Abstract
Homologous recombination provides high-fidelity DNA repair throughout all domains of life. Live cell fluorescence microscopy offers the opportunity to image individual recombination events in real time providing insight into the in vivo biochemistry of the involved proteins and DNA molecules as well as the cellular organization of the process of homologous recombination. Herein we review the cell biological aspects of mitotic homologous recombination with a focus on Saccharomyces cerevisiae and mammalian cells, but will also draw on findings from other experimental systems. Key topics of this review include the stoichiometry and dynamics of recombination complexes in vivo, the choreography of assembly and disassembly of recombination proteins at sites of DNA damage, the mobilization of damaged DNA during homology search, and the functional compartmentalization of the nucleus with respect to capacity of homologous recombination.
Homologous recombination is responsible for high-fidelity DNA repair. Technical advances in protein tagging and microscopy are providing insight into the in vivo biochemistry and cellular organization of this process.
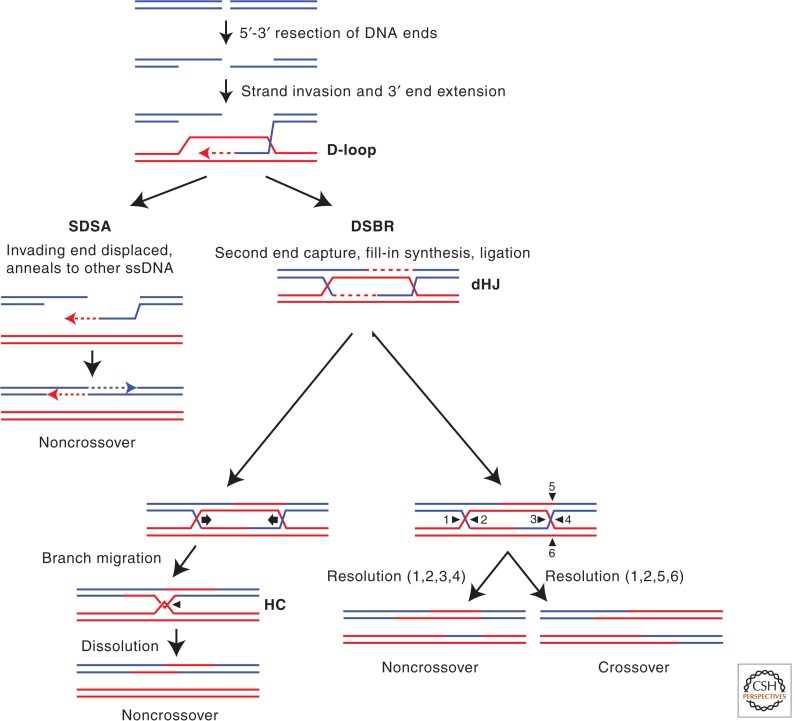
Homologous recombination (HR) is defined as the homology-directed exchange of genetic information between two DNA molecules (Fig. 1). Mitotic recombination is often initiated by single-stranded DNA (ssDNA), which can arise by several avenues (Mehta and Haber 2014). They include the processing of DNA double-strand breaks by 5′ to 3′ resection, during replication of damaged DNA, or during excision repair (Symington 2014). The ssDNA is bound by replication protein A (RPA) to control its accessibility to the Rad51 recombinase (Sung 1994, 1997a; Sugiyama et al. 1997; Morrical 2014). The barrier to Rad51-catalyzed recombination imposed by RPA can be overcome by a number of mediators, such as BRCA2 and Rad52, which serve to replace RPA with Rad51 on ssDNA, and the Rad51 paralogs Rad55-Rad57 (RAD51B-RAD51C-XRCC2-XRCC3) and the Psy3-Csm2-Shu1-Shu2 complex (SHU) (RAD51D-XRCC2-SWS1), which stabilize Rad51 filaments on ssDNA (see Table 1 for homologs of yeast and human HR proteins) (Sung 1997b; Sigurdsson et al. 2001; Martin et al. 2006; Bernstein et al. 2011; Liu et al. 2011; Qing et al. 2011; Amunugama et al. 2013; Zelensky et al. 2014). The Rad51 nucleoprotein filament catalyzes the invasion into a homologous duplex to produce a displacement loop (D-loop) (Fig. 1). At this stage, additional antirecombination functions are exerted by Srs2 (FBH1, PARI), which dissociates Rad51 filaments from ssDNA, and Mph1 (FANCM), which disassembles D-loops (see Daley et al. 2014). Upon Rad51-catalyzed strand invasion, the ATP-dependent DNA translocase Rad54 enables the invading 3′ end to be extended by DNA polymerases to copy genetic information from the intact duplex (Li and Heyer 2009). Ligation of the products often leads to joint molecules (JMs), such as single- or double-Holliday junctions (s/dHJs) or hemicatenanes (HCs), which must be processed to allow separation of the sister chromatids during mitosis. JMs can be dissolved by the Sgs1-Top3-Rmi1 complex (STR) (BTR, BLM-TOP3α-RMI1-RMI2) (see Bizard and Hickson 2014) or resolved by structure-selective nucleases, such as Mus81-Mms4 (MUS81-EME1), Slx1-Slx4, and Yen1 (GEN1) (see Wyatt and West 2014). Mitotic cells favor recombination events that lead to noncrossover events likely to avoid potentially detrimental consequences of loss of heterozygosity and translocations.
Figure 1.
Primary pathways for homology-dependent double-strand break (DSB) repair. Recombinational repair of a DSB is initiated by 5′ to 3′ resection of the DNA end(s). The resulting 3′ single-stranded end(s) invade an intact homologous duplex (in red) to prime DNA synthesis. For DSBs that are repaired by the classical double-strand break repair (DSBR) model, the displaced strand from the donor duplex pairs with the 3′ single-stranded DNA (ssDNA) tail at the other side of the break, which primes a second round of DNA synthesis. After ligation of the newly synthesized DNA to the resected 5′ strands, a double-Holliday junction (dHJ) intermediate is generated. The dHJ can be either dissolved by branch migration (indicated by arrows) into a hemicatenane (HC) leading to noncrossover (NCO) products or resolved by endonucleolytic cleavage (indicated by triangles) to produce NCO (positions 1, 2, 3, and 4) or CO (positions 1, 2, 5, and 6) products. Alternatively to the double-strand break repair (DSBR) pathway, the invading strand is often displaced after limited synthesis and the nascent complementary strand anneals with the 3′ single-stranded tail of the other end of the DSB. After fill-in synthesis and ligation, this pathway generates NCO products and is referred to as synthesis-dependent strand annealing (SDSA).
Table 1.
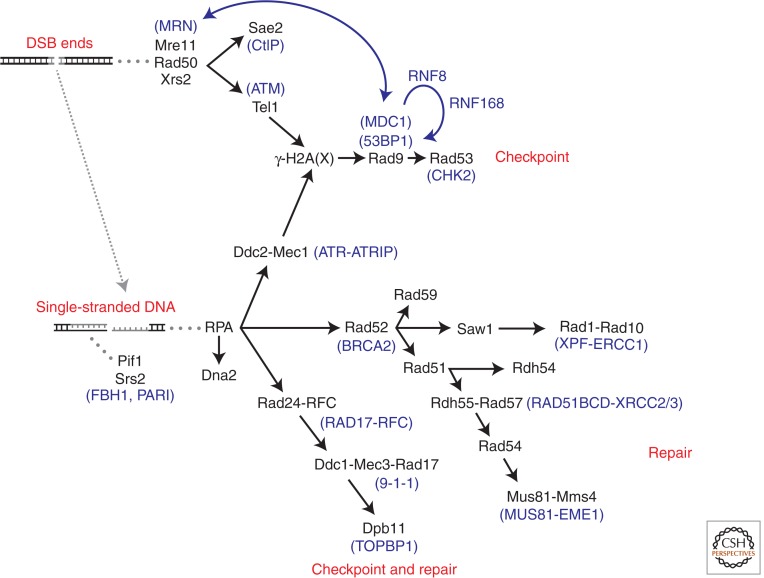
Evolutionary conservation of homologous recombination proteins between Saccharomyces cerevisiae and Homo sapiens
| Functional class | S. cerevisiae | H. sapiens |
|---|---|---|
| End resection | Mre11-Rad50-Xrs2 | MRE11-RAD50-NBS1 |
| Sae2 | CtIP | |
| Exo1 | EXO1 | |
| Dna2-Sgs1-Top3-Rmi1 | DNA2-BLM-TOP3α-RMI1-RMI2 | |
| Adaptors | Rad9 | 53BP1, MDC1 |
| – | BRCA1 | |
| Checkpoint signaling | Tel1 | ATM |
| Mec1-Ddc2 | ATR-ATRIP | |
| Rad53 | CHK2 | |
| Rad24-RFC | RAD17-RFC | |
| Ddc1-Mec3-Rad17 | RAD9-HUS1-RAD1 | |
| Dpb11 | TOPBP1 | |
| Single-stranded DNA binding | Rfa1-Rfa2-Rfa3 | RPA1-RPA2-RPA3 |
| Single-strand annealing | Rad52 | RAD52 |
| Rad59 | – | |
| Mediators | – | BRCA2-PALB2 |
| Rad52 | – | |
| Strand exchange | Rad51 | RAD51 |
| Rad54 | RAD54A, RAD54B | |
| Rdh54 | – | |
| Rad51 paralogs | Rad55-Rad57 | RAD51B-RAD51C-RAD51D-XRCC2-XRCC3 |
| Psy3-Csm2-Shu1-Shu2 | RAD51D-XRCC2-SWS1 | |
| Antirecombinases | Srs2 | FBH1, PARI |
| Mph1 | FANCM | |
| – | RTEL | |
| Resolvases and nucleases | Mus81-Mms4 | MUS81-EME1 |
| Slx1-Slx4 | SLX1-SLX4 | |
| Yen1 | GEN1 | |
| Rad1-Rad10 | XPF-ERCC1 | |
| Dissolution | Sgs1-Top3-Rmi1 | BLM-TOP3α-RMI1-RMI2 |
The vast majority of cell biological studies of mitotic recombination in living cells are performed by tagging of proteins with genetically encoded green fluorescent protein (GFP) or similar molecules (Shaner et al. 2005; Silva et al. 2012). In this context, it is important to keep in mind that an estimated 13% of yeast proteins are functionally compromised by GFP tagging (Huh et al. 2003). By choosing fluorophores with specific photochemical properties, it has been possible to infer biochemical properties, such as diffusion rates, protein–protein interactions, protein turnover, and stoichiometry of protein complexes at the single-cell level. To visualize the location of specific loci within the nucleus, sequence-specific DNA-binding proteins such the Lac and Tet repressors have been used with great success. Specifically, tandem arrays of 100–300 copies of repressor binding sites are inserted within 10–20 kb of the locus of interest in cells expressing the GFP-tagged repressor (Straight et al. 1996; Michaelis et al. 1997). In wild-type budding yeast, such protein-bound arrays are overcome by the replication fork without a cell-cycle delay or checkpoint activation (Dubarry et al. 2011). However, the arrays are unstable in rrm3Δ and other mutants (Dubarry et al. 2011). More pronounced DNA replication blockage by artificial protein-bound DNA tandem arrays has be observed in fission yeast, which is accompanied by increased recombination and formation of DNA anaphase bridges (Sofueva et al. 2011). Likewise, an array of Lac repressor binding sites was reported to induce chromosomal fragility in mouse cells (Jacome and Fernandez-Capetillo 2011). However, these repressor-bound arrays generally appear as a focus with a size smaller than the diffraction limit of light, which is in the range 150–300 nm for wide-field light microscopy.
RECOMBINATION FOCI
In response to DNA damage in eukaryotes, most HR proteins relocalize to nuclear foci of high local concentration at the site of the DNA lesion (Fig. 2) (Lisby et al. 2001, 2003b, 2004; Bekker-Jensen et al. 2006). For example, a single DNA double-strand break (DSB) is sufficient for the formation of a prominent focus containing 600–2100 molecules of Rad52 out of approximately 2300 molecules per haploid cell, yielding a ≥50-fold higher local concentration of Rad52 at the DSB relative to its diffuse nuclear distribution in undamaged cells (Lisby et al. 2003b). Although the minimum number of Rad52 molecules required for mediating a single recombination event is currently unknown (see below), the high local concentration of recombination proteins within these foci may allow constitutively expressed proteins to be active only at the site of DNA damage, and therefore prevent untimely recombination or assembly of recombination complexes at undamaged DNA.
Figure 2.
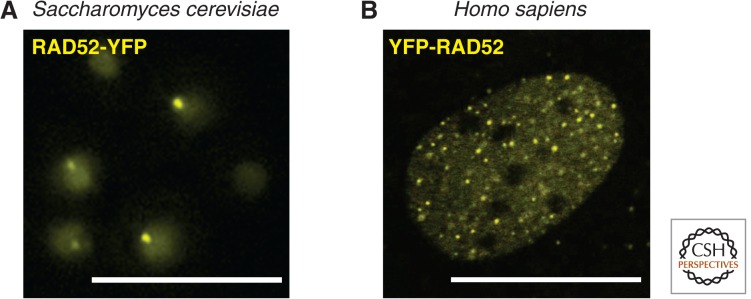
Rad52 foci in yeast and human cells. (A) Rad52 foci in S. cerevisiae. Cells expressing Rad52-YFP (strain W5094-1C) were exposed to 200 µg/mL of zeocin for 1 h at 25°C. (B) RAD52 foci in human cells. The U2OS cell line–expressing YFP-RAD52 were imaged 1 h after exposure to 4 Gy of ionizing radiation, which is equivalent to roughly 50–100 double-strand breaks (DSBs), depending on the phase of the cell cycle. Scale bars, 10 µm. (Image from Bekker-Jensen et al. 2006; reproduced, with permission, from The Rockefeller University Press under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License © 2006.)
Proteins in recombination foci are highly dynamic, and foci can assemble and disassemble within minutes (Altmannova et al. 2010). The dynamic behavior of recombination foci likely reflects a series of opposing processes, such as Rad52 promoting assembly of Rad51 filaments and Srs2 promoting their disassembly (Boundy-Mills and Livingston 1993; Milne and Weaver 1993; Kaytor et al. 1995; Sung 1997a; New et al. 1998; Shinohara and Ogawa 1998; Burgess et al. 2009). Additional dynamics is likely provided by posttranslational modifications (PTMs), which are continuously applied and erased during repair. Furthermore, studies of Rad51, Rad52, and Rad54 foci in mammalian cells indicate that the residence time of individual molecules may vary between proteins and even subpopulations of proteins within foci (Essers et al. 2002). So far, the dynamics of proteins within individual recombination foci has not been studied in yeast.
CHOREOGRAPHY OF FOCUS FORMATION
The order of assembly of recombination factors at foci during recombinational repair is most extensively studied in the context of DSBs, which will be the topic of this section. Key steps of this assembly process are evolutionarily conserved from yeast to mammals. Hence, the section will focus on yeast and highlight some of the additional complexity and differences reported for other experimental systems (Fig. 3). For an extensive review of DSB-induced foci in mammalian cells, see Thompson (2012). In budding yeast, the Mre11-Rad50-Xrs2 complex (MRX) is the first recombination factor recruited to a DSB, likely by associating directly with DNA ends (Chen et al. 2001; Hopfner et al. 2001; Lisby et al. 2004). The Ku (Yku70-Yku80) complex of the nonhomologous end-joining (NHEJ) pathway can delay recruitment of MRX to DSBs (Lisby et al. 2004). To oppose this effect, it appears that MRX actively promotes displacement of Ku to initiate HR (Wu et al. 2008; Mimitou and Symington 2010). Likely, the competition between Ku and MRX for binding to DSBs is important for determining the choice between NHEJ and HR during DSB repair. Moreover, because MRX is also important for NHEJ (Schiestl et al. 1994; Johzuka and Ogawa 1995; Moore and Haber 1996; Tsukamoto et al. 1996), it is possible that Ku and MRX collaborate to direct the repair of DSBs. In mammalian cells, DNA-PKcs provides an additional barrier to HR (Zhu et al. 2011), and even in G2 phase, an estimated 80% of ionizing radiation (IR)-induced DSBs are repaired by NHEJ (Beucher et al. 2009; Shibata et al. 2011).
Figure 3.
Choreography of homologous recombination (HR) foci in yeast and human cells. Order of recruitment of double-strand break (DSB) repair proteins. Nucleation of proteins at foci start at the upper left. Solid black lines represent absolute requirements (see text). Gray dotted lines represent interactions with specific DNA substrates. Arrows indicate protein–protein interactions, modifying functions, and/or feedback loops. Mammalian homologs are indicated in blue.
At the cytological level, the two ends of a DSB are tethered by a mechanism that is partially dependent on MRX and Sae2 (Chen et al. 2001; Lisby et al. 2003a; Kaye et al. 2004; Lobachev et al. 2004; Clerici et al. 2005). As a consequence, the two ends of a DSB give rise to a single Mre11 focus rather than two foci. MRX (MRN) and Sae2 (CtIP) physically interact and collaborate in the initial 5′ to 3′ nucleolytic resection of DSB ends (Mimitou and Symington 2009; Yuan and Chen 2009; Ghodke and Muniyappa 2013), which commits the cell to the HR pathway of repair. The Tel1 (ATM) kinase is recruited to DSBs at all phases of the cell cycle via an interaction with Xrs2 (NBS1) (Nakada et al. 2003; Lisby et al. 2004; Falck et al. 2005; You et al. 2005). The Tel1 kinase phosphorylates oligomeric Sae2 to promote its transition to active monomers/dimers, which allows the protein to be recruited to IR-induced foci (Fu et al. 2014). However, Sae2 also forms spontaneous foci, which are independent of MRX, Tel1, and Mec1 (Lisby et al. 2004; Fu et al. 2014). Similarly, in human cells, IR-induced CtIP foci colocalize with MRE11 and NBS1, but these foci of CtIP and MRN appear to form independently (Chen et al. 2008). Further, Tel1 (ATM) phosphorylates histone H2A (H2AX), which is a chromatin mark specific for damaged DNA in most eukaryotes (Rogakou et al. 1998, 1999; Redon et al. 2003; Stiff et al. 2004). Importantly, the modification of chromatin by H2A phosphorylation facilitates binding of the checkpoint adaptor Rad9 to sites of DNA damage likely through a dual interaction of its BRCA1 carboxy-terminal (BRCT) domains with histone H2A phosphorylated at serine 129 (H2A-S129P), and its Tudor domain with histone H3 methylated at lysine 79 (H3-K79Me) leading to subsequent recruitment and activation of Rad53 (Giannattasio et al. 2005; Javaheri et al. 2006; Toh et al. 2006; Grenon et al. 2007; Hammet et al. 2007; Germann et al. 2011). Although the H2A-S129P mark is induced by DNA damage, the H3-K79Me mark is constitutive and thought to be exposed upon damage (Conde et al. 2009). Notably, Rad53 foci are Rad9-dependent, faint, and short-lived, which is consistent with the notion from mammalian cells that CHK2 (Rad53) must redistribute from the site of DNA damage upon phosphorylation to mediate a pan-nuclear checkpoint response (Lukas et al. 2003). Apparently, yeast Rad9 has diverged into MDC1 and 53BP1 in mammalian cells, where MDC1 is recruited to foci via an interaction with γ-H2AX (Stucki et al. 2005) and 53BP1 binds to H3-K79Me (Huyen et al. 2004) and H4-K20diMe (Botuyan et al. 2006). Recruitment of 53BP1 to foci additionally requires its interaction with MDC1 (Eliezer et al. 2009) and with ubiquitylated H2A(X) (Fradet-Turcotte et al. 2013), which is brought about by phosphorylated MDC1 mediating recruitment of the RNF8-RNF168 ubiquitin ligases (Huen et al. 2007; Mailand et al. 2007; Doil et al. 2009; Stewart et al. 2009; Pinato et al. 2011; Mattiroli et al. 2012; Oestergaard et al. 2012; Delgado-Diaz et al. 2014).
The initial short-range resection of DNA ends by MRX-Sae2 is followed by long-range resection by Exo1 and Dna2-STR (DNA2-BTR) (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008; Cejka et al. 2010; Niu et al. 2010; Nimonkar et al. 2011; Yan et al. 2011; Symington 2014). Interestingly, the proteins involved in short- and long-range resection have distinct focal appearances giving clues to their function and regulation. Mre11 and Sae2 form prominent nuclear foci at all phases of the cell cycle in response to DSBs (Lisby et al. 2004; Barlow et al. 2008). In contrast, Dna2 shuttles between the nucleus and cytoplasm in a cell-cycle-dependent manner, residing in the cytoplasm during G1 phase and relocalizing to the nucleus in S/G2 upon phosphorylation by cyclin-dependent kinase Cdc28 (Kosugi et al. 2009; Chen et al. 2011). Dna2 forms Rad52-colocalizing foci after DSB formation (Zhu et al. 2008). Sgs1 is a low abundance nuclear protein, which forms foci in S/G2/M (Frei and Gasser 2000; M Wagner, pers. comm.). Exo1 levels are cell-cycle regulated gradually increasing through G1 and peaking in late S/G2 phase before it is degraded in anaphase (M Lisby, unpubl.). Resection is accompanied by the dissociation of MRX, Sae2, and Tel1 from DSB-associated foci and binding of RPA to the 3′ single-stranded overhangs (Alani et al. 1992; Lisby et al. 2004; Barlow et al. 2008). In mammalian cells, MRN is retained at DSBs after resection via an interaction with MDC1 (Goldberg et al. 2003; Stewart et al. 2003; Lukas et al. 2004; Lee et al. 2005; Spycher et al. 2008), which binds γ-H2AX, thereby forming a feedback loop for propagation of this histone mark (Stucki et al. 2005). These observations are consistent with the notion that long-range resection is restricted to S/G2 phases of the cell cycle to allow HR only at this time.
The transition to long-range resection appears to be under tighter control in mammalian cells compared with yeast, which may explain the greater usage of NHEJ for DSB repair in higher eukaryotes. Specifically, 53BP1 and RIF1 inhibit long-range resection (Chapman et al. 2013; Zimmermann et al. 2013). Furthermore, the switch to long-range resection is accompanied by a BRCA1- and POH1-dependent repositioning of 53BP1, RAP80, and K63-linked ubiquitin chains to the periphery of enlarged IR-induced foci (Kakarougkas et al. 2013a), indicating that major reorganization of repair foci takes place to accommodate HR. The intensity of RPA foci can be used to estimate the extent of resection. This approach was used to show at the single-cell level that the rate of DSB end resection increases at the G1–S transition (Barlow et al. 2008). RPA is necessary for recruiting a number of checkpoint and HR proteins including the Dna2, Mec1-Ddc2 (ATR-ATRIP) (Costanzo et al. 2003; Zou and Elledge 2003; Mailand et al. 2007), Rad24-RFC (RAD17-RFC) (Zou et al. 2002, 2003; Majka and Burgers 2003), and Ddc1-Mec3-Rad17 (9-1-1) complexes (Bermudez et al. 2003; Lisby et al. 2004; Chen et al. 2013). Further, the recruitment of Ddc1-Mec3-Rad17 (9-1-1) to foci is dependent on Rad24-RFC (RAD17-RFC) (Lisby et al. 2004; Medhurst et al. 2008). Notably, Tel1 and Mec1 have many of the same phosphorylation targets, including histone H2A. As a consequence, Tel1-dependent checkpoint signaling is likely replaced by Mec1-dependent signaling upon resection of DSB ends. The multifunctional Dpb11 (TOPBP1) protein is recruited to foci by the 9-1-1 complex (Greer et al. 2003; Germann et al. 2011), reflecting its role in mediating the DNA damage checkpoint through activation of the Mec1 (ATR) kinase (Kumagai et al. 2006; Mordes et al. 2008; Puddu et al. 2008). In contrast, the DNA replication and recombination functions of Dpb11 appear to be independent of focus formation (Germann et al. 2011).
In S and G2 phase, RPA facilitates the recruitment of Rad52 to DSBs likely via a direct physical interaction (Hays et al. 1995; Lisby et al. 2001, 2004; Plate et al. 2008). The recruitment is independent of DNA replication and requires Cdc28 activity (Barlow and Rothstein 2009). Interestingly, the cell-cycle regulation of Rad52 focus formation can be circumvented at high doses of IR at which Rad52 also forms foci in G1 phase. However, it is unclear whether these foci are productive for recombination (Lisby et al. 2003a). Rad52 interacts with the Rad51 recombinase and Rad59 to recruit these proteins to foci (Milne and Weaver 1993; Davis and Symington 2003; Lisby et al. 2004). In addition, Rad59 also requires Rad52 for its nuclear localization (Lisby et al. 2004). Despite this requirement, recent data suggest that Rad59 can act independently of Rad52 in genome maintenance (Coic et al. 2008; Pannunzio et al. 2008, 2012).
In mammalian cells, BRCA2 serves as the primary mediator for loading RAD51 onto RPA-coated single-stranded DNA and is required for the formation of IR-induced RAD51 foci (Tarsounas et al. 2003; van Veelen et al. 2005; Badie et al. 2010; Jensen et al. 2010; Liu et al. 2010; Thorslund et al. 2010). RAD51 focus formation is additionally regulated by small ubiquitin-like modifier (SUMO) in both yeast and mammalian cells (Bergink et al. 2013; Shima et al. 2013). It remains to be fully understood how BRCA2 is recruited to foci. However, at least two observations are relevant to this question. First, BRCA2 binds directly to ssDNA (Yang et al. 2002; Jensen et al. 2010; Liu et al. 2010; Thorslund et al. 2010). Second, BRCA2 and PALB2 foci colocalize with BRCA1 foci, and their mutual interdependencies suggest that BRCA2-PALB2 recruitment is initiated by the binding of BRCA1 bound to chromatin in the vicinity of a DSB (Xia et al. 2006). Despite vertebrate RAD52 being dispensable for RAD51 focus formation (van Veelen et al. 2005), the synthetic lethality of rad52 mutants with mutants of PALB2, BRCA2, and the RAD51 paralogs RAD51BCD-XRCC2/3 (Fujimori et al. 2001; Feng et al. 2011; Chun et al. 2013; Lok et al. 2013), indicates a role for RAD52 during HR in higher eukaryotes (Rijkers et al. 1998; Stark et al. 2004).
Together with Rad52, the Rad51 paralogs Rad55-Rad57 promote the formation of Rad51 filaments and themselves require Rad51 for focus formation (Sung 1997b; Lisby et al. 2004). Although the SHU complex—composed of Psy3, Csm2, Shu1, and Shu2—does not relocalize to foci in response to DNA damage in mitotically growing cells, its Csm2 and Psy3 subunits are needed for efficient formation of Rad55 foci (Godin et al. 2013). Similarly, human RAD51B-RAD51C stabilizes the RAD51 nucleoprotein filament (Amunugama et al. 2013), which is consistent with RAD51B-RAD51C providing a mediator function for the assembly of the RAD51-ssDNA nucleoprotein filaments (Sigurdsson et al. 2001). In addition, the Psy3 homolog RAD51D is required for RAD51 focus formation after IR exposure in mammalian cells (Chun et al. 2013). Formation of DNA damage-induced foci of Rad54 requires both Rad55-Rad57 and Rad51, suggesting that Rad54 recruitment to the site of DNA damage requires Rad51 nucleoprotein filament formation (Lisby et al. 2004; van Veelen et al. 2005). Interestingly, the yeast Rad54 homolog, Rdh54, is recruited both to DSBs and to the kinetochore, although the functional significance of this dual localization is unknown. Analysis of Rad51 foci suggests that Rad54 promotes the disassembly of Rad51 from damaged DNA, while Rdh54 disassembles Rad51 from undamaged DNA (Shah et al. 2010). The recruitment of Rdh54 to DSBs is Rad52- and Rad51-dependent, whereas its localization to the kinetochore is independent of the recombination machinery (Lisby et al. 2004). Interestingly, Rad54, which does not localize to the kinetochore in wild-type cells, localizes to the kinetochore in an rdh54Δ mutant (Lisby et al. 2004), possibly explaining some of the functional redundancy between these two proteins (Klein 1997; Shinohara et al. 1997). Redundancy is also observed between the RAD54A and RAD54B paralogs in mammalian cells (Tanaka et al. 2000; Wesoly et al. 2006). Furthermore, Rad52 interacts with and recruits Saw1 to foci (Li et al. 2008; Diamante et al. 2014), which in turn facilitates the recruitment of the Rad1-Rad10 endonuclease (Moore et al. 2009). The recruitment of Saw1-Rad1-Rad10 may aid the trimming of recombination structures to promote synthesis-dependent strand annealing (SDSA) (Diamante et al. 2014), a feature that appears to be conserved in mammals (Motycka et al. 2004).
Additional proteins that are recruited to recombination foci include the Pif1 helicase, which forms Rad52-colocalizing foci (Wagner et al. 2006; Pinter et al. 2008), and the Srs2 (FBH1, PARI) helicase and antirecombinase (Krejci et al. 2003; Veaute et al. 2003; Simandlova et al. 2013), which is recruited to two distinct classes of foci (Osman et al. 2005; Burgess et al. 2009; Fugger et al. 2009). During S phase, Srs2 is recruited to sumoylated proliferating cell nuclear antigen (PCNA) speckles, also known as replication foci, and in late S/G2, Srs2 is recruited to recombination foci marked by Rad52. The recruitment of Srs2 to recombination foci is largely independent of its SUMO-interacting motif (Burgess et al. 2009). The Mus81-Mms4 (MUS81-EME1) structure-selective endonuclease forms foci (Nomura et al. 2007; Matulova et al. 2009), which are largely dependent on Rad54, consistent with Mus81-Mms4 acting downstream from the strand-invasion step of HR and a direct physical interaction between Mus81 and Rad54 (Interthal and Heyer 2000; Matulova et al. 2009). The SUMO-targeted ubiquitin ligase Slx5-Slx8 forms foci that partially overlap with Rad52 and Rad9 foci in response to DNA damage (Cook et al. 2009), and likely reflect the extensive sumoylation of repair proteins at a DSB (Cremona et al. 2012; Psakhye and Jentsch 2012).
RECOMBINATION CENTERS
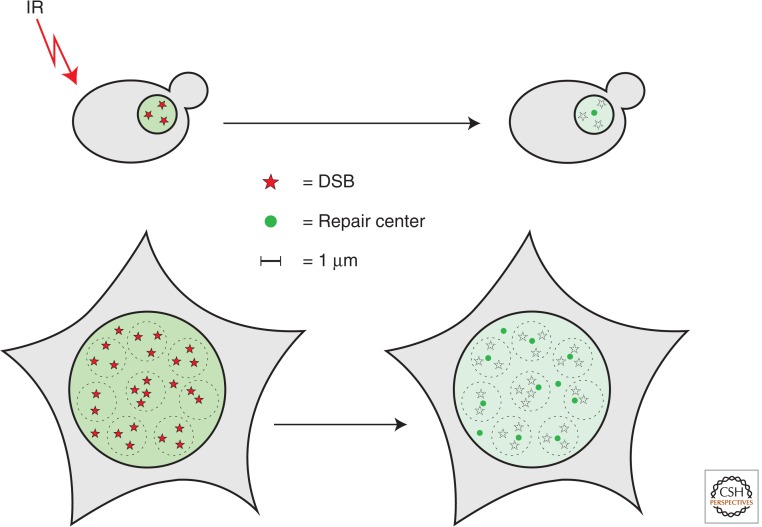
Cells that experience multiple DSBs have the ability to recruit these lesions to the same repair focus (Fig. 4) (Lisby et al. 2003b; Aten et al. 2004; Neumaier et al. 2012). These recombinational repair centers form in both haploid and diploid yeast cells within 20–30 min of γ irradiation as indicated by the observation that irradiated cells initially show many Mre11 foci within 5 min, which transitions to 1–2 Rad52 foci within 20–30 min (M Lisby, unpubl.). In yeast cells, the aggregation of DSBs is global, whereas it appears to be confined to a volume of 1–2 µm in diameter within the nucleus of mammalian cells, although the mobility of DSBs in mammalian cells seems to vary with cell type and experimental conditions (Aten et al. 2004; Soutoglou et al. 2007; Dimitrova et al. 2008; Jakob et al. 2009; Neumaier et al. 2012; Roukos et al. 2013). Likely, the aggregation of multiple DSBs is the result of PTMs, such as sumoylation and phosphorylation acting as molecular glue to regulate protein–protein interactions at repair foci (Cremona et al. 2012; Psakhye and Jentsch 2012) and/or could be the consequence of the attempt to tether DNA ends. Formation of recombination centers shows a partial dependency on Sae2 and the MRX (MRN) complex for tethering ends (Chen et al. 2001; Lisby et al. 2003a; Kaye et al. 2004; Lobachev et al. 2004; Clerici et al. 2005; Roukos et al. 2013), indicating that DSB end resection is likely a prerequisite for holding ends together.
Figure 4.
Formation of repair centers in yeast and human cells. The mobility of double-strand breaks (DSBs) induced by ionizing radiation (IR) is confined within the nucleus to a volume having a diameter of 1–2 µm. Because the yeast nucleus is approximately 2 µm in diameter, most DSBs except centromeric and telomeric are essentially free to roam the nucleus and coalesce into repair centers that can be visualize by green fluorescent protein (GFP) tagging of late-acting nuclear recombination proteins (green). The same principle of spatial confinement applies to subregions of mammalian nuclei indicated by dashed circles 2 µm in diameter. Yeast and mammalian cells are illustrated approximately at the same scale.
LIMITATIONS TO FOCIOLOGY
The study of focus formation (fociology) of recombination proteins has proven a useful tool for analyzing the cellular response to DSBs. However, even if a single DSB is sufficient to trigger focus formation of key recombination proteins (Lisby et al. 2003b), it is likely that some recombination events go undetected by this methodology. For example, recombinational restart of stalled replication forks, some sister chromatid events, and intramolecular recombination may be too fast or require too few molecules of recombination proteins to be detected by current techniques. For example, some proteins (e.g., the SHU complex in mitotic cells) regulate the recruitment of proteins to foci without themselves being detectable at foci (Bernstein et al. 2011). It is worth noting that, in many inducible systems, both sister chromatids are broken, thereby making the DSBs more difficult to repair and allowing more time for proteins to accumulate into foci. Similarly, in mammalian cells, DNA damage-induced protein relocalization after laser microirradiation may be the result of the repair of clusters of DNA breaks in a subnuclear region (Bekker-Jensen et al. 2006).
NUCLEAR COMPARTMENTS
Some genomic sequences are more susceptible to deleterious recombination including repetitive elements, such as the centromeres, telomeres, and Ty elements, and highly transcribed genes, such as the ribosomal gene locus (rDNA) and tRNA genes. Genetic instability at these loci is likely prevented by compartmentalization of the nucleus into domains that suppress recombination and domains that allow or even stimulate recombination. One example is the yeast nucleolus from which late-acting recombination proteins, such as RPA, Rad52, Rad51, Rad59, and Rad55 are largely excluded (Torres-Rosell et al. 2007). DSBs in the rDNA are initially recognized by the MRX complex within the nucleolus and resection is initiated (Torres-Rosell et al. 2007), but they are only bound by Rad52 and downstream factors after relocalization of the break to a position outside the nucleolus. The relocalization of rDNA breaks from the nucleolus to the nucleoplasm requires MRX, the Smc5-Smc6 complex, and SUMO modification of Rad52. Mutations that prevent relocalization lead to Rad52 focus formation inside the nucleolus accompanied by rDNA instability (Torres-Rosell et al. 2007). In human cells, the repair of DSBs in the ribosomal genes has not been studied systematically. However, there are clear indications that most DNA damage response proteins are excluded from the nucleolus (Essers et al. 2002; Bekker-Jensen et al. 2006).
Similarly, budding yeast telomeres are compartmentalized into six to eight clusters (Gotta et al. 1996), which are largely refractory to the DNA damage checkpoint response and recombination (Khadaroo et al. 2009; Ribeyre and Shore 2012). Moreover, telomeres are frequently in the vicinity of the nuclear envelope (Hediger et al. 2002). The anchoring at the nuclear envelope and shielding of telomeres against spontaneous recombination requires the SUN domain protein Mps3 (Antoniacci et al. 2007; Bupp et al. 2007; Oza et al. 2009; Schober et al. 2009; reviewed in Taddei and Gasser 2012). Despite the antirecombinogenic effect of telomere anchoring, their localization at the nuclear periphery is essential for efficient DSB repair in subtelomeric DNA (Therizols et al. 2006). In fact, persistent DSBs, collapsed replication forks, and eroded telomeres associate with the nuclear pore complex (NPC), which stimulates recombinational repair in a Nup84- and Slx8-dependent manner (Nagai et al. 2008; Khadaroo et al. 2009). It was proposed that desumoylation of repair proteins by the SUMO-specific protease Ulp1, which associates with the NPC (Takahashi et al. 2000), could be responsible for the observed stimulation of gene conversion (Nagai et al. 2008). Similarly, in fission yeast, an induced DSB associates with SUN protein Sad1 and KASH protein Kms1 in S/G2 phases of the cell cycle, connecting the DSB to cytoplasmic microtubules. Via this association, Kms1 and the cytoplasmic microtubule regulator Mto1 promote DSB repair by gene conversion (Swartz et al. 2014). In human cells, the genome is organized into discrete domains by 1300 lamin-associated regions (Guelen et al. 2008), which have been proposed to facilitate some of the genome organization provided by the NPC in yeast (Therizols et al. 2006), which do not have lamins (reviewed in Gonzalo 2014). Mouse telomeres were shown to bind to lamins and loss of A-type lamins caused a redistribution of telomeres and telomere shortening (Gonzalez-Suarez et al. 2009). A more general role of lamins in the maintenance of genome integrity in higher eukaryotes has been suggested by the increased DNA damage sensitivity and spontaneous γ-H2AX foci associated with mutation of lamins (Liu et al. 2005; Scaffidi and Misteli 2006; di Masi et al. 2008).
Finally, several lines of evidence suggest that DSB repair is differentially regulated in euchromatin and heterochromatin (reviewed in Chiolo et al. 2013) with a preferential repair of heterochromatin DSBs by HR in higher eukaryotes (Shibata et al. 2011; Kakarougkas et al. 2013b). Notably, DNA DSBs in heterochromatin relocalize to euchromatic regions during repair (Jakob et al. 2011), and as a consequence heterochromatic regions often appear to lack radiation-induced γ-H2AX foci (Cowell et al. 2007; Kim et al. 2007; Goodarzi et al. 2008; Vasireddy et al. 2010; Chiolo et al. 2011; Jakob et al. 2011; Lafon-Hughes et al. 2013). The relocalization of heterochromatin DSBs to the periphery of heterochromatic domains is accompanied by a transient ATM-dependent chromatin relaxation (Ziv et al. 2006; Geuting et al. 2013).
DSB DYNAMICS
Early studies suggested that chromosomes move after DNA damage. First, in budding yeast, multiple DSBs accumulate at one or a few repair centers (Lisby et al. 2001), consistent with their movement to a repair center (Lisby et al. 2003b). Also, studies on budding yeast mating type interconversion show DSB-dependent movement of the donor and recipient loci (Houston and Broach 2006; Oza et al. 2009). Evidence from mammalian cell lines after α-particle-induced damage shows that the γ-H2AX foci recruited to the damage coalesce into a few repair centers (Aten et al. 2004). In mouse cells, uncapped telomeres resulting from the ablation of key shelterin component proteins show increased mobility, which is dependent on the checkpoint and repair protein, 53BP1 (Dimitrova et al. 2008). The 53BP1 protein also plays a role during V(D)J recombination stressing the importance of the proper regulation of chromosome mobility (Difilippantonio et al. 2008). Movement of broken chromosomes within the nucleus may facilitate their relocalization from a compartment of low potential for HR to a prorecombination compartment as detailed in the previous section for rDNA DSBs in budding yeast (Torres-Rosell et al. 2007) and heterochromatic sequences in Drosophila (Chiolo et al. 2011). In both cases, relocation depends on the Smc5-Smc6 complex, and in Drosophila EXO1 and BLM are required, suggesting that relocalization of fly heterochromatic DSBs requires end resection.
Experiments on broken chromosomes in budding yeast as well as in mammalian cells have shown that DNA damage elicits a change such that the volume of the nucleus explored by the broken chromosome more than doubles (up to as much as 10 times) from that seen in the absence of DSBs (Dion et al. 2012; Krawczyk et al. 2012; Mine-Hattab and Rothstein 2012). In yeast, the chromosomes were marked by multiple tandem arrays, whereas in mammalian cells, the movement was measured by tracking 53BP1, a protein that binds to broken ends to promote DNA repair. The change in mobility of the broken locus is termed “local” or “cis.” Interestingly, in budding yeast, unbroken chromosomes also increase the volume that they explore after DNA damage, termed “global” or “trans” (Mine-Hattab and Rothstein 2012; Seeber et al. 2013). The increase in both global and local mobility in haploid and diploid budding yeast cells is genetically controlled. For example, both depend on the Rad51 recombinase (Dion et al. 2012; Mine-Hattab and Rothstein 2012). In haploid budding yeast, the increase in local mobility depends on Rad54 as well as two checkpoint proteins, Rad9 and Mec1 (Dion et al. 2012; Seeber et al. 2013). In diploid cells, deletion of SAE2, which likely causes a delayed appearance of ssDNA, also delays the increase in local chromosome mobility (Mine-Hattab and Rothstein 2012). Unlike the increase in mobility seen after IR or enzymatically induced DSBs, spontaneous Rad52 foci are constrained in haploid cells, possibly reflecting recombination between sister chromatids in the context of DNA replication (Dion et al. 2013), which may not require increased mobility. In diploid yeast cells, after a DSB, homologous pairing takes approximately 20 min before the repair center disassembles and the loci separate again (Mine-Hattab and Rothstein 2012). Thus, increased chromosomal mobility likely facilitates the homology search, which is otherwise restricted by the proximity of donor and recipient loci (Goldman and Lichten 1996; Agmon et al. 2013; Roukos et al. 2013).
GENOME-WIDE CELL BIOLOGY SCREENS
Whole genome approaches to study proteins involved in the DNA damage response have come to the fore in recent years. Two common kinds of genome-wide cell biology screens have been used to examine the cellular response to DNA damage. In the first, a single tagged protein is introduced into the entire nonessential S. cerevisiae gene disruption library to examine the effects of individual deletion mutations on the subcellular behavior of the query protein. In the second, a genome-wide library of GFP-tagged yeast proteins is examined for changes in subcellular localization in response to DNA damage treatment.
In 2007, Alvaro et al. published a screen in S. cerevisiae to identify novel factors that impact Rad52 focus formation (Alvaro et al. 2007). The Rad52 protein in S. cerevisiae is important for DSB repair and HR (Mortensen et al. 2009). The genes governing Rad52 focus formation and maintenance were not well known when this screen was initiated. To find new genes, the formation of spontaneous subnuclear Rad52-YFP foci was monitored in the mutant background of over 4800 nonessential gene disruptions using epifluorescence microscopy. To avoid the potential problem of additional recessive traits that can accumulate in individual strains of the haploid yeast gene disruption library, hybrid diploid strains that are homozygous for each deletion were made by using systematic hybrid loss of heterozygosity (SHyLOH [Alvaro et al. 2006]). Image analysis was performed manually and all images were uploaded into the JCB DataViewer, which allows anyone to access and examine the primary data from that screen (Thorpe et al. 2011). In this screen, more than 80 gene disruptions resulted in increased spontaneous Rad52 foci, including mutations in many genes involved in DNA metabolism and cell-cycle regulation. The identified genes also included 22 uncharacterized open reading frames, IRC2–11, 13–16, 18–25 (increased recombination centers), providing new leads to genes that control the cellular response to DNA damage.
In mammalian cell studies, two laboratories used a similar approach to identify genes that regulate the DNA damage response. The Cimprich group assayed, using siRNA knockdown, approximately 21,000 genes in HeLa cells for increased γ-H2AX focus formation (Paulsen et al. 2009). Similarly, the Cortez group performed a shRNA screen that targeted almost 2300 genes preselected for protein domains associated with nuclear regulatory activities (Lovejoy et al. 2009). In that study, HeLa cells were treated both with and without aphidicolin to induce replication stress and were assayed for KAP1 phosphorylation, a substrate of the apical ATM kinase. The hits were further validated in U2OS cells by assaying for γ-H2AX foci. In the Cortez study, many of the genes identified were involved in DNA metabolism and repair. In the Cimprich study, in addition to nucleotide excision repair, telomere maintenance, DNA replication, and anaphase-promoting complex-dependent processes, they found that the largest group of genes affecting γ-H2AX focus formation was in mRNA processing or related pathways. Only a few genes overlap between these screens as well as with the yeast results using Rad52 foci as the reporter.
For the second approach, Brown and colleagues measured global changes in the localization and levels of the S. cerevisiae library of GFP-tagged proteins in response to DNA damage (Tkach et al. 2012). For their experiments, they tested two drug treatments for the induction of different types of DNA damage. They exposed cells to hydroxyurea (HU), a compound that inhibits DNA replication by limiting the pools of deoxyribonucleotide triphosphates (dNTPs). This resulting dNTP starvation creates DNA damage. They also used methyl methanesulfonate (MMS) to methylate guanine and adenine residues, which can lead to DSBs (Ma et al. 2008), a very toxic form of DNA damage. They compared the effects of these two drug treatments for each protein with untreated controls by monitoring changes in protein levels of all images using the freely available Cellprofiler software (Kamentsky et al. 2011). They also determined changes in localization by visually comparing each fluorescent strain. Of the more than 4000 tagged proteins screened, they found 356 proteins whose abundance changed significantly after DNA damage, and 254 that changed their localization. Surprisingly, only 35 of these proteins showed changes in both abundance and localization. Furthermore they found that proteins that share a location are also enriched for physical and genetic interactions, which predict common functions, encouraging even more global studies of proteome relocalization in response to other cellular stresses. One of the surprises of their study was that cytoplasmic P-body components, which are important for mRNA turnover in the cytoplasm, become elevated in HU-treated cells. To explore this finding, they next used a genome-wide GFP microscopy assay (similar to the Alvaro et al. study described above) to assess the location of Lsm1, a central P-body component. They found that Asc1, a G-protein β subunit involved in glucose sensing, is specifically required for P-body formation after HU-induced DNA damage, but not after other cellular stresses (Tkach et al. 2012). Importantly they also found that the DNA damage checkpoint kinases Tel1 and Mec1 (ATM and ATR in mammalian cells) inhibit P-body formation. Another surprise is that some proteins (Cmr1, Hos2, Apj1, and Pph21) form DNA damage-induced foci in the nucleus that do not colocalize with repair foci of Rad52. A genome-wide screen of GFP-tagged proteins has also been conducted in Schizosaccharomyces pombe (Yu et al. 2013), which identified 51 proteins that were able to form a nuclear focus at a homothallic (HO) switching endonuclease-induced DSB. Eight of these proteins were previously uncharacterized open reading frames (ORFs). All in all, these high-throughput, cell biology-based screens for increased spontaneous DNA damage foci show that repair proteins respond to a variety of genome stress conditions and reveal other pathways not usually associated with the DNA damage response.
PERSPECTIVES
Studies on the cell biology of recombination are still in their infancy. We can look forward to a myriad of technical advances in protein tagging as well as in microscopy (e.g., stochastic optical reconstruction microscopy [STORM], structured illumination microscopy [SIM], HiLo, fluorescence resonance energy transfer [FRET], fluorescence redistribution after photobleaching [FRAP], and single-particle tracking) (Fischer et al. 2011; Lim et al. 2011; Ball et al. 2012; Ishikawa-Ankerhold et al. 2012). These advances when combined with genetic studies will allow a more in-depth look at the architecture of foci as well as provide further insight into the regulation of focus assembly and disassembly. Expansion of genome-wide screens promises to uncover new genes and pathways that impact the cellular response to DNA damage. In the future, more multiple mutant analyses will be undertaken to define the epistasis groups involved. It will be especially important to uncover the genes and regulatory circuitry involved in controlling DNA repair dynamics. Because most live cell–imaging studies only look at a single DNA end, it is important to visualize both ends of a DSB to understand how the homology search is coordinated. Furthermore, DSBs are not the only lesions leading to HR and there is a need to analyze the repair of single-strand nicks and gaps. In addition, methods need to be developed to visualize the underlying changes to the recombining DNA molecules. Another challenge for the future will be to understand which nuclear components are responsible for chromosome territories as well as those that confine diffusion and the mobility of chromosomes. Finally, we can look forward to new insights into the cellular response of recombination processes, especially as they relate to oncogenesis, aging, and other human health issues.
ACKNOWLEDGMENTS
We thank Craig Peterson, Alberto Ciccio, Rune T. Pedersen, Sonia Silva, Eric Bryant, and Michael Smith for insightful comments on the manuscript. We are grateful to the following funding agencies for financial support: Danish Council for Independent Research, the Villum Kann Rasmussen Foundation, and the European Research Council (ERCStG, No. 242905) to M.L., and National Institutes of Health (GM50237 and GM67055) to R.R.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Agmon N, Liefshitz B, Zimmer C, Fabre E, Kupiec M 2013. Effect of nuclear architecture on the efficiency of double-strand break repair. Nat Cell Biol 15: 694–699. [DOI] [PubMed] [Google Scholar]
- Alani E, Thresher R, Griffith JD, Kolodner RD 1992. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol 227: 54–71. [DOI] [PubMed] [Google Scholar]
- Altmannova V, Eckert-Boulet N, Arneric M, Kolesar P, Chaloupkova R, Damborsky J, Sung P, Zhao X, Lisby M, Krejci L 2010. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res 38: 4708–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro D, Sunjevaric I, Reid RJ, Lisby M, Stillman DJ, Rothstein R 2006. Systematic hybrid LOH: A new method to reduce false positives and negatives during screening of yeast gene deletion libraries. Yeast 23: 1097–1106. [DOI] [PubMed] [Google Scholar]
- Alvaro D, Lisby M, Rothstein R 2007. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet 3: e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunugama R, Groden J, Fishel R 2013. The HsRAD51B-HsRAD51C stabilizes the HsRAD51 nucleoprotein filament. DNA Repair (Amst) 12: 723–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniacci LM, Kenna MA, Skibbens RV 2007. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle 6: 75–79. [DOI] [PubMed] [Google Scholar]
- Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303: 92–95. [DOI] [PubMed] [Google Scholar]
- Badie S, Escandell JM, Bouwman P, Carlos AR, Thanasoula M, Gallardo MM, Suram A, Jaco I, Benitez J, Herbig U, et al. 2010. BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol 17: 1461–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball G, Parton RM, Hamilton RS, Davis I 2012. A cell biologist’s guide to high resolution imaging. Methods Enzymol 504: 29–55. [DOI] [PubMed] [Google Scholar]
- Barlow JH, Rothstein R 2009. Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J 28: 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R 2008. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell 30: 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Ammon T, Kern M, Schermelleh L, Leonhardt H, Jentsch S 2013. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51–Rad52 interaction. Nat Cell Biol 15: 526–532. [DOI] [PubMed] [Google Scholar]
- Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A 2003. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci 100: 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Reid RJ, Sunjevaric I, Demuth K, Burgess RC, Rothstein R 2011. The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol Biol Cell 22: 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M 2009. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J 28: 3413–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Bizard AH, Hickson ID 2014. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol 6: a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boundy-Mills KL, Livingston DM 1993. A Saccharomyces cerevisiae RAD52 allele expressing a C-terminal truncation protein: Activities and intragenic complementation of missense mutations. Genetics 133: 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol 179: 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Lisby M, Altmannova V, Krejci L, Sung P, Rothstein R 2009. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J Cell Biol 185: 969–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49: 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell 8: 1105–1115. [DOI] [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu X 2008. Cell cycle-dependent complex formation of BRCA1 · CtIP · MRN is important for DNA double-strand break repair. J Biol Chem 283: 7713–7720. [DOI] [PubMed] [Google Scholar]
- Chen X, Niu H, Chung WH, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G 2011. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol 18: 1015–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lisby M, Symington LS 2013. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell 50: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH 2011. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Tang J, Georgescu W, Costes SV 2013. Nuclear dynamics of radiation-induced foci in euchromatin and heterochromatin. Mutat Res 750: 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Buechelmaier ES, Powell SN 2013. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol Cell Biol 33: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP 2005. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem 280: 38631–38638. [DOI] [PubMed] [Google Scholar]
- Coic E, Feldman T, Landman AS, Haber JE 2008. Mechanisms of Rad52-independent spontaneous and UV-induced mitotic recombination in Saccharomyces cerevisiae. Genetics 179: 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde F, Refolio E, Cordon-Preciado V, Cortes-Ledesma F, Aragon L, Aguilera A, San-Segundo PA 2009. The Dot1 histone methyltransferase and the Rad9 checkpoint adaptor contribute to cohesin-dependent double-strand break repair by sister chromatid recombination in Saccharomyces cerevisiae. Genetics 182: 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook CE, Hochstrasser M, Kerscher O 2009. The SUMO-targeted ubiquitin ligase subunit Slx5 resides in nuclear foci and at sites of DNA breaks. Cell Cycle 8: 1080–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell 11: 203–213. [DOI] [PubMed] [Google Scholar]
- Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ 2007. γH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS ONE 2: e1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, Zhao X 2012. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol Cell 45: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Daley JM, Gaines WA, Kwon YH, Sung P 2014. Regulation of DNA pairing in homologous recombination. Cold Spring Harb Perspect Biol 6: a017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Symington LS 2003. The Rad52-Rad59 complex interacts with Rad51 and replication protein A. DNA Repair (Amst) 2: 1127–1134. [DOI] [PubMed] [Google Scholar]
- Delgado-Diaz MR, Martin Y, Berg A, Freire R, Smits VA 2014. Dub3 controls DNA damage signalling by direct deubiquitination of H2AX. Mol Oncol 8: 884–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamante G, Phan C, Celis AS, Krueger J, Kelson EP, Fischhaber PL 2014. SAW1 is required for SDSA double-strand break repair in S. cerevisiae. Biochem Biophys Res Commun 445: 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 2008. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Masi A, D’Apice MR, Ricordy R, Tanzarella C, Novelli G 2008. The R527H mutation in LMNA gene causes an increased sensitivity to ionizing radiation. Cell Cycle 7: 2030–2037. [DOI] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM 2012. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol 14: 502–509. [DOI] [PubMed] [Google Scholar]
- Dion V, Kalck V, Seeber A, Schleker T, Gasser SM 2013. Cohesin and the nucleolus constrain the mobility of spontaneous repair foci. EMBO Rep 14: 984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446. [DOI] [PubMed] [Google Scholar]
- Dubarry M, Loiodice I, Chen CL, Thermes C, Taddei A 2011. Tight protein-DNA interactions favor gene silencing. Genes Dev 25: 1365–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliezer Y, Argaman L, Rhie A, Doherty AJ, Goldberg M 2009. The direct interaction between 53BP1 and MDC1 is required for the recruitment of 53BP1 to sites of damage. J Biol Chem 284: 426–435. [DOI] [PubMed] [Google Scholar]
- Essers J, Houtsmuller AB, van Veelen L, Paulusma C, Nigg AL, Pastink A, Vermeulen W, Hoeijmakers JH, Kanaar R 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J 21: 2030–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611. [DOI] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN 2011. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci 108: 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RS, Wu Y, Kanchanawong P, Shroff H, Waterman CM 2011. Microscopy in 3D: A biologist’s toolbox. Trends Cell Biol 21: 682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499: 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C, Gasser SM 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev 14: 81–96. [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Chow J, Bernstein KA, Makharashvili N, Arora S, Lee CF, Person MD, Rothstein R, Paull TT 2014. Phosphorylation-regulated transitions in an oligomeric state control the activity of the sae2 DNA repair enzyme. Mol Cell Biol 34: 778–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugger K, Mistrik M, Danielsen JR, Dinant C, Falck J, Bartek J, Lukas J, Mailand N 2009. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J Cell Biol 186: 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori A, Tachiiri S, Sonoda E, Thompson LH, Dhar PK, Hiraoka M, Takeda S, Zhang Y, Reth M, Takata M 2001. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J 20: 5513–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germann SM, Oestergaard VH, Haas C, Salis P, Motegi A, Lisby M 2011. Dpb11/TopBP1 plays distinct roles in DNA replication, checkpoint response and homologous recombination. DNA Repair (Amst) 10: 210–224. [DOI] [PubMed] [Google Scholar]
- Geuting V, Reul C, Lobrich M 2013. ATM release at resected double-strand breaks provides heterochromatin reconstitution to facilitate homologous recombination. PLoS Genet 9: e1003667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodke I, Muniyappa K 2013. Processing of DNA double-stranded breaks and intermediates of recombination and repair by Saccharomyces cerevisiae Mre11 and its stimulation by Rad50, Xrs2, and Sae2 proteins. J Biol Chem 288: 11273–11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem 280: 9879–9886. [DOI] [PubMed] [Google Scholar]
- Godin S, Wier A, Kabbinavar F, Bratton-Palmer DS, Ghodke H, Van Houten B, VanDemark AP, Bernstein KA 2013. The Shu complex interacts with Rad51 through the Rad51 paralogues Rad55-Rad57 to mediate error-free recombination. Nucleic Acids Res 41: 4525–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D’Amours D, Rahman D, Pappin D, Bartek J, Jackson SP 2003. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421: 952–956. [DOI] [PubMed] [Google Scholar]
- Goldman AS, Lichten M 1996. The efficiency of meiotic recombination between dispersed sequences in Saccharomyces cerevisiae depends upon their chromosomal location. Genetics 144: 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez I, Redwood AB, Perkins SM, Vermolen B, Lichtensztejin D, Grotsky DA, Morgado-Palacin L, Gapud EJ, Sleckman BP, Sullivan T, et al. 2009. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J 28: 2414–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S 2014. DNA damage and lamins. Adv Exp Med Biol 773: 377–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177. [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol 134: 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer DA, Besley BD, Kennedy KB, Davey S 2003. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res 63: 4829–4835. [PubMed] [Google Scholar]
- Grenon M, Costelloe T, Jimeno S, O’Shaughnessy A, Fitzgerald J, Zgheib O, Degerth L, Lowndes NF 2007. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast 24: 105–119. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. [DOI] [PubMed] [Google Scholar]
- Hammet A, Magill C, Heierhorst J, Jackson SP 2007. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep 8: 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SL, Firmenich AA, Berg P 1995. Complex formation in yeast double-strand break repair: Participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci 92: 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Dubrana K, Gasser SM 2002. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol 140: 79–91. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105: 473–485. [DOI] [PubMed] [Google Scholar]
- Houston PL, Broach JR 2006. The dynamics of homologous pairing during mating type interconversion in budding yeast. PLoS Genet 2: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411. [DOI] [PubMed] [Google Scholar]
- Interthal H, Heyer WD 2000. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet 263: 812–827. [DOI] [PubMed] [Google Scholar]
- Ishikawa-Ankerhold HC, Ankerhold R, Drummen GP 2012. Advanced fluorescence microscopy techniques—FRAP, FLIP, FLAP, FRET and FLIM. Molecules 17: 4047–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacome A, Fernandez-Capetillo O 2011. Lac operator repeats generate a traceable fragile site in mammalian cells. EMBO Rep 12: 1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Durante M, Taucher-Scholz G 2009. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proc Natl Acad Sci 106: 3172–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Conrad S, Voss KO, Zink D, Durante M, Lobrich M, Taucher-Scholz G 2011. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res 39: 6489–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaheri A, Wysocki R, Jobin-Robitaille O, Altaf M, Cote J, Kron SJ 2006. Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc Natl Acad Sci 103: 13771–13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467: 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K, Ogawa H 1995. Interaction of Mre11 and Rad50: Two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139: 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Ismail A, Katsuki Y, Freire R, Shibata A, Jeggo PA 2013a. Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res 41: 10298–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Ismail A, Klement K, Goodarzi AA, Conrad S, Freire R, Shibata A, Lobrich M, Jeggo PA 2013b. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res 41: 9719–9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE 2011. Improved structure, function and compatibility for CellProfiler: Modular high-throughput image analysis software. Bioinformatics 27: 1179–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP 2004. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol 14: 2096–2106. [DOI] [PubMed] [Google Scholar]
- Kaytor MD, Nguyen M, Livingston DM 1995. The complexity of the interaction between RAD52 and SRS2. Genetics 140: 1441–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadaroo B, Teixeira MT, Luciano P, Eckert-Boulet N, Germann SM, Simon MN, Gallina I, Abdallah P, Gilson E, Geli V, et al. 2009. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat Cell Biol 11: 980–987. [DOI] [PubMed] [Google Scholar]
- Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE 2007. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J Cell Biol 178: 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL 1997. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147: 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H 2009. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci 106: 10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk PM, Borovski T, Stap J, Cijsouw T, ten Cate R, Medema JP, Kanaar R, Franken NA, Aten JA 2012. Chromatin mobility is increased at sites of DNA double-strand breaks. J Cell Sci 125: 2127–2133. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG 2006. TopBP1 activates the ATR-ATRIP complex. Cell 124: 943–955. [DOI] [PubMed] [Google Scholar]
- Lafon-Hughes L, Di Tomaso MV, Liddle P, Toledo A, Reyes-Abalos AL, Folle GA 2013. Preferential localization of γH2AX foci in euchromatin of retina rod cells after DNA damage induction. Chromosome Res 21: 789–803. [DOI] [PubMed] [Google Scholar]
- Lee AC, Fernandez-Capetillo O, Pisupati V, Jackson SP, Nussenzweig A 2005. Specific association of mouse MDC1/NFBD1 with NBS1 at sites of DNA-damage. Cell Cycle 4: 177–182. [DOI] [PubMed] [Google Scholar]
- Li X, Heyer WD 2009. RAD54 controls access to the invading 3′-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res 37: 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Dong J, Pan X, Oum JH, Boeke JD, Lee SE 2008. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol Cell 30: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D, Ford TN, Chu KK, Mertz J 2011. Optically sectioned in vivo imaging with speckle illumination HiLo microscopy. J Biomed Opt 16: 016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci 98: 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Antúnez de Mayolo A, Mortensen UH, Rothstein R 2003a. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle 2: 479–483. [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R 2003b. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol 5: 572–577. [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R 2004. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- Liu B, Wang J, Chan KM, Tjia WM, Deng W, Guan X, Huang JD, Li KM, Chau PY, Chen DJ, et al. 2005. Genomic instability in laminopathy-based premature aging. Nat Med 11: 780–785. [DOI] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD 2010. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17: 1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Wan L, Wu Y, Chen J, Huang J 2011. hSWS1·SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J Biol Chem 286: 41758–41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K 2004. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol 14: 2107–2112. [DOI] [PubMed] [Google Scholar]
- Lok BH, Carley AC, Tchang B, Powell SN 2013. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 32: 3552–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Xu X, Bansbach CE, Glick GG, Zhao R, Ye F, Sirbu BM, Titus LC, Shyr Y, Cortez D 2009. Functional genomic screens identify CINP as a genome maintenance protein. Proc Natl Acad Sci 106: 19304–19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5: 255–260. [DOI] [PubMed] [Google Scholar]
- Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J 2004. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J 23: 2674–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Resnick MA, Gordenin DA 2008. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res 36: 1836–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900. [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM 2003. Yeast Rad17/Mec3/Ddc1: A sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci 100: 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Chahwan C, Gao H, Blais V, Wohlschlegel J, Yates JR 3rd, McGowan CH, Russell P 2006. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J 25: 2564–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK 2012. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150: 1182–1195. [DOI] [PubMed] [Google Scholar]
- Matulova P, Marini V, Burgess RC, Sisakova A, Kwon Y, Rothstein R, Sung P, Krejci L 2009. Cooperativity of Mus81·Mms4 with Rad54 in the resolution of recombination and replication intermediates. J Biol Chem 284: 7733–7745. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Medhurst AL, Warmerdam DO, Akerman I, Verwayen EH, Kanaar R, Smits VA, Lakin ND 2008. ATR and Rad17 collaborate in modulating Rad9 localisation at sites of DNA damage. J Cell Sci 121: 3933–3940. [DOI] [PubMed] [Google Scholar]
- *.Mehta A, Haber JE 2014. Sources of DNA double-strand breaks and models for recombinational DNA repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K 1997. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45. [DOI] [PubMed] [Google Scholar]
- Milne GT, Weaver DT 1993. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev 7: 1755–1765. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2009. DNA end resection: Many nucleases make light work. DNA Repair (Amst) 8: 983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2010. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29: 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine-Hattab J, Rothstein R 2012. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol 14: 510–517. [DOI] [PubMed] [Google Scholar]
- Moore JK, Haber JE 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 16: 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DM, Karlin J, Gonzalez-Barrera S, Mardiros A, Lisby M, Doughty A, Gilley J, Rothstein R, Friedberg EC, Fischhaber PL 2009. Rad10 exhibits lesion-dependent genetic requirements for recruitment to DNA double-strand breaks in Saccharomyces cerevisiae. Nucleic Acids Res 37: 6429–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Nam EA, Cortez D 2008. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci 105: 18730–18734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Morrical SW 2014. DNA pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Lisby M, Rothstein R 2009. Rad52. Curr Biol 19: R676–R677. [DOI] [PubMed] [Google Scholar]
- Motycka TA, Bessho T, Post SM, Sung P, Tomkinson AE 2004. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J Biol Chem 279: 13634–13639. [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Matsumoto K, Sugimoto K 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev 17: 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumaier T, Swenson J, Pham C, Polyzos A, Lo AT, Yang P, Dyball J, Asaithamby A, Chen DJ, Bissell MJ, et al. 2012. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proc Natl Acad Sci 109: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391: 407–410. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y, Adachi N, Koyama H 2007. Human Mus81 and FANCB independently contribute to repair of DNA damage during replication. Genes Cells 12: 1111–1122. [DOI] [PubMed] [Google Scholar]
- Oestergaard VH, Pentzold C, Pedersen RT, Iosif S, Alpi A, Bekker-Jensen S, Mailand N, Lisby M 2012. RNF8 and RNF168 but not HERC2 are required for DNA damage-induced ubiquitylation in chicken DT40 cells. DNA Repair (Amst) 11: 892–905. [DOI] [PubMed] [Google Scholar]
- Osman F, Dixon J, Barr AR, Whitby MC 2005. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol 25: 8084–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23: 912–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannunzio NR, Manthey GM, Bailis AM 2008. RAD59 is required for efficient repair of simultaneous double-strand breaks resulting in translocations in Saccharomyces cerevisiae. DNA Repair (Amst) 7: 788–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannunzio NR, Manthey GM, Liddell LC, Fu BX, Roberts CM, Bailis AM 2012. Rad59 regulates association of Rad52 with DNA double-strand breaks. MicrobiologyOpen 1: 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinato S, Gatti M, Scandiuzzi C, Confalonieri S, Penengo L 2011. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol Cell Biol 31: 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter SF, Aubert SD, Zakian VA 2008. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol 28: 6594–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plate I, Hallwyl SC, Shi I, Krejci L, Muller C, Albertsen L, Sung P, Mortensen UH 2008. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J Biol Chem 283: 29077–29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S 2012. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820. [DOI] [PubMed] [Google Scholar]
- Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M 2008. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol 28: 4782–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing Y, Yamazoe M, Hirota K, Dejsuphong D, Sakai W, Yamamoto KN, Bishop DK, Wu X, Takeda S 2011. The epistatic relationship between BRCA2 and the other RAD51 mediators in homologous recombination. PLoS Genet 7: e1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon C, Pilch DR, Rogakou EP, Orr AH, Lowndes NF, Bonner WM 2003. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep 4: 678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeyre C, Shore D 2012. Anticheckpoint pathways at telomeres in yeast. Nat Struct Mol Biol 19: 307–313. [DOI] [PubMed] [Google Scholar]
- Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A 1998. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol 18: 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T 2013. Spatial dynamics of chromosome translocations in living cells. Science 341: 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T 2006. Lamin A-dependent nuclear defects in human aging. Science 312: 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Zhu J, Petes TD 1994. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol 14: 4493–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM 2009. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev 23: 928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber A, Dion V, Gasser SM 2013. Checkpoint kinases and the INO80 nucleosome remodeling complex enhance global chromatin mobility in response to DNA damage. Genes Dev 27: 1999–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Zheng X, Epshtein A, Carey JN, Bishop DK, Klein HL 2010. Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol Cell 39: 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY 2005. A guide to choosing fluorescent proteins. Nat Methods 2: 905–909. [DOI] [PubMed] [Google Scholar]
- Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G, Lobrich M, et al. 2011. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J 30: 1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H, Suzuki H, Sun J, Kono K, Shi L, Kinomura A, Horikoshi Y, Ikura T, Ikura M, Kanaar R, et al. 2013. Activation of the SUMO modification system is required for the accumulation of RAD51 at sites of DNA damage. J Cell Sci 126: 5284–5292. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391: 404–407. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, Shinohara A 1997. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P 2001. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev 15: 3308–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S, Gallina I, Eckert-Boulet N, Lisby M 2012. Live cell microscopy of DNA damage response in Saccharomyces cerevisiae. Methods Mol Biol 920: 433–443. [DOI] [PubMed] [Google Scholar]
- Simandlova J, Zagelbaum J, Payne MJ, Chu WK, Shevelev I, Hanada K, Chatterjee S, Reid DA, Liu Y, Janscak P, et al. 2013. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J Biol Chem 288: 34168–34180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Osman F, Lorenz A, Steinacher R, Castagnetti S, Ledesma J, Whitby MC 2011. Ultrafine anaphase bridges, broken DNA and illegitimate recombination induced by a replication fork barrier. Nucleic Acids Res 39: 6568–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T 2007. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9: 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M 2008. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol 181: 227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M 2004. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol 24: 9305–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434. [DOI] [PubMed] [Google Scholar]
- Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA 2004. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res 64: 2390–2396. [DOI] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC 1997. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem 272: 7940–7945. [DOI] [PubMed] [Google Scholar]
- Sung P 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265: 1241–1243. [DOI] [PubMed] [Google Scholar]
- Sung P 1997a. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem 272: 28194–28197. [DOI] [PubMed] [Google Scholar]
- Sung P 1997b. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev 11: 1111–1121. [DOI] [PubMed] [Google Scholar]
- Swartz RK, Rodriguez EC, King MC 2014. A role for nuclear envelope bridging complexes in homology-directed repair. Mol Biol Cell 25: 2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Symington LS 2014. End resection at DNA double-strand breaks: Mechanism and regulation. Cold Spring Harb Perspect Biol 6: a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Gasser SM 2012. Structure and function in the budding yeast nucleus. Genetics 192: 107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Mizoi J, Toh EA, Kikuchi Y 2000. Yeast Ulp1, an Smt3-specific protease, associates with nucleoporins. J Biochem 128: 723–725. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hiramoto T, Fukuda T, Miyagawa K 2000. A novel human rad54 homologue, Rad54B, associates with Rad51. J Biol Chem 275: 26316–26321. [DOI] [PubMed] [Google Scholar]
- Tarsounas M, Davies D, West SC 2003. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene 22: 1115–1123. [DOI] [PubMed] [Google Scholar]
- Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, Fabre E 2006. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol 172: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH 2012. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: The molecular choreography. Mutat Res 751: 158–246. [DOI] [PubMed] [Google Scholar]
- Thorpe PH, Alvaro D, Lisby M, Rothstein R 2011. Bringing Rad52 foci into focus. J Cell Biol 194: 665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC 2010. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol 17: 1263–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, et al. 2012. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol 14: 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh GW, O’Shaughnessy A M, Jimeno S, Dobbie IM, Grenon M, Maffini S, O’Rorke A, Lowndes NF 2006. Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair (Amst) 5: 693–703. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M 2007. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9: 923–931. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Kato J, Ikeda H 1996. Effects of mutations of RAD50, RAD51, RAD52, and related genes on illegitimate recombination in Saccharomyces cerevisiae. Genetics 142: 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veelen LR, Essers J, van de Rakt MW, Odijk H, Pastink A, Zdzienicka MZ, Paulusma CC, Kanaar R 2005. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat Res 574: 34–49. [DOI] [PubMed] [Google Scholar]
- Vasireddy RS, Karagiannis TC, El-Osta A 2010. γ-Radiation-induced γH2AX formation occurs preferentially in actively transcribing euchromatic loci. Cell Mol Life Sci 67: 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312. [DOI] [PubMed] [Google Scholar]
- Wagner M, Price G, Rothstein R 2006. The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics 174: 555–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesoly J, Agarwal S, Sigurdsson S, Bussen W, Van Komen S, Qin J, van Steeg H, van Benthem J, Wassenaar E, Baarends WM, et al. 2006. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol Cell Biol 26: 976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Topper LM, Wilson TE 2008. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wyatt HDM, West SC 2014. Holliday junction resolvases. Cold Spring Harb Perspect Biol 6: a023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–729. [DOI] [PubMed] [Google Scholar]
- Yan H, Toczylowski T, McCane J, Chen C, Liao S 2011. Replication protein A promotes 5′→3′ end processing during homology-dependent DNA double-strand break repair. J Cell Biol 192: 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297: 1837–1848. [DOI] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 25: 5363–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ren JY, Zhang JM, Suo F, Fang XF, Wu F, Du LL 2013. A proteome-wide visual screen identifies fission yeast proteins localizing to DNA double-strand breaks. DNA Repair (Amst) 12: 433–443. [DOI] [PubMed] [Google Scholar]
- Yuan J, Chen J 2009. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J Biol Chem 284: 31746–31752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zelensky A, Kanaar R, Wyman C 2014. Mediators of homologous DNA pairing. Cold Spring Harb Perspect Biol 6: a016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM 2011. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339: 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 8: 870–876. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev 16: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci 100: 13827–13832. [DOI] [PMC free article] [PubMed] [Google Scholar]