Abstract
Over the past decade the zebrafish (Danio rerio) has become an attractive new vertebrate model organism for studying mycobacterial pathogenesis. The combination of medium-throughput screening and real-time in vivo visualization has allowed new ways to dissect host pathogenic interaction in a vertebrate host. Furthermore, genetic screens on the host and bacterial sides have elucidated new mechanisms involved in the initiation of granuloma formation and the importance of a balanced immune response for control of mycobacterial pathogens. This article will highlight the unique features of the zebrafish–Mycobacterium marinum infection model and its added value for tuberculosis research.
Zebrafish can be infected by Mycobacterium marinum, a close genetic relative of Mycobacterium tuberculosis. Improved genetic tools, imaging techniques, and high-throughput work flows have made this system attractive for studying tuberculosis.
Why would one use zebrafish (Danio rerio) to study tuberculosis (TB)? Although zebrafish are vertebrates, they do not have lungs, an obvious caveat for studying a pulmonary disease. Furthermore, at present it is unclear whether Mycobacterium tuberculosis can give rise to successful infections in cold-blooded animals. Robert Koch tried to infect cold-blooded animals, including a turtle, a goldfish, three eels, and five frogs. After two months, none of them showed any sign of disease, whereas most mammals were either clearly ill or showed tubercles upon autopsy (Koch 1884). Despite these drawbacks, zebrafish have emerged as a valuable organism to study infectious diseases and especially TB (Grunwald and Eisen 2002; Meeker and Trede 2008; Ramakrishnan 2013). The power of the model, real-time imaging of biological processes, was first exploited for TB by the group of Ramakrishnan (Davis et al. 2002), leading the way to study mycobacterial virulence factors and host characteristics in real time in a living vertebrate animal. In recent years, the strength of the zebrafish model has been greatly extended with the increasing availability of transgenic zebrafish lines, improved imaging techniques, and a growing list of genetic tools and large-scale mutant analysis. This article will highlight the unique features of the zebrafish–Mycobacterium marinum infection model and its added value for TB research.
WHAT IS THE ZEBRAFISH–M. marinum MODEL OF TUBERCULOSIS?
To appreciate the zebrafish—M. marinum model of TB, it is important to discuss the basic traits and tools of both the zebrafish and its natural pathogen M. marinum.
General Properties of Zebrafish
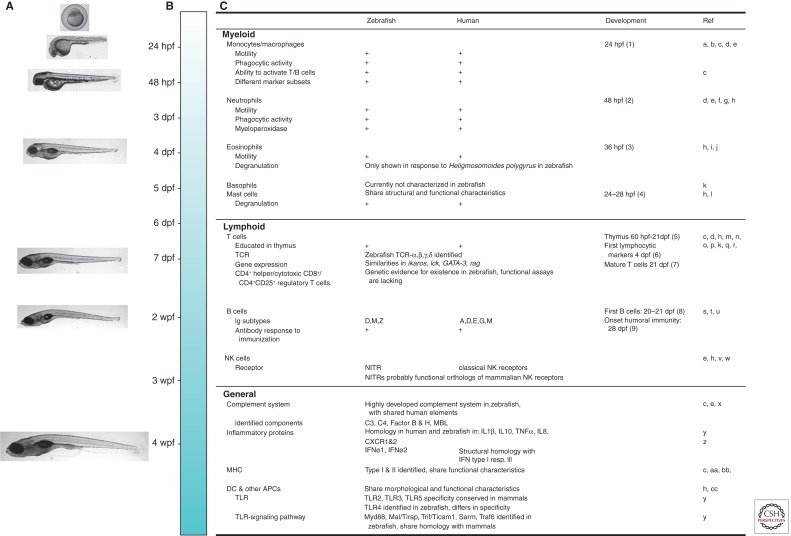
Advantageous features of the zebrafish include their small size (adults are 3- to 5-cm long), the possibility of keeping them at high population density (5 fish/L), and their ease of breeding—a single female can lay up to 300 eggs a week (Meijer and Spaink 2011). Zebrafish embryos develop externally and are transparent during embryo and larval stages, making it possible to follow host–pathogen interaction in real time. In contrast to other animal models, the zebrafish can be studied during the first weeks of development. In this period, the embryo solely relies on the innate immune system (Fig. 1) (van der Sar et al. 2004b; Meeker and Trede 2008; Novoa and Figueras 2012; van der Vaart et al. 2012), which provides the opportunity to study the contribution of innate immunity to disease in an isolated fashion. Furthermore, it allows for distinguishing between this arm of immunity and a combined innate and adaptive immune response in the context of infection, like in adult fish, which have a complex adaptive immune system akin to that of mammals (Fig. 1) (Traver et al. 2003; Meeker and Trede 2008; Renshaw and Trede 2012; van der Vaart et al. 2012). The finalized whole-genome sequence of zebrafish (Howe et al. 2013) reveals that ∼70% of human genes have at least one obvious zebrafish ortholog.
Figure 1.
Development of zebrafish immunity in comparison with the human immune system. Zebrafish possess a complex immune system, similar to that of humans. Development of zebrafish larvae is shown in A and a time line is shown in B. The appearance of components of the immune system is shown in C, and comparison is made with human immune components. Components of the innate immune system are detectable and active in the first days postfertilization (dpf) (e.g., macrophages, neutrophils, eosinophils, and mast cells). Adaptive immunity takes longer to develop and starts with thymus development at 60 hours postfertilization (hpf) and the appearance of the first lymphocytic markers at ∼4 dpf. At 21 dpf, the thymus is fully matured, and the first mature T cells and B cells are detected; humoral immunity is functional at 28 dpf. wpf, weeks postfertilization; TCR, T-cell receptor; NK, natural killer; MHC, major histocompatibility complex; DC, dendritic cell; APC, antigen-presenting cell; TLR, Toll-like receptor; NITR, novel immune-type receptor; TNF-α, tumor necrosis factor-α; IFN, interferon. References cited in figure as follows: a, Herbomel et al. 1999; b, Herbomel et al. 2001; c, Traver et al. 2003; d, Meijer and Spaink 2011; e, Novoa and Figueras 2012; f, Renshaw et al. 2006; g, Le Guyader et al. 2008; h, Renshaw and Trede 2012; i, Bertrand et al. 2007; j, Balla et al. 2010; k, Meeker and Trede 2008; l, Dobson et al. 2008; m, Lam et al. 2002; n, Trede et al. 2004; o, Danilova et al. 2004; p, Schorpp et al. 2006; q, Meeker et al. 2010; r, Laing and Hansen 2011; s, Lam et al. 2004; t, Danilova et al. 2005; u, Page et al. 2013; v, Yoder 2009; w, Yoder et al. 2010; x, van der Sar et al. 2004b; y, van der Vaart et al. 2012; z, Palha et al. 2013; aa, de Jong et al. 2011; bb, de Jong and Zon 2012; cc, Lugo-Villarino et al. 2010.
Because of the genetic possibilities (Amsterdam and Hopkins 2006; Lesley and Ramakrishnan 2008; Meijer and Spaink 2011; Blackburn et al. 2013) and the specimen availability and size (and therefore screening options), zebrafish are often seen as a bridge between cell culture systems and mammals (Brittijn et al. 2009).
General Properties of M. marinum
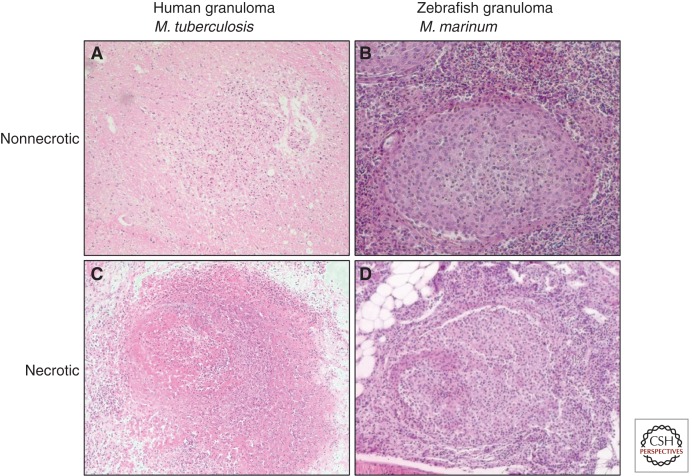
Because M. tuberculosis does not seem to cause disease in cold-blooded animals, an alternative pathogen is used. Zebrafish are susceptible to a number of mycobacterial pathogens, of which M. marinum is the most interesting candidate (Watral and Kent 2007). M. marinum naturally inhabits aquatic environments and is the causative agent of a tuberculosis-like disease in cold-blooded animals (Tobin and Ramakrishnan 2008). Furthermore, this species is a close genetic relative of M. tuberculosis. At 6.6 Mb, the genome of M. marinum is ∼1.5 times the size of that of M. tuberculosis, which likely reflects its expanded host range and capabilities to survive in the environment. Orthologous coding sequences share an average amino acid identity of 85% (Stinear et al. 2008). Furthermore, the two species share different mechanisms for intracellular growth and host survival. M. tuberculosis genes can usually complement mutations in M. marinum orthologs and vice versa (Gao et al. 2003; Stinear et al. 2008; Tobin and Ramakrishnan 2008; Stoop et al. 2011). Similar to M. tuberculosis, specific genotypic lineages of M. marinum are associated with variability in virulence (van der Sar et al. 2004a; Ostland et al. 2008; Hernandez-Pando et al. 2012). Apart from the free-living stage, another clear difference for M. marinum is its restricted growth temperature, which lies between 28°C and 30°C. Growth is normally halted at 37°C, which is considered as one of the main factors that limits M. marinum infections to cooler surface of the skin (Kent et al. 2006). M. marinum is primarily associated with human skin lesions called fish tank granulomas. Interestingly, these local granulomas are often histopathologically indistinguishable from M. tuberculosis dermal granulomas (Travis et al. 1985; MacGregor 1995) (Fig. 2). M. marinum has several other advantages over working with M. tuberculosis, including fewer biosafety restrictions (BSL2 instead of BSL3) and a relatively short replication time (4 h) (Tobin and Ramakrishnan 2008).
Figure 2.
Pathology in adult fish compared with human granulomas. Zebrafish granulomas caused by M. marinum show great similarities with human granulomas formed after infection with M. tuberculosis. Panels represent nonnecrotic early granuloma in human (A) and zebrafish (B) and granulomas with a necrotic center in human (C) and zebrafish (D). Human granulomas obtained from a neuropathology study in our Department of Pediatric Infectious Diseases and Immunology (D Zaharie, S Roest, M van der Kulp, AM van Furth, pers. comm.).
ROUTES OF INFECTION
The natural infection route for M. marinum has not been fully elucidated, but the available evidence strongly indicates that the gastrointestinal tract is the port of entry (Harriff et al. 2007). Furthermore, transmission was significantly enhanced when the bacteria were supplied within free-living unicellular eukaryotes, including amoeba and paramecium (Harriff et al. 2007; Peterson et al. 2013). However, these more natural routes of transmission are not really applicable for infection experiments, as the infection dose and timing cannot be easily controlled. Therefore, to study mycobacterial pathogenesis in vivo, zebrafish are infected with M. marinum via different inoculation routes (Fig. 3). Adult zebrafish are usually infected by intraperitoneal or intramuscular injection, whereas the most commonly used infection route in embryos is injection into the caudal vein at 28 hpf (Meijer and Spaink 2011; Benard et al. 2012). Local inoculation routes (e.g., via the hindbrain ventricle, muscle, notochord, or otic vesicle [Fig. 3]) can be used to study macrophage and neutrophil chemotaxis. Alternatively, yolk injection at the one- to four-cell stage can be applied for early infections in a high-throughput setting (Meijer and Spaink 2011).
Figure 3.
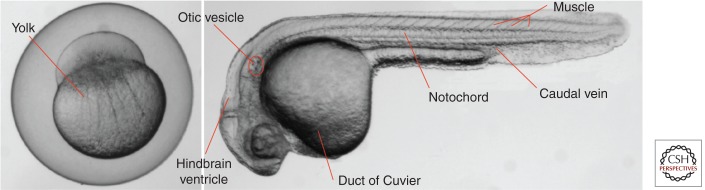
Routes of infection. Zebrafish are infected with M. marinum at different time points and via different inoculation routes. Systemic infection is achieved by injection into the caudal vein at 28 hpf or inoculation via the duct of Cuvier in embryos at 2–3 dpf (Benard et al. 2012). Local injection routes are the hindbrain ventricle, muscle, notochord (Alibaud et al. 2011), or otic vesicle. In addition to intravenous injection at 24–28 hpf, yolk injection can be applied at the one- to four-cell stage in a high-throughput setting (Benard et al. 2012).
KEY FEATURES OF M. marinum INFECTION IN ZEBRAFISH
Actually two zebrafish infection models exist, the adult and the embryonic-larval model. Each has its own characteristics and benefits and both will be discussed.
Pathology in Adult Fish
Adult zebrafish develop on intraperitoneal injection with M. marinum, a chronic infection with necrotic (caseating) granulomas, a key feature of human TB (Pozos and Ramakrishnan 2004; van der Sar et al. 2004a; Berg and Ramakrishnan 2012). These granulomas are preferentially formed in fatty tissue and are most commonly found in the pancreas, adipose tissue, liver, spleen, and gonads (Swaim et al. 2006; Parikka et al. 2012; Oksanen et al. 2013; Stoop et al. 2013). The first granulomas can already be found in the first weeks postinfection (Swaim et al. 2006; Parikka et al. 2012). Even the first signs of necrosis, consisting of cytoplasmic and nuclear debris, are present at this time. The induction of a latent, chronic, or active mycobacterial disease depends on the infection dose and the M. marinum strain used (van der Sar et al. 2004a; Swaim et al. 2006; Parikka et al. 2012). A low infection dose results in a latent disease with stable numbers of granulomas over time, whereas a high-dose infection leads to a more progressive and active disease (Parikka et al. 2012). During a chronic disease course in zebrafish, bacterial growth seems to mimic growth curves of various other animal models of TB—growth for the first 3–4 wk and reaching a plateau when adaptive immunity develops (North and Jung 2004; van der Sar et al. 2004a; Swaim et al. 2006; Parikka et al. 2012; Ramakrishnan 2012). At 16–20 wk postinfection, most granulomas contain a necrotic center, which is also the location where the bacteria are predominantly present. Most granulomas form a fibrotic and/or cellular cuff, which separates them from the surrounding tissue at this time point (Swaim et al. 2006; Parikka et al. 2012; Ramakrishnan 2012).
As in human TB, maximal control of M. marinum infection in zebrafish is dependent on an intact adaptive immune system (Swaim et al. 2006; Parikka et al. 2012; Ramakrishnan 2012). Because of the lack of immune markers, characterization of the immune response of zebrafish during mycobacterial infection is mainly based on transcriptome and deep sequencing studies (Meijer et al. 2005; Hegedus et al. 2009; Meijer and Spaink 2011; van der Vaart et al. 2012). These studies show a modest but complex host response in the early stages of infection (Meijer et al. 2005; Hegedus et al. 2009). Detailed analysis of immune factors involved in mycobacterial disease depends on the generation of more knockout zebrafish and development of specific antibodies directed against immune cells and chemokines/cytokines.
Pathology in Embryos
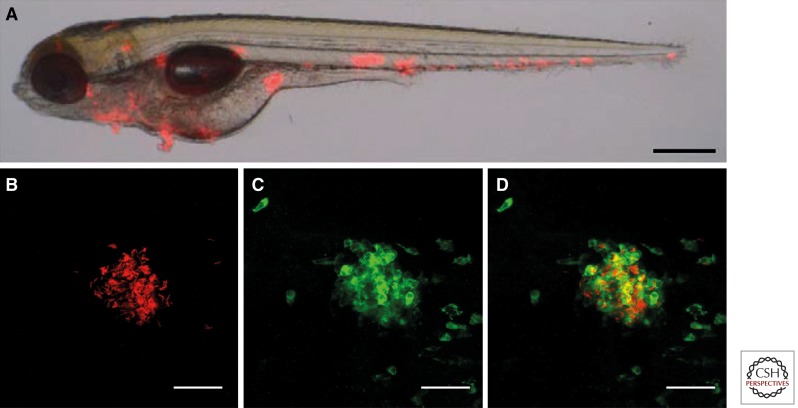
Pathology in zebrafish embryos is, because of practical/ethical reasons, usually only studied for 5–6 d. Within this short time frame early granuloma formation can be studied by real-time imaging (Fig. 4). This allows visualization of early steps in mycobacterial pathogenesis in the context of innate immunity. On infection, M. marinum is readily phagocytosed by macrophages (Lesley and Ramakrishnan 2008; Yang et al. 2012; Ramakrishnan 2013), which traverse endothelial and epithelial barriers and form infectious clusters in deeper tissue within 4 d (Davis et al. 2002; Lesley and Ramakrishnan 2008; Tobin and Ramakrishnan 2008). Once early granulomas form, macrophages adopt a distinctive epithelioid morphology. Within these clusters mycobacteria activate genes that are known to be specifically activated within mature granulomas in adults, confirming that these infectious clusters actually resemble granulomas (Tobin and Ramakrishnan 2008). This means that innate immune determinants are sufficient to drive M. marinum granuloma formation/initiation (Tobin and Ramakrishnan 2008; Meijer and Spaink 2011; Ramakrishnan 2013).
Figure 4.
Pathology in embryos. (A) Merged bright-field and fluorescent image of a zebrafish embryo infected with red fluorescent M. marinum and photographed at 5 dpi. (Adapted with permission from Stoop et al. 2011.) Clustering of mycobacteria and early granuloma formation is shown as red spots. (B–D) Higher magnification of an early granuloma at 5 dpi formed after bloodstream infection, derived from analysis using confocal imaging by our research group. (B) M. marinum E11 (in red), (C) phagocytes stained with anti-L-Plastin (in green), (D) merge of B and C confirming the colocalization of these cells in early granulomas in zebrafish embryos. Scale bar, 35 µm.
LESSONS LEARNED FROM THE ZEBRAFISH INFECTION MODEL
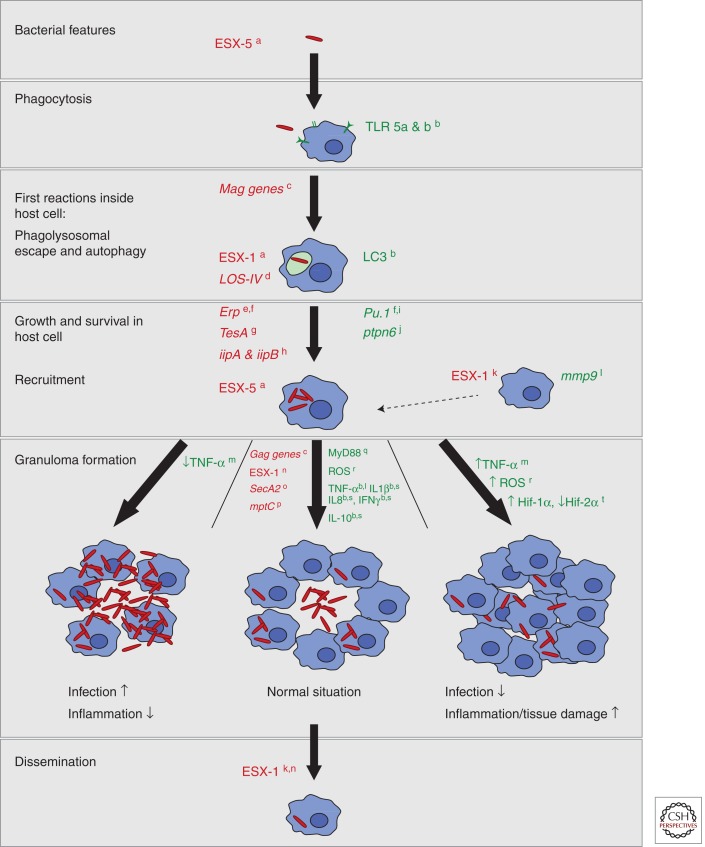
We will discuss a number of bacterial features and host characteristics important during the early steps of mycobacterial infection that have been discovered using the zebrafish model (Fig. 5).
Figure 5.
Graphical summary. This summary shows bacterial and host characteristics important in mycobacterial pathogenesis derived from and validated in the zebrafish infection model. The different hallmarks of pathogenesis are shown. For each step, red shows identified mycobacterial factors important for the pathogen to survive and cope with the immune system of the host, and green shows host factors required for an appropriate immune response. Three types of granuloma are described and schematically depicted in this summary. The granuloma in the middle is the normal granuloma with a balance between inflammation and infection; at the left and right, granulomas without the right balance are depicted with high infection and high inflammation, respectively. Labels in the figure refer to the following references: a, Stoop et al. 2012; b, van der Vaart et al. 2012; c, Davis et al. 2002; d, van der Woude et al. 2012; e, Cosma et al. 2006; f, Meijer and Spaink 2011; g Alibaud et al. 2011; h, Gao et al. 2006; I, Clay et al. 2007; j, Kanwal et al. 2013; k, Davis and Ramakrishnan 2009; l, Volkman et al. 2010; m, Tobin et al. 2010; n, Volkman et al. 2004; o, van der Woude et al. 2013; p, Stoop et al. 2013; q, van der Vaart et al. 2013; r, Roca and Ramakrishnan 2013; s, van der Sar et al. 2009; t, Elks et al. 2013.
Dynamic Granulomas
Classically the granuloma is regarded as a static structure “walling off” bacteria from the rest of the body and therefore critical for host protection (Ulrichs and Kaufmann 2006; Rubin 2009; Schaaf and Zumla 2009). This idea was changed upon observation of the early stages of granuloma formation in zebrafish embryos, which revealed the dynamics of this process (Ramakrishnan 2012). Elegant studies with photobleaching of distinctive clusters in zebrafish embryos and reinfection experiments showed that infected macrophages can detach from the established granuloma and wander off to new locations to form secondary granulomas, thereby disseminating M. marinum (Lesley and Ramakrishnan 2008; Ramakrishnan 2013). Furthermore, macrophages attracted to existing granulomas consume damaged/apoptotic infected cells and their bacterial content in the center of the granuloma, leading to expansion of the early aggregate. These experiments revealed two things: (1) Granuloma formation might actually aid bacterial proliferation, because accelerated bacterial proliferation coincides with granuloma formation (Lesley and Ramakrishnan 2008); and (2) early granulomas are not fixed in size and location. Subsequently, TB studies in mice and nonhuman primates further supported the notion that granulomas are actually highly dynamic structures (Egen et al. 2008; Lin et al. 2013; Ramakrishnan 2013).
Genetic Susceptibility to TB
A broad variation in TB susceptibility and differences between individuals is a long-understood concept.
In the search for candidates for host susceptibility, the zebrafish model has contributed by using forward genetic screens. Tobin et al. (2010) used this method to identify mutant zebrafish with increased susceptibility to M. marinum. Genetic analysis of one such mutant showed that the lta4h locus was affected. This locus controls the balance between pro- and anti-inflammatory eicosanoids. Also in humans LTA4H polymorphisms seem to play a role in the control of infection and inflammation during TB (Tobin et al. 2010). This characterization led to the conclusion that inflammation must be balanced, and misbalance can result in either an inadequate inflammatory or tissue-destructive hyperinflammatory state. Additional research (Tobin et al. 2012) showed that therapies directed to a specific profile could favor disease outcome (Berg and Ramakrishnan 2012), highlighting how well the zebrafish model resembles aspects of human TB and how useful this model can be to study features of this disease.
In addition to the lta4h locus, other genes seem to be required to maintain the balance of mycobacterial infection. For instance, ptpn6 morphant embryos, in which the gene is temporarily knocked down, show a hyperinflammation phenotype (Kanwal et al. 2013). The ptpn6 gene is associated with chronic inflammatory disease in human and plays an important role as a negative regulator of the innate immune system, probably by regulating the induction levels of several kinases in TLR signaling (Kanwal et al. 2013).
Complementary Features of the Embryo and Adult Systems
An illustrative example in which virulence patterns showed large differences in the embryo model compared with the adult model is described by Stoop et al. (2013). In this study they examined the effect of a knockout in the mycobacterial mptC gene, which is required for mannan core branching of lipomannan and lipoarabinomannan. This modification has been linked to TLR-2 activation (Nigou et al. 2008). Interestingly, although this mutant is clearly attenuated in embryos, the effect is only minor in the context of the adaptive immune system. The reverse is also possible, as was shown by Weerdenburg et al. (2012). An M. marinum mutant disrupted in ESX-5 secretion was slightly attenuated in embryos, but showed increased virulence in adult zebrafish, characterized by highly increased bacterial loads and early onset of granuloma formation. The molecular basis for this difference has not been identified yet, but seems to be independent of the adaptive immune response, as the hypervirulence phenotype was also observed in zebrafish rag mutants. These studies highlight the importance of studying both the embryo and adult systems.
Mycobacterial Virulence Factors
The zebrafish embryo is an excellent model to study the importance of different mycobacterial virulence factors in different steps of infection. The erp (pirG) gene, coding for a cell wall–associated protein with unknown function, was first identified as required for virulence in M. tuberculosis (Berthet et al. 1998; Cosma et al. 2006). Using microscopic examination of infected zebrafish embryos it could be shown that M. marinum lacking Erp failed to grow and survive upon phagocytosis, an event very early in granuloma pathogenesis (Cosma et al. 2006; Meijer and Spaink 2011). Subsequently, macrophages were eliminated in zebrafish embryos by injection of pu.1 morpholino, thereby knocking down the pu.1 transcription factor, which is required for myeloid development (Meijer and Spaink 2011). Now, growth of the erp mutant was restored, indicating that in vivo attenuation was specifically linked to defective growth inside macrophages (Lesley and Ramakrishnan 2008).
A number of studies have used different setups to identify M. marinum virulence factors, most of which seem to underscore the similarities between M. marinum and M. tuberculosis. The most elaborate screen was performed by Stoop et al. (Stoop et al. 2011, 2013; van der Woude et al. 2013), who screened in total 1000 random transposon mutants for early granuloma formation and virulence. With nearly half of the highly attenuated mutants, the most prominent virulence locus identified in these experiments was esx-1. This is not entirely surprising, as the esx-1 locus is probably the most extensively studied virulence locus in pathogenic mycobacteria. The esx-1 locus is coding for components of a protein secretion system and its substrates, and although the actual mechanism is still not completely resolved, the most compelling data suggests that ESX-1 effector proteins are required for phagolysosomal escape (Stamm et al. 2003; Houben et al. 2012). In addition to the phagosome escape phenotype, macrophage recruitment and dissemination of disease (Volkman et al. 2004; Davis and Ramakrishnan 2009; Stoop et al. 2011) have also been attributed to the ESX-1 system, although these effects could be indirect because esx-1-deficient M. marinum does not reach its normal location within the phagocytosing cell. Importantly, phagosomal escape of pathogenic mycobacteria was first convincingly shown for M. marinum (Stamm et al. 2003) and only later for M. tuberculosis, underscoring the importance of this model.
In conclusion, the combination of real-time imaging and high-throughput settings seem ideally suited to screen for bacterial factors involved in the establishment of a successful infection.
Using Zebrafish to Identify New Antimycobacterial Compounds
The search for new antimicrobial compounds or therapies can be accelerated using the zebrafish model. Activity and dosage of antimycobacterial compounds in zebrafish closely resemble characteristics in humans (Adams et al. 2011). In addition, the zebrafish model has helped to challenge the model that persistence is linked to arrested growth (Adams et al. 2011; Philips and Ernst 2011). Using the zebrafish model, it was shown, by spatial monitoring of the behavior of fluorescent bacteria after treatment with antibiotics, that both macrophages and granulomas play a role in the induction and dissemination of drug-tolerant bacteria. The intramacrophage-mediated oxidative stress induces the expression of bacterial efflux pumps in actively replicating bacteria. It was also shown that bacterial efflux pump inhibitors (e.g., verapamil) can be added to the standard antibiotic treatment to reduce macrophage-induced drug tolerance and possibly shorten treatment (Adams et al. 2011; Philips and Ernst 2011; Berg and Ramakrishnan 2012; Zumla et al. 2013).
Another example of using zebrafish embryos in the identification of new antimycobacterial drugs is a recent study by Makarov, who produced and analyzed a new generation of benzathiozinones (Makarov et al. 2014). These compounds bind DprE1 and thereby selectively inhibit the biosynthesis of crucial cell wall components. The most effective second-generation compound (i.e., PBTZ169) was compared with the first-generation lead compound in a zebrafish embryo infection model. Although both compounds reduced bacterial load in zebrafish embryos, this model showed an important difference in toxicity, whereas the original compound led to developmental abnormalities, like deposits in the notochord and subsequent shortening of the anteroposterior axes, and PBTZ169 did not. These examples show that effectiveness and toxicity of antimycobacterial compounds can be assessed accurately using zebrafish embryos.
CONCLUDING REMARKS
The use of zebrafish larvae for studying microbial infection has led to important new insights in host defense mechanisms, which often appear to be common for higher vertebrates (Table 1). However, we still need to extend our comparison of zebrafish model with the mammalian systems to show the translational value for biomedical applications. The rapid increase of available high-throughput technologies in the zebrafish toolbox, such as advances in robotic injection and automated readouts of zebrafish embryos (Spaink et al. 2013), will lead to new approaches for TB research. In addition, new reporter lines of zebrafish that provide readouts for activation of the immune system are highly useful tools for even better in vivo visualization of mycobacterial infections (Kanther and Rawls 2010; Palha et al. 2013). What we still need are specific antibodies for distinguishing immune cell types and technologies for generating cell-specific and conditional knockout mutants.
Table 1.
Top five advantages of the zebrafish model in mycobacterial research
| 1 | Fast model, small animal, ease of breeding, ease of genetic manipulation |
| 2 | Transparency and availability of transgenic zebrafish lines make real-time imaging possible |
| 3 | Innate and adaptive immunity can be studied separately |
| 4 | Mycobacterium marinum is strongly related to Mycobacterium tuberculosis and causes granulomatous disease in zebrafish with shared characteristics to human granulomatous disease |
| 5 | Screens possible for (i) mycobacterial virulence factors; (ii) host factors; (iii) therapeutic compounds, like antibiotics |
Zebrafish provide an excellent opportunity to address questions that are difficult to solve in mammalian systems. In return, discoveries in zebrafish must be confirmed in mammalian systems to maximize their translational impact.
Footnotes
Editors: Stefan H.E. Kaufmann, Eric J. Rubin, and Alimuddin Zumla
Additional Perspectives on Tuberculosis available at www.perspectivesinmedicine.org
REFERENCES
- Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L 2011. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145: 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibaud L, Rombouts Y, Trivelli X, Burguiere A, Cirillo SL, Cirillo JD, Dubremetz JF, Guerardel Y, Lutfalla G, Kremer L 2011. A Mycobacterium marinum TesA mutant defective for major cell wall-associated lipids is highly attenuated in Dictyostelium discoideum and zebrafish embryos. Mol Microbiol 80: 919–934. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Hopkins N 2006. Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet 22: 473–478. [DOI] [PubMed] [Google Scholar]
- Balla KM, Lugo-Villarino G, Spitsbergen JM, Stachura DL, Hu Y, Banuelos K, Romo-Fewell O, Aroian RV, Traver D 2010. Eosinophils in the zebrafish: Prospective isolation, characterization, and eosinophilia induction by helminth determinants. Blood 116: 3944–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard EL, van der Sar AM, Ellett F, Lieschke GJ, Spaink HP, Meijer AH 2012. Infection of zebrafish embryos with intracellular bacterial pathogens. J Vis Exp 61: e3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RD, Ramakrishnan L 2012. Insights into tuberculosis from the zebrafish model. Trends Mol Med 18: 689–690. [DOI] [PubMed] [Google Scholar]
- Berthet FX, Lagranderie M, Gounon P, Laurent-Winter C, Ensergueix D, Chavarot P, Thouron F, Maranghi E, Pelicic V, Portnoi D, et al. 1998. Attenuation of virulence by disruption of the Mycobacterium tuberculosis erp gene. Science 282: 759–762. [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D 2007. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development 134: 4147–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn PR, Campbell JM, Clark KJ, Ekker SC 2013. The CRISPR system—Keeping zebrafish gene targeting fresh. Zebrafish 10: 116–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittijn SA, Duivesteijn SJ, Belmamoune M, Bertens LF, Bitter W, de Bruijn JD, Champagne DL, Cuppen E, Flik G, Vandenbroucke-Grauls CM, et al. 2009. Zebrafish development and regeneration: New tools for biomedical research. Int J Dev Biol 53: 835–850. [DOI] [PubMed] [Google Scholar]
- Clay H, Davis JM, Beery D, Huttenlocher A, Lyons SE, Ramakrishnan L 2007. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe 2: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma CL, Klein K, Kim R, Beery D, Ramakrishnan L 2006. Mycobacterium marinum Erp is a virulence determinant required for cell wall integrity and intracellular survival. Infect Immun 74: 3125–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Hohman VS, Sacher F, Ota T, Willett CE, Steiner LA 2004. T cells and the thymus in developing zebrafish. Dev Comp Immunol 28: 755–767. [DOI] [PubMed] [Google Scholar]
- Danilova N, Bussmann J, Jekosch K, Steiner LA 2005. The immunoglobulin heavy-chain locus in zebrafish: Identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol 6: 295–302. [DOI] [PubMed] [Google Scholar]
- Davis JM, Ramakrishnan L 2009. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L 2002. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17: 693–702. [DOI] [PubMed] [Google Scholar]
- de Jong JL, Zon LI 2012. Histocompatibility and hematopoietic transplantation in the zebrafish. Adv Hematol 2012: 282318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JL, Burns CE, Chen AT, Pugach E, Mayhall EA, Smith AC, Feldman HA, Zhou Y, Zon LI 2011. Characterization of immune-matched hematopoietic transplantation in zebrafish. Blood 117: 4234–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson JT, Seibert J, Teh EM, Da’as S, Fraser RB, Paw BH, Lin TJ, Berman JN 2008. Carboxypeptidase A5 identifies a novel mast cell lineage in the zebrafish providing new insight into mast cell fate determination. Blood 112: 2969–2972. [DOI] [PubMed] [Google Scholar]
- Egen JG, Rothfuchs AG, Feng CG, Winter N, Sher A, Germain RN 2008. Macrophage and T cell dynamics during the development and disintegration of mycobacterial granulomas. Immunity 28: 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elks PM, Brizee S, van der Vaart M, Walmsley SR, van Eeden FJ, Renshaw SA, Meijer AH 2013. Hypoxia inducible factor signaling modulates susceptibility to mycobacterial infection via a nitric oxide dependent mechanism. PLoS Pathog 9: e1003789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao LY, Groger R, Cox JS, Beverley SM, Lawson EH, Brown EJ 2003. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect Immun 71: 922–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao LY, Pak M, Kish R, Kajihara K, Brown EJ 2006. A mycobacterial operon essential for virulence in vivo and invasion and intracellular persistence in macrophages. Infect Immun 74: 1757–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald DJ, Eisen JS 2002. Headwaters of the zebrafish—Emergence of a new model vertebrate. Nat Rev Genet 3: 717–724. [DOI] [PubMed] [Google Scholar]
- Harriff MJ, Bermudez LE, Kent ML 2007. Experimental exposure of zebrafish, Danio rerio (Hamilton), to Mycobacterium marinum and Mycobacterium peregrinum reveals the gastrointestinal tract as the primary route of infection: A potential model for environmental mycobacterial infection. J Fish Dis 30: 587–600. [DOI] [PubMed] [Google Scholar]
- Hegedus Z, Zakrzewska A, Agoston VC, Ordas A, Racz P, Mink M, Spaink HP, Meijer AH 2009. Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol Immunol 46: 2918–2930. [DOI] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C 1999. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126: 3735–3745. [DOI] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C 2001. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol 238: 274–288. [DOI] [PubMed] [Google Scholar]
- Hernandez-Pando R, Marquina-Castillo B, Barrios-Payan J, Mata-Espinosa D 2012. Use of mouse models to study the variability in virulence associated with specific genotypic lineages of Mycobacterium tuberculosis. Infect Genet Evol 12: 725–731. [DOI] [PubMed] [Google Scholar]
- Houben D, Demangel C, van Ingen J, Perez J, Baldeon L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, et al. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol 14: 1287–1298. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Rawls JF 2010. Host-microbe interactions in the developing zebrafish. Curr Opin Immunol 22: 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwal Z, Zakrzewska A, den Hertog J, Spaink HP, Schaaf MJ, Meijer AH 2013. Deficiency in hematopoietic phosphatase ptpn6/Shp1 hyperactivates the innate immune system and impairs control of bacterial infections in zebrafish embryos. J Immunol 190: 1631–1645. [DOI] [PubMed] [Google Scholar]
- Kent ML, Watral V, Wu M, Bermudez LE 2006. In vivo and in vitro growth of Mycobacterium marinum at homoeothermic temperatures. FEMS Microbiol Lett 257: 69–75. [DOI] [PubMed] [Google Scholar]
- Koch R 1884. Die Ätiologie der Tuberkulose, Mitteilungen aus dem Kaiserl. Gesundheitsamte 2: 1–88. [Google Scholar]
- Laing KJ, Hansen JD 2011. Fish T cells: Recent advances through genomics. Dev Comp Immunol 35: 1282–1295. [DOI] [PubMed] [Google Scholar]
- Lam SH, Chua HL, Gong Z, Wen Z, Lam TJ, Sin YM 2002. Morphologic transformation of the thymus in developing zebrafish. Dev Dyn 225: 87–94. [DOI] [PubMed] [Google Scholar]
- Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM 2004. Development and maturation of the immune system in zebrafish, Danio rerio: A gene expression profiling, in situ hybridization and immunological study. Dev Comp Immunol 28: 9–28. [DOI] [PubMed] [Google Scholar]
- Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, Mordelet E, Zapata A, Shinomiya H, Herbomel P 2008. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood 111: 132–141. [DOI] [PubMed] [Google Scholar]
- Lesley R, Ramakrishnan L 2008. Insights into early mycobacterial pathogenesis from the zebrafish. Curr Opin Microbiol 11: 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PL, Coleman T, Carney JP, Lopresti BJ, Tomko J, Fillmore D, Dartois V, Scanga C, Frye LJ, Janssen C, et al. 2013. Radiologic responses in cynomolgous macaques for assessing tuberculosis chemotherapy regimens. Antimicrob Agents Chemother 10.1128/AAC.00277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo-Villarino G, Balla KM, Stachura DL, Banuelos K, Werneck MB, Traver D 2010. Identification of dendritic antigen-presenting cells in the zebrafish. Proc Natl Acad Sci 107: 15850–15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor RR 1995. Cutaneous tuberculosis. Clin Dermatol 13: 245–255. [DOI] [PubMed] [Google Scholar]
- Makarov V, Lechartier B, Zhang M, Neres J, van der Sar AM, Raadsen SA, Hartkoorn RC, Ryabova OB, Vocat A, Decosterd LA, et al. 2014. Towards a new combination therapy for tuberculosis with next generation benzothiazinones. EMBO Mol Med 6: 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker ND, Trede NS 2008. Immunology and zebrafish: Spawning new models of human disease. Dev Comp Immunol 32: 745–757. [DOI] [PubMed] [Google Scholar]
- Meeker ND, Smith AC, Frazer JK, Bradley DF, Rudner LA, Love C, Trede NS 2010. Characterization of the zebrafish T cell receptor β locus. Immunogenetics 62: 23–29. [DOI] [PubMed] [Google Scholar]
- Meijer AH, Spaink HP 2011. Host-pathogen interactions made transparent with the zebrafish model. Curr Drug Targets 12: 1000–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AH, Verbeek FJ, Salas-Vidal E, Corredor-Adamez M, Bussman J, van der Sar AM, Otto GW, Geisler R, Spaink HP 2005. Transcriptome profiling of adult zebrafish at the late stage of chronic tuberculosis due to Mycobacterium marinum infection. Mol Immunology 42: 1185–1203. [DOI] [PubMed] [Google Scholar]
- Nigou J, Vasselon T, Ray A, Constant P, Gilleron M, Besra GS, Sutcliffe I, Tiraby G, Puzo G 2008. Mannan chain length controls lipoglycans signaling via and binding to TLR2. J Immunol 180: 6696–6702. [DOI] [PubMed] [Google Scholar]
- North RJ, Jung YJ 2004. Immunity to tuberculosis. Annu Rev Immunol 22: 599–623. [DOI] [PubMed] [Google Scholar]
- Novoa B, Figueras A 2012. Zebrafish: Model for the study of inflammation and the innate immune response to infectious diseases. Adv Exp Med Biol 946: 253–275. [DOI] [PubMed] [Google Scholar]
- Oksanen KE, Halfpenny NJ, Sherwood E, Harjula SK, Hammaren MM, Ahava MJ, Pajula ET, Lahtinen MJ, Parikka M, Ramet M 2013. An adult zebrafish model for preclinical tuberculosis vaccine development. Vaccine 31: 5202–5209. [DOI] [PubMed] [Google Scholar]
- Ostland VE, Watral V, Whipps CM, Austin FW, St-Hilaire S, Westerman ME, Kent ML 2008. Biochemical, molecular, and virulence characteristics of select Mycobacterium marinum isolates in hybrid striped bass Morone chrysops × M. saxatilis and zebrafish Danio rerio. Dis Aquat Organ 79: 107–118. [DOI] [PubMed] [Google Scholar]
- Page DM, Wittamer V, Bertrand JY, Lewis KL, Pratt DN, Delgado N, Schale SE, McGue C, Jacobsen BH, Doty A, et al. 2013. An evolutionarily conserved program of B-cell development and activation in zebrafish. Blood 122: e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palha N, Guivel-Benhassine F, Briolat V, Lutfalla G, Sourisseau M, Ellett F, Wang CH, Lieschke GJ, Herbomel P, Schwartz O, et al. 2013. Real-time whole-body visualization of Chikungunya Virus infection and host interferon response in zebrafish. PLoS Pathog 9: e1003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikka M, Hammaren MM, Harjula SK, Halfpenny NJ, Oksanen KE, Lahtinen MJ, Pajula ET, Iivanainen A, Pesu M, Ramet M 2012. Mycobacterium marinum causes a latent infection that can be reactivated by γ irradiation in adult zebrafish. PLoS Pathog 8: e1002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TS, Ferguson JA, Watral VG, Mutoji KN, Ennis DG, Kent ML 2013. Paramecium caudatum enhances transmission and infectivity of Mycobacterium marinum and M. chelonae in zebrafish Danio rerio. Dis Aquat Organ 106: 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips JA, Ernst JD 2011. Directly observing therapy: A new view of drug tolerance in tuberculosis. Cell 145: 13–14. [DOI] [PubMed] [Google Scholar]
- Pozos TC, Ramakrishnan L 2004. New models for the study of mycobacterium-host interactions. Curr Opin Immunol 16: 499–505. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan L 2012. Revisiting the role of the granuloma in tuberculosis. Nat Rev Immunol 12: 352–366. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan L 2013. Looking within the zebrafish to understand the tuberculous granuloma. Adv Exp Med Biol 783: 251–266. [DOI] [PubMed] [Google Scholar]
- Renshaw SA, Trede NS 2012. A model 450 million years in the making: Zebrafish and vertebrate immunity. Dis Model Mech 5: 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK 2006. A transgenic zebrafish model of neutrophilic inflammation. Blood 108: 3976–3978. [DOI] [PubMed] [Google Scholar]
- Roca FJ, Ramakrishnan L 2013. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell 153: 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin EJ 2009. The granuloma in tuberculosis—Friend or foe? N Engl J Med 360: 2471–2473. [DOI] [PubMed] [Google Scholar]
- Schaaf HS, Zumla A 2009. Tuberculosis: A comprehensive clinical reference. Saunders, Edinburgh. [Google Scholar]
- Schorpp M, Bialecki M, Diekhoff D, Walderich B, Odenthal J, Maischein HM, Zapata AG, Boehm T 2006. Conserved functions of Ikaros in vertebrate lymphocyte development: Genetic evidence for distinct larval and adult phases of T cell development and two lineages of B cells in zebrafish. J Immunol 177: 2463–2476. [DOI] [PubMed] [Google Scholar]
- Spaink HP, Cui C, Wiweger MI, Jansen HJ, Veneman WJ, Marin-Juez R, de Sonneville J, Ordas A, Torraca V, van der Ent W, et al. 2013. Robotic injection of zebrafish embryos for high-throughput screening in disease models. Methods 62: 246–254. [DOI] [PubMed] [Google Scholar]
- Stamm LM, Morisaki JH, Gao LY, Jeng RL, McDonald KL, Roth R, Takeshita S, Heuser J, Welch MD, Brown EJ 2003. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med 198: 1361–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, et al. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18: 729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoop EJ, Schipper T, Huber SK, Nezhinsky AE, Verbeek FJ, Gurcha SS, Besra GS, Vandenbroucke-Grauls CM, Bitter W, van der Sar AM 2011. Zebrafish embryo screen for mycobacterial genes involved in the initiation of granuloma formation reveals a newly identified ESX-1 component. Dis Model Mech 4: 526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoop EJ, Bitter W, van der Sar AM 2012. Tubercle bacilli rely on a type VII army for pathogenicity. Trend Microbiol 20: 477–484. [DOI] [PubMed] [Google Scholar]
- Stoop EJ, Mishra AK, Driessen NN, van Stempvoort G, Bouchier P, Verboom T, van Leeuwen LM, Sparrius M, Raadsen SA, van Zon M, et al. 2013. Mannan core branching of lipo(arabino)mannan is required for mycobacterial virulence in the context of innate immunity. Cell Microbiol 15: 2093–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaim LE, Connolly LE, Volkman HE, Humbert O, Born DE, Ramakrishnan L 2006. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect Immun 74: 6108–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin DM, Ramakrishnan L 2008. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol 10: 1027–1039. [DOI] [PubMed] [Google Scholar]
- Tobin DM, Vary JC Jr., Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King MC, Hawn TR, et al. 2010. The lta4 h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 140: 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin DM, Roca FJ, Oh SF, McFarland R, Vickery TW, Ray JP, Ko DC, Zou Y, Bang ND, Chau TT, et al. 2012. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell 148: 434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Herbomel P, Patton EE, Murphey RD, Yoder JA, Litman GW, Catic A, Amemiya CT, Zon LI, Trede NS 2003. The zebrafish as a model organism to study development of the immune system. Adv Immunol 81: 253–330. [PubMed] [Google Scholar]
- Travis WD, Travis LB, Roberts GD, Su DW, Weiland LW 1985. The histopathologic spectrum in Mycobacterium marinum infection. Arch Pathol Lab Med 109: 1109–1113. [PubMed] [Google Scholar]
- Trede NS, Langenau DM, Traver D, Look AT, Zon LI 2004. The use of zebrafish to understand immunity. Immunity 20: 367–379. [DOI] [PubMed] [Google Scholar]
- Ulrichs T, Kaufmann SH 2006. New insights into the function of granulomas in human tuberculosis. J Pathol 208: 261–269. [DOI] [PubMed] [Google Scholar]
- van der Sar AM, Abdallah AM, Sparrius M, Reinders E, Vandenbroucke-Grauls CM, Bitter W 2004a. Mycobacterium marinum strains can be divided into two distinct types based on genetic diversity and virulence. Infect Immun 72: 6306–6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sar AM, Appelmelk BJ, Vandenbroucke-Grauls CM, Bitter W 2004b. A star with stripes: Zebrafish as an infection model. Trends Microbiol 12: 451–457. [DOI] [PubMed] [Google Scholar]
- van der Sar AM, Spaink HP, Zakrzewska A, Bitter W, Meijer AH 2009. Specificity of the zebrafish host transcriptome response to acute and chronic mycobacterial infection and the role of innate and adaptive immune components. Mol Immunol 46: 2317–2332. [DOI] [PubMed] [Google Scholar]
- van der Vaart M, Spaink HP, Meijer AH 2012. Pathogen recognition and activation of the innate immune response in zebrafish. Adv Hematol 2012: 159807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart M, van Soest JJ, Spaink HP, Meijer AH 2013. Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Dis Model Mech 6: 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude AD, Sarkar D, Bhatt A, Sparrius M, Raadsen SA, Boon L, Geurtsen J, van der Sar AM, Luirink EN, et al. 2012. Unexpected link between lipooligosaccharide biosynthesis and surface protein release in Mycobacterium marinum. J Biol Chem 287: 20417–20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude AD, Stoop EJ, Stiess M, Wang S, Ummels R, van Stempvoort G, Piersma SR, Cascioferro A, Jimenez CR, Houben EN, et al. 2013. Analysis of SecA2-dependent substrates in Mycobacterium marinum identifies protein kinase G (PknG) as a virulence effector. Cell Microbiol 16: 280–295. [DOI] [PubMed] [Google Scholar]
- Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L 2004. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol 2: e367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman HE, Pozos TC, Zheng J, Davis JM, Rawls JF, Ramakrishnan L 2010. Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science 327: 466–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watral V, Kent ML 2007. Pathogenesis of mycobacterium spp. in zebrafish (Danio rerio) from research facilities. Comp Biochem Physiol C Toxicol Pharmacol 145: 55–60. [DOI] [PubMed] [Google Scholar]
- Weerdenburg EM, Abdallah AM, Mitra S, de Punder K, van der Wel NN, Bird S, Appelmelk BJ, Bitter W, van der Sar AM 2012. ESX-5-deficient Mycobacterium marinum is hypervirulent in adult zebrafish. Cell Microbiol 14: 728–739. [DOI] [PubMed] [Google Scholar]
- Yang CT, Cambier CJ, Davis JM, Hall CJ, Crosier PS, Ramakrishnan L 2012. Neutrophils exert protection in the early tuberculous granuloma by oxidative killing of mycobacteria phagocytosed from infected macrophages. Cell Host Microbe 12: 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA 2009. Form, function and phylogenetics of NITRs in bony fish. Dev Comp Immunol 33: 135–144. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Turner PM, Wright PD, Wittamer V, Bertrand JY, Traver D, Litman GW 2010. Developmental and tissue-specific expression of NITRs. Immunogenetics 62: 117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A, Nahid P, Cole ST 2013. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov 12: 388–404. [DOI] [PubMed] [Google Scholar]