gene defects may be associated with adverse pregnancy outcomes, the absolute risk of adverse outcomes remains low. Thus, current recommendations do not support universal screening for inherited thrombophilias in women with a history of pregnancy loss (Robertson et al. 2006; de Jong et al. 2011; Middeldorp 2011). Both the specific thrombophilia and the type of pregnancy loss (isolated vs. recurrent; early vs. late) contribute to the vast spectrum of documented associations and the underlying pathophysiologic mechanisms used to support these associations (de Jong et al. 2013). Despite the present recommendations, however, many practitioners will screen for prevalent inherited thrombophilias (factor V Leiden, prothrombin gene mutations and methylenetetrahydrofolate reductase in Caucasian patients; protein C, protein S and antithrombin deficiencies in some patients of eastern Asian descent) in RPL patients who have a first-degree relative with a known or suspected thrombophilia or who report a personal history of venous thromboembolism (Practice Committee of the American Society for Reproductive Medicine 2012). That said, data supporting treatment of a diagnosed gene defect associated with an inherited thrombophilia in the absence of a personal thrombotic history are weak. Although observational studies have shown a decrease in pregnancy complications in women with the Factor V Leiden mutation or mutation in the prothrombin promoter region that are prophylactically treated with low molecular weight heparin (Bouvier et al. 2014), there is a lack of randomized control studies that confirm these same findings. Recently, two randomized controlled trials investigated the empiric use of aspirin alone or a combination of aspirin with low molecular weight heparin in women with RPL and found no improvement in live birth rate when compared with placebo (Clark et al. 2010; Kaandorp et al. 2010). Based on current evidence, therefore, the empiric use of antithrombotic agents in women with unexplained RPL is not recommended. (Royal College of Obstetricians and Gynaecologists 2011; Kaandorp et al. 2014). More randomized placebo-control trials are needed to show both a mechanistic association between inherited thrombophilias and RPL and successful treatment options to improve live birth rates in couples suffering from recurrent miscarriage.
GENETIC TESTING AS THERAPY IN RPL
Preimplantation Genetic Diagnosis and Preimplantation Genetic Screening
Because chromosomal errors, some recurrent, may be responsible for a significant number of RPL losses, increasing attention is being paid to the possible utility of assisted reproductive technologies (ART) with or without PGD or PGS in the management of RPL. There are two related approaches to genetic analysis of embryos created through assisted reproduction. PGD tests for specific genetic abnormalities known to be heritable and present in one or both of the parents. PGS uses more global genetic assessment of the embryo to detect a wide variety of genetic abnormalities; when used, PGS is more commonly used in couples with “idiopathic” recurrent pregnancy loss. Both allow for selection of embryos for transfer based on genetic criteria; affected embryos are generally not transferred back into the uterus.
PGD was first used in humans in 1990, when single cells (blastomeres) were biopsied from six to eight cell cleavage stage embryos to determine the gender of those at risk for specific X-linked disorders (Handyside et al. 1990), an indication not typical in an RPL population. The use of PGD has subsequently expanded to include the diagnosis of autosomal dominant, autosomal recessive, and X-linked single gene disorders and unbalanced chromosome translocations that would negatively affect a child at birth or in early childhood. Diagnosable heritable disorders are numerous, and include autosomal dominant Huntington’s disease, myotonic dystrophy, neurofibromatosis, and Charcot–Marie–Tooth disease, autosomal recessive beta thalassemia, sickle cell disease, spinal muscular atrophy, cystic fibrosis, and X-linked disorders such as fragile X and muscular dystrophy (Harper et al. 1994; Harper 2010). PGD is most commonly exploited in carrier couples without infertility or RPL. Its use in RPL couples is mainly limited to translocation carriers, because most single gene disorders do not result in sporadic or recurrent pregnancy loss.
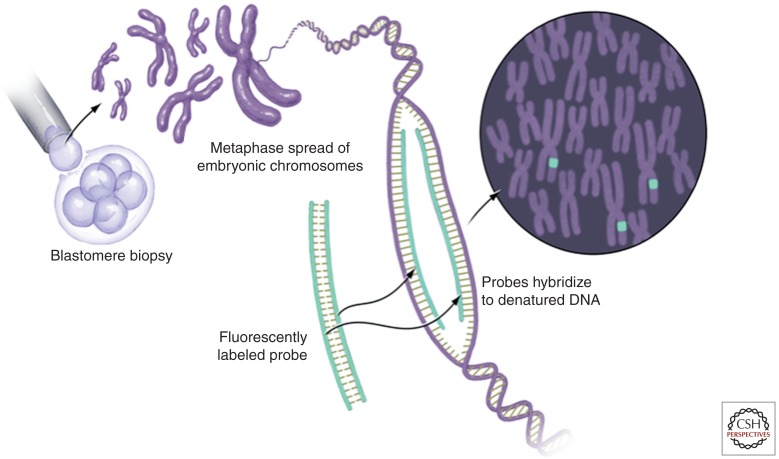
The use of PGS in couples with RPL is more controversial than the use of PGD to detect transmission of diagnosed parental karyotype abnormalities. Some of this controversy is likely secondary to the introduction of PGS into the management of RPL at a time when genetic analytic tools were fairly rudimentary. Most reports on the utility of PGS in infertility and in RPL have used fluorescence in situ hybridization (FISH) for diagnosis of aneuploidy (Fig. 2). Like other cytogenetic approaches, FISH assays against metaphase spread preparations and requires live dividing cells. In FISH, fluorescently labeled probes are used to hybridize to the metaphase spread. The most frequently used probes identify specific chromosomes and readily show the presence of additional chromosomes or the absence of a chromosome, as seen in unbalanced translocations, trisomies, and monosomies. FISH can also be used to detect structural chromosome abnormalities, such as deletions (Fridstrom et al. 2001). When used for PGS rather than PGD, FISH technology is limited by the number of probes that can be differentially labeled and discriminated at one time. Although the technique itself is quite specific, only a subgroup of chromosomes can be analyzed simultaneously using this screening technique. False negative results with multichromosome FISH can therefore be unacceptably high, particularly when the result of misdiagnosis could be the transfer of an aneuploid embryo and subsequent failed implantation, miscarriage or even an affected live–born child. Some studies have shown that when compared with newer techniques, the results of multichromosome FISH may falsely diagnose up to 25% of aneuploid embryos as normal because the particular chromosome pair or pairs showing the abnormality were not among those tested using the chosen probes (Wilton et al. 2003). In 2008, the American Society of Assisted Reproductive Medicine (ASRM), the European Society for Human Reproduction and Embryology (ESHRE), and the British Fertility Society ruled that the routine implementation of PGS was ineffective in improving IVF pregnancy rates and in reducing miscarriage rates (Practice Committee of ASRM 2008b). Leading up to this ruling, most PGS studies used day 3 blastomere (see below) and FISH analysis. The most common subgroup of probes used in multicolor FISH labeled of chromosomes X, Y, 13, 15, 16, 17, 18, 21, and 22; and most studies reported on assisted reproductive technology outcomes. Two studies investigated the utility of PGS methodology in unexplained RPL (Munné et al. 2005; Platteau et al. 2005). When compared with a control group of patients with idiopathic recurrent miscarriage undergoing conservative therapy, live-birth rates were similar among the PGS and control groups; however, pregnancy rates were significantly lower among patients undergoing genetic testing (Brigham et al. 1999; Munné et al. 2005; Platteau et al. 2005). PGS was found to decrease take-home baby rates while subjecting patients and couples to invasive and costly therapy. Instead, a conservative approach focusing on supportive care and early pregnancy monitoring by ultrasound every two weeks was encouraged (Practice Committee of ASRM 2012).
Figure 2.
Cytogenetic analysis of blastomere DNA using fluorescence in situ hybridization (FISH). DNA from a single cell (e.g., a blastomere) is collected at metaphase and the chromosomes are affixed to a surface for labeling and microscopic examination. Fluorescently labeled probes specific to known segments of a given chromosome are used to label the prepared chromosomes. Multicolor FISH uses between three and 11 differently colored probes to distinguish specific chromosomes. Gain or loss of the segment of a given chromosome labeled by a probe can be determined by simply counting the number of times a given color labels the DNA within the metaphase spread.
Biopsy Methods
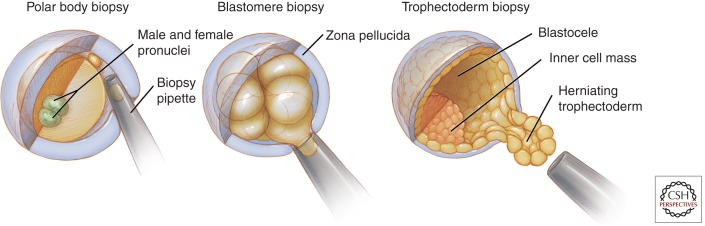
Improvements in biopsy and genetic screening techniques (discussed below) have replaced FISH-based approaches with methods that permit aneuploidy analysis of all 24 chromosomal pairs. Obtaining appropriate genetic material for testing by PGD or PGS presently uses one of three well-established procedures: polar body biopsy of the oocyte; blastomere biopsy at the eight cell stage of development (typically day 3 postfertilization, pf), or trophectoderm biopsy of a blastocyst on day 5 or 6 pf (Fig. 3). Although these biopsy procedures differ significantly from each other, all involve risk of loss of the embryo. A fourth method, blastocentesis of day 5 or 6 pf blastocysts, is currently being investigated as a less invasive method of gathering genetic material for analysis of the preimplantation embryo (Gianaroli et al. 2014). Access to embryo-derived DNA in the blastocoele cavity using this method might allow diagnosis without the removal of a blastomere or trophectoderm.
Figure 3.
Biopsy methods for preimplantation genetic analysis. There are several stages at which material can be collected for preimplantation genetic analysis. Collection at the earliest stage involves removal of a polar body after the completion of meiosis I or meiosis II. Depicted is collection after fertilization (two pronuclei) and extrusion of the second polar body. Blastomere biopsy at the cleavage stage is typically performed when the embryo contains six to eight blastomeres; one to two blastomeres are retrieved from a single embryo. Trophectoderm biopsy is most frequently performed just before spontaneous embryo hatching from the zona pellucida on day 5 postfertilization (pf) and retrieves several trophectoderm cells. All biopsy methods involve manipulation of the zona pellucida. Using mechanical, chemical or laser-assisted techniques, a breach in the zona pellucida is made at the time of collection in polar body and cleavage stage blastomere biopsies. With trophectoderm biopsy, the zona pellucida is thinned on approximately day 3 pf, creating an area for preferential herniation of the trophectoderm on day 5 pf, just before spontaneous hatching. Trophectoderm cells are typically retrieved using laser-assisted techniques.
Polar body biopsy
Biopsy of the oocyte near the time of polar body extrusion gives the earliest access to material for genetic testing. One benefit of polar body biopsy is the ability to detect meiotic errors in the oocyte, which primarily occur in meiosis I and are specifically associated with advanced maternal age. As errors can also occur during meiosis II, biopsy of the second polar body should be performed to prevent misdiagnosis. Because the polar body will not become a part of the fetus or placenta, polar body biopsy limits risk. The most significant limitation to polar body biopsy is its inability to detect paternally derived mutations and errors occurring after fertilization. Therefore, polar body biopsy is only useful when the risk of transmission of monogenetic diseases or aneuploidy is strictly of maternal origin (Montag et al. 2009).
Blastomere biopsy
Until very recently, the most frequently used method to access the genome of the developing embryo was blastomere biopsy. This technique is typically performed at or near the 8 cell stage of embryonic growth. During the course of early embryo development, the blastomeres transition from fairly weak to fairly robust intercellular connections, so biopsy at a significantly earlier stage dramatically increases the risk for embryo destruction, whereas biopsy at a later stage becomes technically more difficult. Blastomere biopsy requires dissolution of or excision through the zona pellucida surrounding the blastocyst at the 8 cell stage to allow removal of one or two cells from the embryo. Unlike polar body biopsy, sampling of the embryo at this stage enables analysis of both parental genetic contributions (Coco 2014). Unfortunately, a high rate of mosaicism (>25%; possibly as high as 50%, Delhanty 1997) is present among the blastomeres at this stage of embryonic development, thereby limiting the reliability of blastomere biopsy. Because the chromosomal content of the blastomeres that were not sampled is inferred from the chromosomal content of the single cell that was, false positive and false negative results are frequent. Although removing a second blastomere at the time of sampling may improve accuracy, this comes at the cost of decreases in embryo survival (Harper et al. 1995; Delhanty 1997; Munné et al. 1999; De Vos et al. 2009; Harton et al. 2011). A significant disadvantage of cleavage stage blastomere biopsy is the associated decrease in implantation rates for biopsied embryos (Coco 2014). A final drawback of both polar body and blastomere biopsies is the paucity of collected materials, typically only a single cell or its equivalent. This necessitates that some form of whole genome amplification be used to make use of several of the existing and evolving genetic diagnostic approaches (Macauley and Voet 2014).
Trophectoderm biopsy
In contrast to the cleavage stage embryo, the day 5 blastocyst contains more than 100 cells and consists of three distinct components: an inner cell mass containing the totipotent stem cells of the fetus; an outer cell layer or trophectoderm; and a blastocele. Most of the cells in the blastocyst will go on to form the placenta and other extraembryonic structures, whereas only a small number of cells comprise the inner cell mass that will develop into the embryonic components after implantation. Although the blastocyst will not usually emerge from the zona pellucida until days 6–7 pf, the outer trophectoderm layer will preferentially herniate on day 5 pf through an area that has been chemically or mechanically thinned on day 3 pf, making it readily accessible to biopsy. Trophectoderm biopsy at the blastocyst stage is rapidly becoming the most common sampling modality for PGD and PGS. One advantage of trophectoderm biopsy over cleavage stage biopsy is its ability to improve genetic screening accuracy by safely sampling a greater number of cells per biopsy (Yang et al. 2012b; Coco 2014). Because only extraembryonic material is sampled (similar to sampling of a chorionic villus later in the first trimester of pregnancy), a further advantage is a reduced risk of harm to the future fetus. Recent studies have shown that trophectoderm biopsy, unlike blastomere biopsy, does not affect implantation rates (Scott et al. 2013a,b; Yang et al. 2013). Like cleavage stage biopsy, mosaicism can also yield misleading results when using trophectoderm biopsy for sampling. This problem is partially mitigated by the developmental stage of the embryo (some abnormal embryos may not survive 5 d in culture, Adler et al. 2014) and the ability to access and analyze several trophectoderm cells (Fragouli et al. 2011; Capalbo et al. 2013).
Until recently, the use of trophectoderm biopsy was hampered by time-sensitive parameters; sampling is performed on day 5 of culture and transfer of fresh embryos needs to occur by day 6 pf. Time-consuming (>24 h) genetic analytics preclude the ability to transfer fresh embryos because testing results are not available to direct selection of embryos for transfer. Advances in embryo cryopreservation and the resultant switch from less efficient slow-freeze methods to ultrafast vitrification have dramatically improved embryo survival rates and clinical post-thaw pregnancy rates (Zheng et al. 2005; Keskintepe et al. 2009). Embryo cryopreservation now allows a nearly unlimited window for genetic testing to occur without concomitant decreases in clinical pregnancy and implantation rates (Keskintepe et al. 2009). Efficient and effective new molecular diagnostic tools continue to be developed at a very rapid pace. As these techniques become clinically available, they will be embraced by the ART community and their introduction into standard IVF practice should allow for fresh transfer of embryos that have been genetically screened using trophectoderm biopsy to become routine. Note that, although experts in the field have suggested that biopsy at the blastocyst stage is currently the optimal method for PGS of embryos (Scott et al. 2013a), these recommendations are based on the infertility literature; no investigations of adequate size and design nor systematic reviews are available to make similar recommendations for patients with RPL.
FUTURE DIRECTIONS
In the wake of the completion of the human genome project, future preimplantation genetic testing is predicted to include affordable sequencing of individual embryonic cells for comprehensive chromosomal and single-gene disorder analysis (Martin et al. 2013). Two studies have investigated the use of such next-generation sequencing (NGS) for preimplantation embryo assessment. One study investigated the use of NGS to identify aneuploidy and chromosomal rearrangements, while the other used NGS in the PGD of monogenic diseases (Treff et al. 2013; Yin et al. 2013). Both studies reported similar overall costs and similar or increased diagnostic accuracy when compared with current methods. Additional studies with large sample sizes are needed, however, before NGS-based preimplantation testing can be implemented in routine practice. Further, NGS offers a level of genetic detail that may identify genetic abnormalities that, while present, may not have been assessed for clinical relevance.
INCORPORATION OF GENETIC TESTING INTO THE CARE OF COUPLES WITH RPL
As the field of genetic testing continues to evolve quickly, recommendations concerning the use of genetic testing in couples with RPL are confounded by the fact that the optimal definition, diagnostic testing, and therapeutic approaches to couples experiencing RPL remain controversial. At present, the following approach to the use of genetic testing in couples experiencing RPL is prudent.
Diagnosis:
-
To aid in diagnosis and prognostic counseling, obtain karyotypes from prior losses if results or tissues are available and karyotype POCs from the second and subsequent pregnancy losses.
Because only 50%–60% of couples with RPL will have a causative factor identified after extensive testing, a thorough and cost-efficient approach should be instituted in the investigation of RPL. A recent study has shown that the use of selective RPL evaluation, defined as RPL evaluation if the second miscarriage is euploid, decreases health care costs when compared with universal RPL evaluation, defined as evaluation after the second miscarriage of <10 weeks' size (Bernardi et al. 2012). This proved true even in cases of advanced maternal age. Along with the benefits of decreased financial expenditure, both patients and physicians gain valuable information regarding the current miscarriage and subsequent reproductive potential when genetic testing of POCs is included. When patients know why a miscarriage occurred, it may be easier to overcome the grief associated with pregnancy loss. This allows patients to make a more educated decision in pursuing future pregnancies.
Karyotype the couple experiencing RPL. This aids in counseling couples about future risks and may allow for well-informed consideration of the use of PGD to identify embryos that are affected by the parental genetic abnormality.
Unfortunately, there is still an absence of adequate data to support the concept that utilization of new PGS techniques improves outcomes in recurrent miscarriage (Gleicher et al. 2014). Although the newest PGS methodologies improve the accuracy of aneuploidy diagnosis when compared with older methods, investigations that examine how these new variables affect the target population of patients who would most benefit from PGS are lacking (Gleicher and Barad 2012).
CONCLUSIONS
Genetic variables appear to play a complex role in the efficiency of human reproduction. Classically, high rates of chromosomal errors have been among the leading etiologies for fetal loss and more recent studies have begun to highlight the important role that specific single gene defects may play in pregnancy maintenance. Overall, the prognosis for a patient with RPL is good and most women with a history of RPL are less likely to miscarry in subsequent pregnancy than to deliver a live–born. It is only after a large number of sequential losses that this ratio reverses. To help aid couples struggling with RPL, limited and focused genetic testing is recommended as part of the diagnostic approach. PGD may be indicated in a small proportion of couples with defined translocations or select single gene disorders. Although great strides have been made to increase the accuracy and practicality of PGS for couples with RPL, such investigations are not presently indicated outside of clinical studies. Still, they hold much promise for future incorporation into the treatment of couples with RPL.
ACKNOWLEDGMENTS
The authors thank Linda Graziadei for her outstanding contributions to formatting and editing the final manuscript.
Footnotes
Editors: Diana W. Bianchi and Errol R. Norwitz
Additional Perspectives on Molecular Approaches to Reproductive and Newborn Medicine available at www.perspectivesinmedicine.org
REFERENCES
- Adler A, Lee HL, McCulloh DH, Ampeloquio E, Clarke-Williams M, Wertz BH, Grifo J 2014. Blastocyst culture selects for euploid embryos: Comparison of blastomere and trophectoderm biopsies. Fertil Steril 28: 485–491. [DOI] [PubMed] [Google Scholar]
- Alberman E 1988. The epidemiology of repeated abortion. In Early pregnancy loss: Mechanisms and treatment, (ed. Beard RW and Sharp F), pp. 9–17 Springer-Verlag, New York. [Google Scholar]
- Aldrich CL, Stephenson MD, Karrison T, Odem RR, Branch DW, Scott JR, Schreiber JR, Ober C 2001. HLA-G genotypes and pregnancy outcome in couples with unexplained recurrent miscarriage. Mol Hum Reprod 7: 1167–1172. [DOI] [PubMed] [Google Scholar]
- Angell RR 1994. Aneuploidy in older women. Higher rates of aneuploidy in oocytes from older women. Hum Reprod 9: 1199–1200. [DOI] [PubMed] [Google Scholar]
- Asadpor U, Totonchi M, Sabbaghian M, Hoseinifar H, Akhound MR, Zari Moradi Sh, Haratian K, Sadighi Gilani MA, Gourabi H, Mohseni Meybodi A 2013. Ubiquitin-specific protease (USP26) gene alterations associated with male infertility and recurrent pregnancy loss (RPL) in Iranian infertile patients. J Assist Reprod Genet 30: 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzato PC, Daher S, Traina E, Torioni MR, Gueuvoghlanian-Silva BY, Puccini RF, Pendeloski KP, Mattar R 2013. FAS and FAS-L genotype and expression in patients with recurrent pregnancy loss. Reprod Sci 20: 1111–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett IJ, Lomax BL, Loukianova T, Tang SS, Lestou VS, Kalousek DK 2001. Comparative genomic hybridization; a new tool for reproductive pathology. Arch Pathol Lab Med 125: 81–84. [DOI] [PubMed] [Google Scholar]
- Bell KA, Van Deerlin PG, Haddad BR, Reinberg RF 1999. Cytogenetic diagnosis of “normal 46,XX” karyotypes in spontaneous abortions frequently may be misleading. Fertil Steril 71: 334–341. [DOI] [PubMed] [Google Scholar]
- Bell KA, Van Deerlin PG, Feinberg RF, du Manoir S, Haddad BR 2001. Diagnosis of aneuploidy in archival, paraffin-embedded pregnancy loss tissues by comparative genomic hybridization. Fertil Steril 75: 374–379. [DOI] [PubMed] [Google Scholar]
- Benkhalifa M, Kasakyan S, Clement P, Baldi M, Tachdjian G, Demirol A, Gurgan T, Fiorentino F, Mohammed M, Qumsiyeh MB 2005. Array comparative genomic hybridization profiling of first-trimester spontaneous abortions that fail to grow in vitro. Prenatal Diagn 25: 894–900. [DOI] [PubMed] [Google Scholar]
- Bernardi LA, Plunkett BA, Stephenson MD 2012. Is chromosome testing of the second miscarriage cost saving? A decision analysis of selective versus universal recurrent pregnancy loss evaluation. Fertil Steril 98: 156–161. [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Horst J, Chlystun M, Croucher PJP, Nebel A, Bohring A, Markoff A 2007. A common haplotype of the annexin A5 (ANXA5) gene promoter is associated with recurrent pregnancy loss. Hum Mol Genet 16: 573–578. [DOI] [PubMed] [Google Scholar]
- Bouvier S, Cochery-Nouvellon E, Lavigne-Lissalde G, Mercier E, Fabbro-Peray P, Balducchi JP, Mares P, Gris JC 2014. Comparative incidence of pregnancy outcomes in thrombophilis-positive women from the NOH-APS observational study. Blood 123: 414–421. [DOI] [PubMed] [Google Scholar]
- Brezina PR, Benner A, Rechitsky S, Kuliev A, Pomerantseva E, Pauling D, Kearns WG 2011. Single-gene testing combined with single nucleotide polymorphism microarray preimplantation genetic diagnosis for aneuploidy: A novel approach in optimizing pregnancy outcome. Fertil Steril 95: 1786 e5–e8. [DOI] [PubMed] [Google Scholar]
- Brigham SA, Conlon C, Farquharson RG 1999. A longitudinal study of pregnancy outcome following idiopathic recurrent miscarriage. Hum Reprod 14: 2868–2871. [DOI] [PubMed] [Google Scholar]
- Brook JD, McCurrack ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. 1992. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 69: 799–808. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Li M, Hu M, Shi Y, Gu W 2007. The p53-Mdm2-HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene 26: 7262–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne JLB, Ward K 1994. Genetic factors in recurrent abortion. Clinical Obstet Gynecol 37: 693–704. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Wright G, Elliott T, Ubaldi FM, Rienzi L, Nagy ZP 2013. FISH reanalysis of inner cell mass and trophectoderm samples of previously array-CGH screened blastocysts shows high accuracy of diagnosis and no major impact of mosaicism at the blastocyst stage. Hum Reprod 28: 2298–2307. [DOI] [PubMed] [Google Scholar]
- Carp HJ 2008. Recurrent miscarriage: Genetic factors and assessment of the embryo. Isr Med Assoc J 10: 229–231. [PubMed] [Google Scholar]
- Carp H, Toder V, Aviram A, Daniely M, Mashiach S, Barkai G 2001. Karyotype of the abortus in recurrent miscarriage. Fertil Steril 75: 678–682. [DOI] [PubMed] [Google Scholar]
- Carp H, Guetta E, Dorf H, Soriano D, Barkai G, Schiff E 2006. Embryonic karyotype in recurrent miscarriage with parental karyotypic aberrations. Fertil Steril 85: 446–450. [DOI] [PubMed] [Google Scholar]
- Carrell D, Wilcox AL, Lowy L, Peterson CM, Jones KP, Erickson L, Campbell B, Branch DW, Hatasaka HH 2003. Elevated sperm chromosome aneuploidy and apoptosis in patients with unexplained recurrent pregnancy loss. Obstet Gynecol 101: 1229–1235. [DOI] [PubMed] [Google Scholar]
- Chamley LW, McKay EJ, Pattison NS 1993. Inhibition of heparin/antithrombin III cofactor activity by anticadiolipin antibodies: A mechanism for thrombosis. Thromb Res 71: 103–111. [DOI] [PubMed] [Google Scholar]
- Christiansen OB, Mathiesen O, Lauritsen JG, Grunnet N 1990. Idiopathic recurrent spontaneous abortion. Evidence of a familial predisposition. Acta Obstet Gynecol Scand 69: 597–601. [DOI] [PubMed] [Google Scholar]
- Clark P, Walker ID, Langhorne P, Crichton L, Thomsom A, Greaves M, Whyte S, Greer IA 2010. SPIN: The Scottish Pregnancy Intervention Study: A multicentre randomised controlled trial of low molecular weight heparin and low dose aspirin in women with recurrent miscarriage. Blood 115: 4162–4167. [DOI] [PubMed] [Google Scholar]
- Clifford K, Rai R, Regan L 1997. Future pregnancy outcome in unexplained recurrent first trimester miscarriage. Hum Reprod 12: 387–389. [DOI] [PubMed] [Google Scholar]
- Coco R 2014. Reprogenetics: Preimplantational genetics diagnosis. Genet Mol Biol 37: 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniely M, Aviram-Goldring , Barkai G, Goldman B 1998. Detection of chromosomal aberration in fetuses arising from recurrent spontaneous abortion by comparative genomic hybridization. Hum Reprod 13: 805–809. [DOI] [PubMed] [Google Scholar]
- Dawood F, Mountford R, Farquharson R, Quenby S 2007. Genetic polymorphisms on the factor V gene in women with recurrent miscarriage and acquired APCR. Hum Reprod 22: 546–553. [DOI] [PubMed] [Google Scholar]
- de Jong PG, Goddijn M, Middeldorp S 2011. Testing for inherited thrombophilia in recurrent miscarriage. Sem Reprod Med 29: 540–545. [DOI] [PubMed] [Google Scholar]
- de Jong PG, Goddijn M, Middeldorp S 2013. Antithrombic therapy for pregnancy loss, Hum Reprod 19: 656–673. [DOI] [PubMed] [Google Scholar]
- de la Rochebrochard E, de Mouzon J, Thépot F, Thonneau P 2006. Fathers over 40 and increased failure to conceive: The lessons of in vitro fertilization in France. Fertil Steril 85: 1420–1424. [DOI] [PubMed] [Google Scholar]
- De Vos A, Staessen C, De Rycke M, Verpoest W, Haentjens P, Devroey P, Liebaers I, Van de Velde H 2009. Impact of cleavage-stage embryo biopsy in view of PGD on human blastocyst implantation: A prospective cohort of single embryo transfers. Hum Reprod 24: 2988–2996. [DOI] [PubMed] [Google Scholar]
- Delhanty JD 1997. Chromosome analysis by FISH in human preimplantation genetics. Hum Reprod 12: 153–155. [PubMed] [Google Scholar]
- Dizon-Townson DS, Melin L, Nelson LM, Varner M, Ward K 1997. Fetal carriers of the factor V Leiden mutation are prone to miscarriage and plancental infarction. Amer J Obstet Gynecol 177: 402–405. [DOI] [PubMed] [Google Scholar]
- Dizon-Townson D, Miller C, Sibai B, Spong CY, Thom E, Wendel G Jr, Wenstrom K, Samuels P, Cotroneo MA, Moawad A, et al. 2005. The relationship of the factor V Leiden mutation and pregnancy outcomes for mother and fetus. Obstet Gynecol 106: 517–524. [DOI] [PubMed] [Google Scholar]
- Dumont P, Leu JIJ, Della Pietra AC, George DL, Murphy M 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33: 357–365. [DOI] [PubMed] [Google Scholar]
- Edmonds DK, Lindsay KS, Miller JF, Williamson E, Wood PJ 1982. Early embryonic mortality in women. Fertil Steril 38: 447–453. [PubMed] [Google Scholar]
- Eiben B, Bartels I, Bähr-Porsch S, Borgmann S, Gatz G, Gellert G, Osmers R 1990. Cytogenetic analysis of 750 spontaneous abortions with the direct-preparation method of chorionic villi and its implications for studying genetic causes of pregnancy wastage. Amer J Hum Genet 47: 656–663. [PMC free article] [PubMed] [Google Scholar]
- Ford WCL, North K, Taylor H, Farrow A, Hull MGR, Golding J 2000. Increasing paternal age is associated with delayed conception in a large population of fertile couples: Evidence for declining fecundity in older men. The ALSPAC Study Team (Avon Longitudinal Study of Pregnancy and Childhood). Hum Reprod 15: 1703–1708. [DOI] [PubMed] [Google Scholar]
- Fraga LR, Dutra CG, Boquett JA, Vianna FS, Goncalves RO, Paskulin DD, Costa OL, Ashton-Prolla P, Sanseverino MT, Schuler-Faccini L 2014. P53 signaling pathway polymorphisms associated to recurrent pregnancy loss. Mol Biol Reprod 41: 1871–1877. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Alfarwati S, Daphnis DD, Goodall N, Mania A, Griffiths T, Gordon A, Wells D 2011. Cytogenetic analysis of human blastocysts with FISH, CGH and aCGH: Scientific data and technical evaluation. Hum Reprod 26: 480–490. [DOI] [PubMed] [Google Scholar]
- Fridstrom M, Ahrlund-Richter L, Iwarsson E, Malmgren H, Inzunza J, Rosenlund B, Sjoblom P, Nordenskjold M, Biennow E, Hovatta O 2001. Clinical outcome of treatment cycles using preimplantation genetic diagnosis for structural chromosomal abnormalities. Prenat Diagn 21: 781–787. [DOI] [PubMed] [Google Scholar]
- Geraedts JPM 1996. Chromosomal anomalies and recurrent miscarriage. Infertil Reprod Med Clin North Am 7: 677–688. [Google Scholar]
- Gianaroli l, Magli MC, Pomante A, Crivello AM, Cafueri G, Valerio M, Ferraretti AP 2014. Blastocentesis: A source of DNA for preimplantation genetic testing. Results from a pilot study. Fertil Steril 102: 1692–1699. [DOI] [PubMed] [Google Scholar]
- Gleicher N, Barad DH 2012. A review of, and commentary on the ongoing second clinical introduction of preimplantation genetic screening (PGS) to routine IVF practice. J Assist Reprod Genet 12: 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleicher N, Kushnir VA, Barad DH 2014. Preimplantation genetic screening (PGS) still in search of a clinical application: A systematic review. Reprod Biol Endocrinol 12: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman C, Jeyendran RS, Coulam CB 2008. Vascular endothelial growth factor gene polymorphism and implantation failure. Reprod Biomed 16: 720–723. [DOI] [PubMed] [Google Scholar]
- Gopel W, Ludwig M, Junge AK, Kohlman T, Diedrich K, Moller 2001. Selection pressure for the factor-V-Leiden mutation and embryo implantation. Lancet 358: 1238–1239. [DOI] [PubMed] [Google Scholar]
- Grandone E, Margaglione M, Colaizzo D, d’Addedda M, Cappucci G, Vecchione G, Scianname N, Pavone G, Di Minno G 1997. Factor V Leiden is associated with repeated and recurrent unexplained fetal losses. Thromb Homeost 77: 822–824. [PubMed] [Google Scholar]
- Handyside AH, Kontogianni EH, Hardy K, Winston RML 1990. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature 344: 768–770. [DOI] [PubMed] [Google Scholar]
- Harper JC 2010. ESHRE PGD Consortium data collection X: Cycles from January to December 2007 with pregnancy follow-up to October 2008. Hum Reprod 25: 2685–2707. [DOI] [PubMed] [Google Scholar]
- Harper JC, Coonen E, Ramaekers FC, Delhanty JD, Handyside AH, Winston RM, Hopman AH 1994. Identification of the sex of human preimplantation embryos in two hours using an improved spreading method and fluorescent in-situ hybridization (FISH) using directly labelled probes. Hum Reprod 9: 721–724. [DOI] [PubMed] [Google Scholar]
- Harper JC, Coonen E, Handyside AH, Winston RM, Hopman AH, Delhanty JD 1995. Mosaicism of autosomes and sex chromosomes in morphologically normal, monospermic preimplantation human embryos. Prenat Diagn 15: 41–49. [DOI] [PubMed] [Google Scholar]
- Harton GL, Magli MC, Lundin K, Montag M, Lemmen J, Harper JC, European Society for Human Reproduction, Embryology (ESHRE), PGD Consortium/Embryology Special Interest Group. 2011. ESHRE PGD Consortium/Embryology Special Interest Group—Best practice guidelines for polar body and embryo biopsy for preimplantation genetic diagnosis/screening (PGD/PGS). Hum Reprod 26: 41–46. [DOI] [PubMed] [Google Scholar]
- Hassold TJ 1980. A cytogenetic study of repeated spontaneous abortions. Am J Hum Genet 32: 723–730. [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Chiu D 1985. Maternal age-specific rates of numerical chromosome abnormalities with special reference to trisomy. Hum Genet 70: 11–17. [DOI] [PubMed] [Google Scholar]
- Hassold T, Jacobs P, Kline J, Stein Z, Warburton D 1980. Effect of maternal age on autosomal trisomies. Ann Hum Genet 44: 29–36. [DOI] [PubMed] [Google Scholar]
- Hassold T, Warburton D, Kline J, Stein Z 1984. The relationship of maternal age and trisomy among trisomic spontaneous abortions. Am J Hum Genet 36: 1349–1356. [PMC free article] [PubMed] [Google Scholar]
- Hawley R, Frazier J, Rasooly R 1994. Separation anxiety: The etiology of nondisjunction in flies and people. Hum Mol Genet 3: 1521–1528. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Sasaki H, Suzuki S, Nishiyama T, Kitaori T, Mizutani E, Sugiura-Ogasawara M 2013. Genotyping analyses for polymorphisms of ANXA5 gene in patients with recurrent pregnancy loss. Fertil Steril 100: 1018–1024. [DOI] [PubMed] [Google Scholar]
- Hogge WA, Byrnes AL, Lanasa MC, Surti U 2003. The clinical use of karyotyping spontaneous abortions. Am J Obstet Gynecol 189: 397–402. [DOI] [PubMed] [Google Scholar]
- Hu Y, Chen X, Chen LL, Xu ZF, Wang X, Cui H 2006. Comparative genomic hybridization analysis of spontaneous abortion. Int J Gynaecol Obstet 92: 52–57. [DOI] [PubMed] [Google Scholar]
- Hu WW, Feng ZH, Teresky AK, Levine AJ 2007. p53 regulates maternal reproduction through LIF. Nature 450: 721–724. [DOI] [PubMed] [Google Scholar]
- Jarrett KL, Michaelis RC, Phelan MC, Vincent VA, Best RG 2001. Microsatellite analysis reveals a high incidence of maternal cell contamination in 46,XX products of conception consisting of villi or a combination of villi and membranous material. Am J Obstet Gynecol 185: 198–203. [DOI] [PubMed] [Google Scholar]
- Kaandorp SP, Goddijn M, van der Post JA, Hutten BA, Verhoeve HR, Hamulyak K, Mol BW, Folkeringa N, Nahuis M, Papatsonis DN, et al. 2010. Aspirin plus heparin or aspirin alone in women with recurrent miscarriage. N Engl J Med 362: 1586–1596. [DOI] [PubMed] [Google Scholar]
- Kaandorp S, Di Nisio M, Goddijn M, Middelldorp S 2014. Aspirin or anticoagulants for treating recurrent miscarriage in women without antiphospholipid syndrome. Cochrane Database Syst Rev 7: CD004734. [DOI] [PubMed] [Google Scholar]
- Keskintepe L, Sher G, Machnicka A, Tortoriello D, Bayrak A, Fisch J, Agca Y 2009. Vitrification of human embryos subjected to blastomere biopsy for pre-implantation genetic screening produces higher survival and pregnancy rates than slow freezing. J Assist Reprod Genet 26: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinhaus K, Perrin M, Friedlander Y, Paltiel O, Malaspina D, Harlap S 2006. Paternal age and spontaneous abortion. Obstet Gynecol 108: 369–377. [DOI] [PubMed] [Google Scholar]
- Kotzot D 1999. Abnormal phenotypes in uniparental disomy (UPD): Fundamental aspects and a critical review with bibliography of UPD other than 15. Am J of Med Genet 82: 265–274. [PubMed] [Google Scholar]
- Kudesia R, Li M, Smith J, Patel A, Williams Z 2014. Rescue karyotyping: A case series of array-based comparative genomic hybridization evaluation of archival conceptual tissue. Reprod Biol Endocrinol 12: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir VA, Scott RT, Frattarelli JL 2010. The impact of paternal age on aneuploidy rates in first trimester pregnancy loss. J Med Genet Genomics 2: 38–43. [Google Scholar]
- Lathi RB, Milki AA 2002. Tissue sampling technique affects accuracy of karyotype from missed abortions. J Assist Reprod Genet 19: 536–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy B, Sigurjonsson S, Pettersen B, Maisenbacher MK, Hall MP, Demko Z, Lathi RB, Tao R, Aggarwal V, Rabinowitz M 2014. Genomic imbalance in products of conception: Single-nucleotide polymorphism chromosomal microarray analysis. Obstet Gynecol 124: 202–209. [DOI] [PubMed] [Google Scholar]
- Lomax B, Tang S, Separovic E, Phillips D, Hillard E, Thomson T 2000. Comparative genomic hybridization in combination with flow cytometry improves results of cytogenetic analysis of spontaneous abortions. Am J Hum Genet 66: 1516–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley IC, Voet T 2014. Single cell genomics: Advances and future perspectives. PLoS Genet 10: e1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquard K, Westphal LM, Milki AA, Lathi RB 2010. Etiology of recurrent pregnancy loss in women over the age of 35 years. Fertil Steril 94: 1473–1477. [DOI] [PubMed] [Google Scholar]
- Martin J, Cervero A, Mir P, Martinez-Conejero JA, Pellicer A, Simon C 2013. The impact of next-generation sequencing technology on preimplantation genetic diagnosis and screening. Fertil and Steril 99: 1054–1061. [DOI] [PubMed] [Google Scholar]
- Mathur N, Triplett L, Stephenson MD 2014. Miscarriage chromosome testing: Utility of comparative genomic hybridization with reflex microsatellite analysis in preserved miscarriage tissue. Fertil Steril 101: 1349–1352. [DOI] [PubMed] [Google Scholar]
- Middeldorp S 2011. Is thrombophilia testing useful? Hematol Am Soc Hematol Educ Program 2011: 150–155. [DOI] [PubMed] [Google Scholar]
- Mohamed MA, El Moaty MA, El Kholy AF, Mohamed SA, Ali AL 2010. Thrombophilic gene mutations in women with repeated spontaneous miscarriage. Genet Test Mol Biomarkers 14: 593–597. [DOI] [PubMed] [Google Scholar]
- Montag M, van der Ven K, van der Ven H 2009. Polar body biopsy. In Preimplantation genetic diagnosis, 2nd ed. (ed. Harper JC), pp. 166–174 Cambridge University Press, Cambridge, MA. [Google Scholar]
- Munné S, Alikani M, Tomkin G, Grifo J, Cohen J 1995. Embryo morphology, developmental rates, and maternal age are correlated with chromosome abnormalities. Fertil Steril 64: 382–391. [PubMed] [Google Scholar]
- Munné S, Magli C, Cohen J, Morton P, Sadowy S, Gianaroli L, Tucker M, Marquez C, Sable D, Ferraretti AP, et al. 1999. Positive outcome after preimplantation diagnosis of aneuploidy in human embryos. Hum Reprod 14: 2191–2199. [DOI] [PubMed] [Google Scholar]
- Munné S, Chen S, Fischer J, Colls P, Zheng X, Stevens J, Escudero T, Oter M, Schoolcraft B, Simpson JL, et al. 2005. Preimplantation genetic diagnosis reduces pregnancy loss in women aged 35 years and older with a history of recurrent miscarriages. Fertil Steril 84: 331–335. [DOI] [PubMed] [Google Scholar]
- Ogasawara M, Aoki K, Okada S, Suzumori K 2000. Embryonic karyotype of abortuses in relation to the number of previous miscarriages. Fertil Steril 73: 300–304. [DOI] [PubMed] [Google Scholar]
- Pfeiffer KA, Fimmers R, Engels G, van Der Ven H, van Der Ven K 2001. The HLA-G genotype is potentially associated with idiopathic recurrent spontaneous abortion. Mol Hum Reprod 7: 373–378. [DOI] [PubMed] [Google Scholar]
- Platteau P, Staessen C, Michiels A, Van Steirteghem A, Liebaers I, Devroey P 2005. Preimplantation genetic diagnosis for aneuploidy screening in patients with unexplained recurrent miscarriages. Fertil Steril 82: 393–397. [DOI] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine. 2008a. Definitions of infertility and recurrent pregnancy loss. Fertil Steril 89: 1603. [DOI] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine. 2008b. Preimplantation genetic testing: A Practice Committee opinion. Fertil Steril 90: S136–S143. [DOI] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine. 2012. Evaluation and treatment of recurrent pregnancy loss: A committee opinion. Fertil Steril 98: 1103–1111. [DOI] [PubMed] [Google Scholar]
- Puscheck EE, Jeyendran RS 2007. The impact of male factor on recurrent pregnancy loss. Curr Opin Obstet Gynecol 19: 222–228. [DOI] [PubMed] [Google Scholar]
- Rajcan-Separovic E, Diego-Alvarez D, Robinson WP, Tyson C, Qiao Y, Harvard C, Fawcett C, Kalousek D, Philipp T, Somerville MJ, et al. 2010. Identification of copy number variants in miscarriages from couples with idiopathic recurrent pregnancy loss. Hum Reprod 25: 2913–2922. [DOI] [PubMed] [Google Scholar]
- Ren A, Wang J 2006. Methylenetetrahydrofolate reductase C677T polymorphism and the risk of unexplained recurrent pregnancy loss: A meta-analysis. Fertil Steril 86: 1716–1722. [DOI] [PubMed] [Google Scholar]
- Ridker PM, Miletich JP, Buring JE, Ariyo AA, Price DT, Manson JE, Hill JA 1998. Factor V Leiden mutation as a risk factor for recurrent pregnancy loss. Ann Intern Med 128: 1000–1003. [DOI] [PubMed] [Google Scholar]
- Robertson L, Wu O, Langhorne P, Twaddle S, Clark P, Lowe GD, Walker ID, Greaves M, Brenkel I, Regan L, Grear IA, et al. The Thrombosis: Risk and Economic Assessment of Thrombophilia Screening (TREATS) Study. 2006. Thrombophilia in pregnancy: A systematic review. Br J Haematol 132: 171–196. [DOI] [PubMed] [Google Scholar]
- Roque H, Paidas MJ, Funai EF, Kuczynski E, Lockwood CJ 2004. Maternal thrombophilias are not associated withearly pregnancy loss. Thromb Haemost 91: 290–295. [DOI] [PubMed] [Google Scholar]
- Regan L, Backos M, Rai R 2011. Green-top Guideline No 17. The investigation and treatment of couples with recurrent first-trimester and second-trimester miscarriage. Royal College of Obstetricians and Gynaecologists (RCOG), London. [Google Scholar]
- Rubio C, Gil-Salom M, Simon C, Vidal F, Rodrigo L, Minguez Y, Remohi J, Pellicer A 2001. Incidence of sperm chromosomal abnormalities in a risk population: Relationship with sperm quality and ICSI outcome. Hum Reprod 16: 2084–2092. [DOI] [PubMed] [Google Scholar]
- Scott KL, Hong KH, Scott RT Jr 2013a. Selecting the optimal time to perform biopsy for preimplantation genetic testing. Fertil Steril 100: 608–614. [DOI] [PubMed] [Google Scholar]
- Scott RT, Upham KM, Forman EJ, Zhao T 2013b. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: A randomized and paired clinical trial. Fertil Steril 100: 624–630. [DOI] [PubMed] [Google Scholar]
- Sierra S, Stephenson MD 2006. Genetics of recurrent pregnancy loss. Semin Reprod Med 24: 17–24. [DOI] [PubMed] [Google Scholar]
- Silver RM, Branch D 2007. Sporadic and Recurrent Pregnancy Loss. In Clinical obstetrics: The fetus & mother, 3rd ed (ed. Reece EA, Hobbins JC), pp. 143–160 Blackwell Publishing, Boston. [Google Scholar]
- Silver RM, Zhao Y, Spong CY, Sibai B, Wendel G Jr, et al. 2010. Prothrombin gene G20210A mutation and obstetric complications. Obstet Gynecol 115: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón C, Rubio C, Vidal F, Gimenez C, Moreno C, Parrilla JJ, Pellicer A 1998. Increased chromosome abnormalities in human preimplantation embryos after in-vitro fertilization in patients with recurrent miscarriage. Reprod Fertil Dev 10: 87–92. [DOI] [PubMed] [Google Scholar]
- Slama R, Bouyer J, Windham G, Fenster L, Werwatz A, Swan SH 2005. Influence of paternal age on the risk of spontaneous abortion. Amer J Epidemiol 161: 816–823. [DOI] [PubMed] [Google Scholar]
- Sloter E, Nath J, Eskenazi B, Wyrobek AJ 2004. Effects of male age on the frequencies of germinal and heritable chromosomal abnormalities in humans and rodents. Fertil Steril 81: 925–943. [DOI] [PubMed] [Google Scholar]
- Steck T, Giess R, Suetterlin MW, Bolland M, Wiest S, Poehls UG, Dietl J 2004. Leukaemia inhibitory factor (LIF) gene mutations in women with unexplained infertility and recurrent failure of implantation after IVF and embryo transfer. Eur J Obstet Gynecol Reprod Biol 112: 69–73. [DOI] [PubMed] [Google Scholar]
- Stephenson M 1996. Frequency of factors associated with habitual abortion in 197 couples. Fertil Steril 66: 24–29. [PubMed] [Google Scholar]
- Stephenson M, Kutteh W 2007. Evaluation and management of recurrent early pregnancy loss. Clin Obstet Gynecol 50: 132–145. [DOI] [PubMed] [Google Scholar]
- Stephenson MD, Awartani K, Robinson WP 2002. Cytogenetic analysis of miscarriages from couples with recurrent miscarriage: A case-control study. Hum Reprod 17: 446–451. [DOI] [PubMed] [Google Scholar]
- Stern JJ, Dorfmann AD, Gutiérrez-Najar AJ, Cerrillo M, Coulam CB 1996. Frequency of abnormal karyotypes among abortuses from women with and without a history of recurrent spontaneous abortion. Fertil Steril 65: 250–253. [DOI] [PubMed] [Google Scholar]
- Sullivan AE, Silver RM, LaCoursiere DY, Porter TF, Branch DW 2004. Recurrent fetal aneuploidy and recurrent miscarriage. Obstet Gynecol 104: 784–788. [DOI] [PubMed] [Google Scholar]
- Suthers G 1996. Mutations, malformations and mortality. J Ped Child Health 32: 10–15. [DOI] [PubMed] [Google Scholar]
- Suzumori N, Sugiura-Ogasawara M 2010. Genetic factors as a cause of miscarriage. Curr Med Chem 17: 3431–3437. [DOI] [PubMed] [Google Scholar]
- Thiesen A 2008. Microarray-based comparative genomic hybridization (aCGH). Nat Educ 1: 45. [Google Scholar]
- Treff NR, Fedick A, Tao X, Devkota B, Taylor D, Scott RT Jr 2013. Evaluation of targeted next-generation sequencing-based preimplantation genetic diagnosis of monogenic disease. Fertil Steril 99: 1377–1384. [DOI] [PubMed] [Google Scholar]
- Vagnini L, Baruffi RLR, Mauri AL, Petersen CG, Massaro FC, Pontes A, Franco JG 2007. The effects of male age on sperm DNA damage in an infertile population. Reprod Biomed Online 15: 514–519. [DOI] [PubMed] [Google Scholar]
- Vatin M, Bouvier S, Bellazi L, Montagutelli X, Laissue P, Ziyyat A, Serres C, De Mazancourt P, Dieudonne MN, Mornet E, et al. 2014. Polymorphisms of human placental alkaline phosphatase are associated with in vitro fertilization success and recurrent pregnancy loss. Am J Pathol 184: 362–368. [DOI] [PubMed] [Google Scholar]
- Viaggi CD, Cavani S, Malacarne M, Floriddia F, Zerega G, Baldo C, Mogni M, Castagnetta M, Piombo G, Covielli DA, et al. 2013. First-trimeter euploid miscarriages analyzed by array-CGH. J Appl Genet 54: 353–359. [DOI] [PubMed] [Google Scholar]
- Vidal F, Giménez C, Rubio C, Simón C, Pellicer A, Santaló J, Egozcue J 1998. FISH preimplantation diagnosis of chromosome aneuploidy in recurrent pregnancy wastage. J Assist Reprod Genet 15: 310–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang P, Wang X, He X, Wang Z, Xu D, Hu J, Wang B 2013. Significant association between angiotensin-converting enzyme gene insertion/deletion polymorphism and risk of recurrent miscarriage: A systematic review and meta-analysis. Metabolism 62: 1227–1238. [DOI] [PubMed] [Google Scholar]
- Wapner RJ, Lewis D 2002. Genetics and metabolic causes of stillbirth. Semin Perinatol 26: 70–74. [DOI] [PubMed] [Google Scholar]
- Warburton D, Kline J, Stein Z, Hutzler M, Chin A, Hassold T 1987. Does the karyotype of a spontaneous abortion predict the karyotype of a subsequent abortion? Evidence from 273 women with two karyotyped spontaneous abortions. Amer J Hum Genet 41: 465–483. [PMC free article] [PubMed] [Google Scholar]
- Warren JE, Silver RM 2008. Genetics of pregnancy loss. Clin Obstet Gynecol 51: 84–95. [DOI] [PubMed] [Google Scholar]
- Wilcox AJ, Weinberg CR, O’Connor JF, Baird DD, Schlatterer JP, Canfield RE, Armstrong EG, Nisula BC 1988. Incidence of early loss of pregnancy. N Engl J Med 319: 189–194. [DOI] [PubMed] [Google Scholar]
- Wilton L, Voullaire L, Sargeant P, Williamson R, McBain J 2003. Preimplantation aneuploidy screening using comparative genomic hybridization or fluorescence in situ hybridization of embryos from patients with recurrent implantation failure. Fertil Steril 80: 860–868. [DOI] [PubMed] [Google Scholar]
- Wolf CC, Horger EO III 1995. Indications for examination of spontaneous abortion specimens: A reassessment. Am J Obstet Gynecol 173: 1364–1368. [DOI] [PubMed] [Google Scholar]
- Yang C, Fangfang W, Jie L, Yanlong Y, Jie W, Xuefei L, Xuerong Z, Yanling H 2012a. Angiotensin-converting enzyme insertion/deletion (I/D) polymorphisms and recurrent pregnancy loss: A meta-analysis. J Assist Reprod Genet 29: 1167–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, Peck AC, Sills SE, Salem RD 2012b. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: Results from a randomized pilot study. Mol Cytogenet 5: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Salem SA, Liu X, Kuang Y, Salem RD, Liu J 2013. Selection of euploid blastocysts for cryopreservation with array comparative genomic hybridization (aCGH) results in increased implantation rates in subsequent frozen and thawed embryo transfer cycles. Mol Cytogenet 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Tan K, Vajta G, Jiang H, Tan Y, Zhang C, Chen F, Chen S, Zhang C, Pan X, et al. 2013. Massively parallel sequencing for chromosomal abnormality testing in trophectoderm cells of human blastocysts. Biol Reprod 88: 69. [DOI] [PubMed] [Google Scholar]
- Zheng WT, Zhuang GL, Zhou CQ, Fang C, Ou JP, Li T, Zhang MF, Liang XY 2005. Comparison of the survival of human biopsied embryos after cryopreservation with four different methods using non-transferable embryos. Hum Reprod 20: 1615–1618. [DOI] [PubMed] [Google Scholar]