Abstract
Vacuolar type rotary H+-ATPases (VoV1) couple ATP synthesis/hydrolysis by V1 with proton translocation by Vo via rotation of a central rotor apparatus composed of the V1-DF rotor shaft, a socket-like Vo-C (eukaryotic Vo-d) and the hydrophobic rotor ring. Reconstitution experiments using subcomplexes revealed a weak binding affinity of V1-DF to Vo-C despite the fact that torque needs to be transmitted between V1-DF and Vo-C for the tight energy coupling between V1 and Vo. Mutation of a short helix at the tip of V1-DF caused intramolecular uncoupling of VoV1, suggesting that proper fitting of the short helix of V1-D into the socket of Vo-C is required for tight energy coupling between V1 and Vo. To account for the apparently contradictory properties of the interaction between V1-DF and Vo-C (weak binding affinity but strict requirement for torque transmission), we propose a model in which the relationship between V1-DF and Vo-C corresponds to that between a slotted screwdriver and a head of slotted screw. This model is consistent with our previous result in which the central rotor apparatus is not the major factor for the association of V1 with Vo (Kishikawa and Yokoyama, J Biol Chem. 2012 24597-24603).
Introduction
The Vacuole-type ATPases (VoV1) are found in many organisms and are involved in a variety of physiological processes [1–3]. V-ATPases in eukaryotic cells (eukaryotic VoV1) translocate protons across the membrane consuming ATP. Most prokaryotic VoV1 (also referred to as A-ATPase or AoA1 [1, 4]) produce ATP using the energy stored in a transmembrane electrochemical proton gradient [3, 5], while the VoV1 of some anaerobic bacteria, such as Enterococcus hirae, function as a sodium pump [6].
The VoV1 and FoF1 ATPases/synthases are evolutionarily related and share a rotary mechanism to perform their specific functions [3, 7–9]. The basic structures of the ATPases/synthases are conserved among species [1–3]. The soluble, cytoplasmic portion of FoF1 and VoV1 (called F1 and V1, respectively), responsible for ATP hydrolysis/synthesis, is connected via the central rotor stalk and the peripheral stator stalk to the transmembrane portion (Fo and Vo) that houses the ion transporting pathway [1–3]. In Thermus thermophilus VoV1, the V1 portion is composed of a hexameric A3B3 cylinder and a central shaft comprised of the D and F subunits [3, 10, 11]. The Vo portion is composed of 5 different subunits with a stoichiometry of C1E2G2I1L12 (see Fig. 1A). In FoF1, a γ -subunit (equivalent to subunits D and F of VoV1) binds directly to the rotor ring [12]. In contrast, at the boundary surface of VoV1, Vo-C forms a socket-like structure which accommodates the V1-DF central shaft [13], indicating that V1-DF does not contact the rotor ring directly. Thus, the boundary surface of VoV1 is significantly different from that in FoF1. VoV1 also has a more complex peripheral stalk structure than FoF1. The stator structure of FoF1 consists of a single peripheral stalk, while V1 is connected with Vo by two or three peripheral stalks [4, 14, 15].
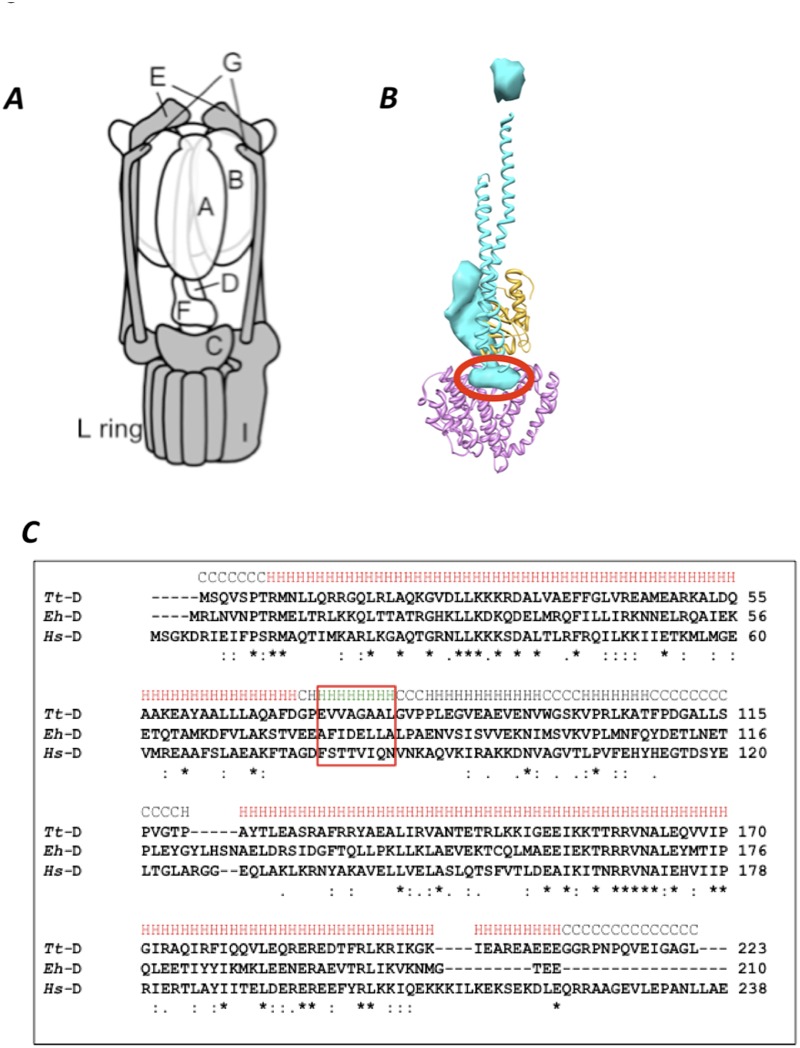
Fig 1. VoV1 and the short helix of V1-D subunit.
A, Schematic representation of T. thermophirus VoV1 [16]. Subunits in Vo and V1 are shown in gray and white, respectively. B, The structure of the central rotor apparatus of VoV1 obtained by EM density map (PDBID; 3J0J) with the short helix in V1-D subunit circled in red. The V1-D, V1-F and Vo-C subunits are represented in blue, yellow, and pink, respectively. C, Sequence alignment of V1-D subunit of T. thermophilus (Tt), E. hirae (Eh) and H. sapiens (Hs). Identical amino acid residues are represented by asterisks. The sequences of the short helix of the V1-D subunit are surrounded by the red box.
Recent reconstitution studies of T. thermophilus VoV1 have demonstrated that the A3B3 domain is tightly associated with the two EG peripheral stalks of Vo, even in the absence of the central shaft subunits [16]. In other words, the peripheral stalks are the major factor mediating association of V1 and Vo, consistent with the unique boundary surface between V1-DF and Vo-C in VoV1. This arrangement is highly relevant for the detachment of V1-DF from Vo-C [13, 16]. However torque needs to be transmitted between V1-DF and Vo-C for tight energy coupling between V1 and Vo [16]. Thus a sticky interaction which also allows detachment of V1-DF from Vo-C is required. How the protein maintains these two somewhat contradictory properties has yet to be investigated.
Lau et al. reported a sub-nanometer resolution structure of T. thermophilus VoV1 by single particle cryo-electron microscopy [11] showing that the rod like structure of V1-DF is positioned in the cavity of Vo-C. This suggests that the rod like structure might play an important role in binding of V1-DF and Vo-C. A crystal structure of V1-DF isolated from E. hirae VoV1 suggested that the rod like structure might be a short helix at the tip of the V1-DF (Fig. 1B, [17]).
In this study, reconstitution and fluorescence resonance energy transfer (FRET) analysis of VoV1 subcomplexes reveal that the binding affinity of V1-DF with Vo-C subunit is weak. Further investigations indicated that the short helix of the V1-DF subunit has important roles in both reconstitution of VoV1 and torque transmission. We propose a structural model accounting for both the detachable and sticky nature of the interaction between V1-DF and Vo-C.
Materials and Methods
Proteins isolation of Vo and CL12
Wild-type or mutant VoV1 (C-T105C/C-C268S/C-C323S) from T. thermophilus strains incorporating a His3 tag on the C terminus of subunit L were generated by the integration vector system [18]. Culture of the modified T. thermophilus strains, membrane preparation, solubilization of His-tagged VoV1 and purification of VoV1, Vo and CL12 were carried out as described previously [19]. The mutated Vo and CL12 (C-T105C/C-C268S/C-C323S) were used for the FRET experiments.
Preparation of V1 (A3B3DF) and V1-DF
Escherichia coli strain BL21-CodonPlus-RP (Stratagene) was used for expression of V1 (A3B3DF) and V1-DF. These recombinant subcomplexes were isolated as described previously [20, 21]. The expression plasmids for V1 containing DF from H. sapiens or E. hirae were constructed by the same method as described in ref. [21]. The genes encoding the D and F subunits were amplified from human cDNA and pCemtp18 [22], containing the complete E. hirae VoV1 (ntp) operon. The D and F genes of the T. thermophilus V1 expression plasmid were replaced with the amplified genes. The expression plasmids for mutated V1 lacking the short helix or containing the swapped short helix of E. hirae were constructed by PCR mutagenesis. To introduce the swapped short helix of E. hirae, complementary oligonucleotide primers containing the sequences encoding the 8 amino acids of this region were used to amplify the gene fragment. The fragment was then digested with appropriate restriction enzymes, and inserted into the corresponding region of the T. thermophilus V1 expression plasmid [20]. The mutant V1 (A-His8/ΔCys, A-C255A/A-S232A/A-T235S, F-S54C) and mutant DF (F- His6, S54C) were used for either reconstitution or FRET experiments [16].
Reconstitution of VoV1
The purity of each subcomplex was confirmed by SDS-PAGE. V1 or V1-DF (each 1 mg/ml) in MOPDM buffer (50 mM MOPS (pH 7.0), 150 mM NaCl, 0.03% n-dodecyl-β-D-maltoside) was mixed with 1 mg/ml Vo or CL12 at an equal volume ratio. The mixtures were incubated for 1 h at 25°C and then applied onto the Superdex HR-200 column equilibrated with the same buffer. The reconstituted VoV1 were collected and used for further analysis immediately.
FRET analysis
FRET analysis was carried out as described previously [16]. The purified V1 (A-His8/ΔCys, A- S232A/A-T235S, F-S54C) or DF (F-His6, S54C) was immediately labeled with an excess amount of Cy3-maleimide (GE healthcare, used as a donor molecule) in MOPDM buffer. Following a 60 min incubation at 25°C, proteins were separated from unbound reagent with a PD-10 column (GE Healthcare). The mutated Vo (C-T105C/C-C268S/C-C323S) was labeled with Cy5-maleimide (GE Healthcare, used as an acceptor molecule) by the same method described above. The specific labeling of subunit F in V1 or DF, and subunit C in Vo or CL12 was checked by measurement of subunit fluorescence. FRET, as a result of reconstitution of VoV1, was monitored with a fluorimeter using an excitation wavelength of 532 nm and an emission wavelength of 570 nm (FP-6200, JASCO). A cuvette was filled with 1.2 ml of MOPDM buffer containing 2 nM labeled V1 or DF and incubated at 25°C until the fluorescence intensity reached a constant level. For measurement of binding kinetics, 8.0 μl of labeled Vo or CL12 was added into the cuvette at a final concentration of 10 nM.
Mesurements of ATP synthesis of the VoV1
The reconstituted complexes were incorporated into liposomes using a freeze-thaw method [23]. Acidification of the proteoliposomes and measurement of ATP synthesis were carried out at 25°C. To acidify the interior of proteoliposomes, 30 μl of the proteoliposome solution was mixed with 15 μl of acidification buffer (300 mM MES pH4.7) and then incubated for 5 min at 25°C. The ATP was measured as the increase of intensity of luminescence in a Luminescencer-PSN (ATTO). The ATP synthesis reaction was initiated by injection of 30 μl of the acidified proteoliposomes into 0.5 ml of base buffer containing 100 mM Tricin-sodium (pH 8.5), 2.5 mM MgSO4, 10 mM phosphate, 2.2 mg of luciferin/luciferase compound, 0.5 mM ADP, 36 nM valinomycin, and 100 mM KCl. For calibration, ATP was injected into the base buffer.
Measurements of proton channel and proton pump activity by the VoV1
Crude soybean L-α-Phosphatidylcholine (Sigma) was washed with 20 mM Tricine-sodium (pH 8.5), 2.5 mM MgSO4 to remove K+ as described [24]. K+-loaded proteoliposomes containing enzyme were prepared as follows; aliquots containing reconstituted complex were diluted to 0.5 mg/ml in 20 mM Tricine, 2.5 mM MgCl2 20 mM MES pH8.0, 4% n-Octyl-β-D-glucoside (Sigma), the washed 20 mg/ml lipid and 150 mM KCl for the measurement of proton pump or 500mM KCl for the measurement of proton channel activity. Bio-beads SM-2 (Bio-Rad) were added to remove the detergent and incubated for 2 h at 25°C. Resultant proteoliposomes were centrifuged and subjected to the proton pump and proton channel analysis.
The acidification of liposomes was measured by the quenching of 9-amino-6-chloro-2-methoxyacridine, (ACMA) (Sigma) fluorescence [25]. Aliquots containing 5 μg of protein were suspended in 1.2 ml of 20 mM Tricine, 2.5 mM MgCl2, 40 mM MES pH8.0, 500 mM NaCl for the measurement of proton channel or 110 mM NaCl /40 mM KCl for the measurement of proton pump activity, in the presence of 15 ng of ACMA. The time course of fluorescence quenching was monitored using a fluorimeter (FP-6200, JASCO).
Other assays
Protein concentrations of V1 were determined from UV absorbance calibrated by quantitative amino acid analysis; 1 mg/ml gives an optical density of 0.88 at 280 nm. Protein concentrations of Vo and VoV1 were determined by BCA protein assay, with BSA used as the protein standard. ATPase activity was measured at 25°C with an enzyme-coupled ATP regenerating system [5]. Polyacrylamide gel electrophoresis in the presence of SDS or AES was carried out as described previously [5]. The proteins were stained with Coomassie Brilliant Blue.
Results
Weak interaction at the boundary surface between V1-DF and CL12
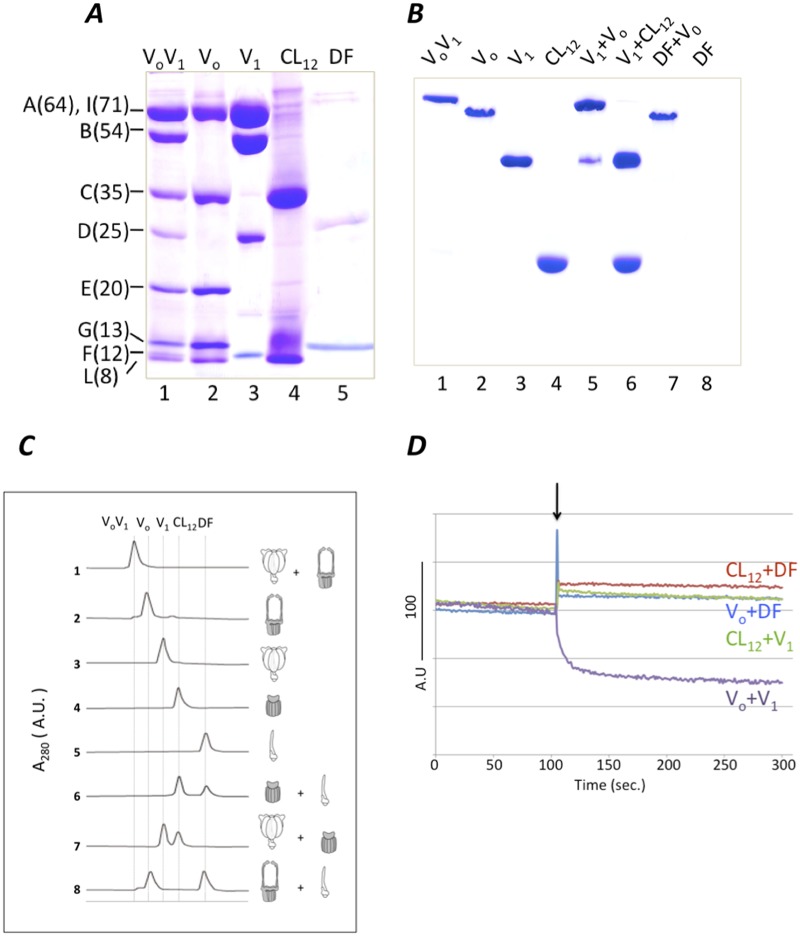
Reconstitution of A3B3 and Vo has indicated that the interaction between A3B3 and two EG peripheral stalks is rigid [16]. However the precise nature of the interaction between V1-DF and Vo-C has not been experimentally characterized. In order to examine the interaction the isolated V1-DF subunits or subcomplexes from V1 with Vo or CL12, were mixed with Vo or CL12. The purity and subunit stoichiometry of all subcomplexes were confirmed by both SDS-PAGE and AES-PAGE (Fig. 2A and B). The band corresponding to V1-DF was not detected on AES-PAGE gel. Reconstituted VoV1 was identified as a single band on AES-PAGE when V1 was incubated with Vo at a molar ratio of 1:1 (Fig. 2B, lane 5). However attempts to reconstitute V1 with CL12 were unsuccessful as assessed by both AES-PAGE and gel permeation analysis (Fig. 2B, lane 6 and Fig. 2C, line 7). The same was also true for V1-DF and CL12 or Vo (Fig. 2B, lane 7 and Fig. 2C, line 6 and 8). Together, these results strongly suggest that the interaction of the boundary surface between V1-DF and Vo-C in the complex is insufficient for reconstitution of a stable complex.
Fig 2. Analysis of reconstitution of complexes and subcomplexes.
A, 15% SDS-PAGE analysis. Lane 1, VoV1; lane 2, Vo; lane 3 V1; lane 4, CL12; lane 5, DF. Molecular weights of each subunit are indicated in parentheses. B, 6% AES-PAGE. The mixtures containing the subcomplexes were incubated for 1h at 25°C respectively, prior to analysis. Lane 1, VoV1; lane 2, Vo; lane 3, V1; lane 4, CL12; lane 5, Vo and V1; lane 6, V1 and CL12; lane 7, DF and Vo, lane 8; DF. The band of DF complex was not detected on AES-PAGE gel. C, Gel permeation analysis of complexes and subcomplexes. The mixtures containing subcomplexes indicated by the scheme were incubated for 1h at 25°C respectively, followed by analysis. The molecular weights of each complex are, VoV1 (659kDa), Vo (268kDa), V1 (391 kDa), CL12 (131 kDa) and V1-DF (37 kDa). D, FRET analysis of reconstituted complexes and subcomplexes. Fluorescence of 3 nM V1-Cy3 (purple and green lines) or 3 nM V1-DF-Cy3 (blue and red lines) was recorded (excitation at 532nm) before and after addition of Vo-Cy5 or CL12-Cy5. The Vo-Cy5 or CL12-Cy5 was added at the time indicated by the arrow.
Further analysis of the interaction at the boundary surface between V1-DF and Vo-C was carried out by fluorescent resonance of energy transfer (FRET), a powerful method for detecting protein-protein interaction/association [16]. For FRET analysis, each component was labeled with Cy3 or Cy5 (fluorescent dyes as described in ref. [16]). A mutated V1-DF or V1 incorporating a single cysteine residue (F/S54C) was labeled with Cy3 as a donor, while a mutated Vo or CL12 (C/T105C) was labeled with Cy5 as an acceptor. Reconstitutions were carried out in a cuvette containing 1.2 ml of the 2 nM V1 or V1-DF labeled with Cy3. The reconstitution efficiency was evaluated by the decrease in donor emission (Fluorescence at 570 nm, ref. [16]). As shown in Fig. 2D, the fluorescence of V1 decreased sharply upon addition of Vo into the cuvette, indicating reconstitution of V1 and Vo complex (purple line). In contrast, addition of 8.0 μl of 1.5 mM of CL12 or Vo into a cuvette containing V1-DF exhibited no decrease in fluorescence at 570 nm (red or blue lines). Furthermore the addition of 8.0 μl of 1.5 mM of CL12 into a cuvette containing V1 showed no decrease in fluorescence (green line). These results clearly indicate low binding affinity between V1-DF and Vo-C, consistent with the findings from the reconstitution experiments [16].
Effect of exogenous V1-DF on reconstitution of V1 and Vo
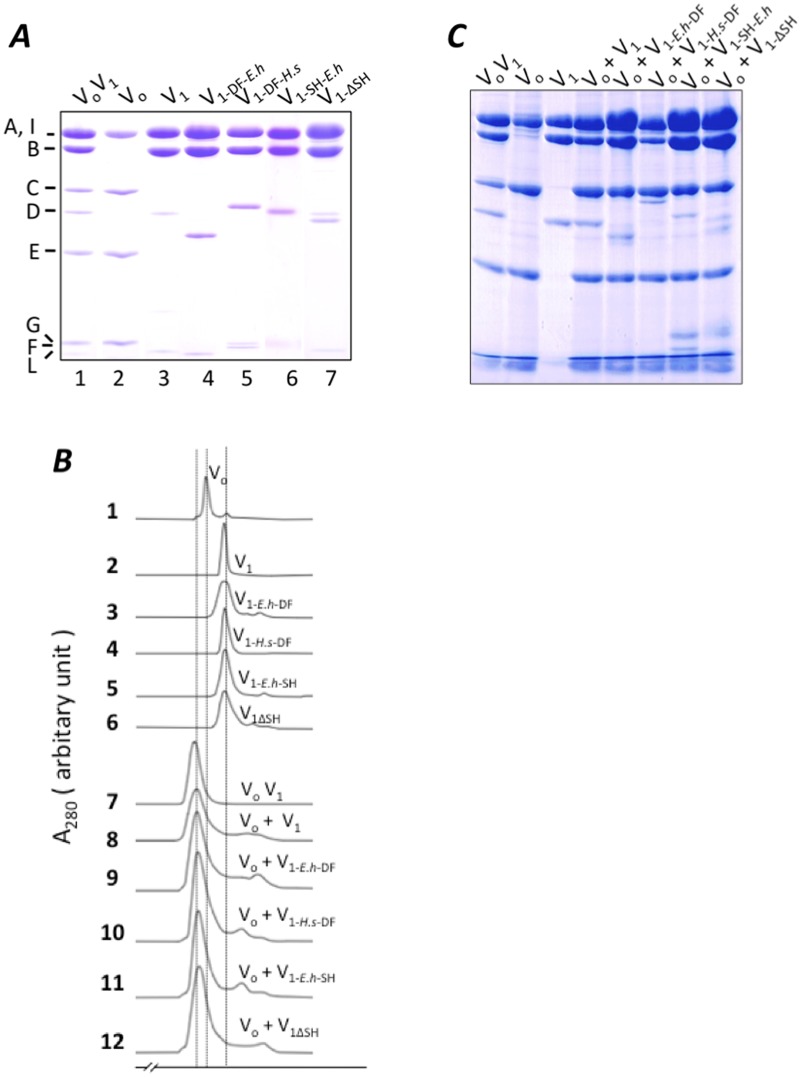
The amino acid sequence of V1-DF is not highly conserved among species (Fig. 1C). To investigate the effect of theses differences on reconstitution of V1 and Vo, expression constructs of V1 containing V1-DF from Homo sapiens (V1-DF-H.s) or E. hirae (V1-DF-E.h) were generated. These chimeric V1 were purified and subjected to ATPase analysis as described in Materials and Methods. Subunit stoichiometry of each chimeric V1 was confirmed by SDS-PAGE analysis (Fig. 3A). These chimeric V1 were ATPase active (Table 1, S1 Fig.), indicating that the exogenous V1-DF functions as a rotor in A3B3 of T. thermophilus. As shown in Table 1, the presence of the exogenous DF shaft from H. sapiens markedly enhanced the ATPase activity of V1. The enhanced ATPase activity of chimeric V1 is likely due to primary sequence difference between the DF of T. thermophilus and that of H. sapiens. We will discuss the effect of the H. sapiens DF on the activity of V1 fully elsewhere.
Fig 3. Analysis of reconstituted V1 with Vo.
Isolated V1 and Vo were analyzed by SDS-PAGE (A). Lane 1, VoV1; lane 2, Vo; lane 3, V1; lane 4, chimeric V1 containing E. hirae V1-DF (V1-Eh-DF); lane 5, chimeric V1 containing H. sapiens V1-DF (V1Hs-DF); lane 6, mutated V1 containing the exogenous short helix of E. hirae (V1SH-Eh); lane 7, the mutated V1 lacking the short helix (V1ΔSH). Analysis of reconstituted mutated V1 and Vo by gel permeation (B). The mixtures containing the mutated V1 and Vo were incubated for 1 h at 25°C, prior to analysis. Subunit stoichiometry of the fraction containing each mutated VoV1 was confirmed by SDS-PAGE analysis (C).
Table 1. Kinetics parameters of the mutated V1 for ATPase activity.
| V1 | V1-E.h-DF | V1-H.s-DF | V1-SH-E.h | V1ΔSH | |
|---|---|---|---|---|---|
| K m [mM] | 0.23 ± 0.05 | 0.76 ± 0.04 | 0.48 ± 0.04 | 0.22 ± 0.05 | 0.24 ± 0.06 |
| V max [s-1] | 30.6 ± 1.8 | 38.0 ± 0.80 | 132 ± 3.0 | 27.5 ± 1.5 | 21.8 ± 1.4 |
K m and V max values represent means ± SD (n = 3).
Each chimeric V1 was mixed with an excess amount of Vo from T. thermophilus and incubated for 1 hour. Analysis by gel permeation analysis revealed peaks corresponding to reconstituted VoV1 containing the exogenous V1-DF (Fig. 3B, lines 9 and 10). Subunit stoichiometry of the fraction containing each chimeric VoV1 was confirmed by SDS-PAGE analysis (Fig. 3C). These results indicate that the chimeric V1 incorporating the exogenous V1-DF can assemble with Vo.
Role of the short helix of V1-D subunit in interaction of V1 and Vo
The EM density structure of intact VoV1 of T. thermophilus and the crystal structure of V1-DF suggested that an α-helix of V1-D (a.a. 73–80) is key for interaction between V1-DF and Vo-C ([11,17] and Fig. 1B and C). This helix is referred to as the short helix hereafter. To investigate the role of the short helix in reconstitution of V1 and Vo, V1-SH-E.h containing the exogenous short helix of E. hirae (A74FIDELLA81, Fig. 1C), and V1ΔSH lacking the short helix of the V1-DF were constructed. The subunit stoichiometry and purity of the mutated V1 constructs were confirmed by SDS-PAGE analysis (Fig. 3A). Reconstitution of Vo and V1-SH-E.h or V1ΔSH was confirmed by gel permeation and FRET analysis (Fig. 3B, line 11, 12). Subunit stoichiometry of the fraction containing each mutated VoV1 was confirmed by SDS-PAGE analysis (Fig. 3C). These findings indicate that the short helix is not essential for reconstitution of V1 with Vo.
Role of the short helix of V1-D subunit in energy coupling between V1 and Vo
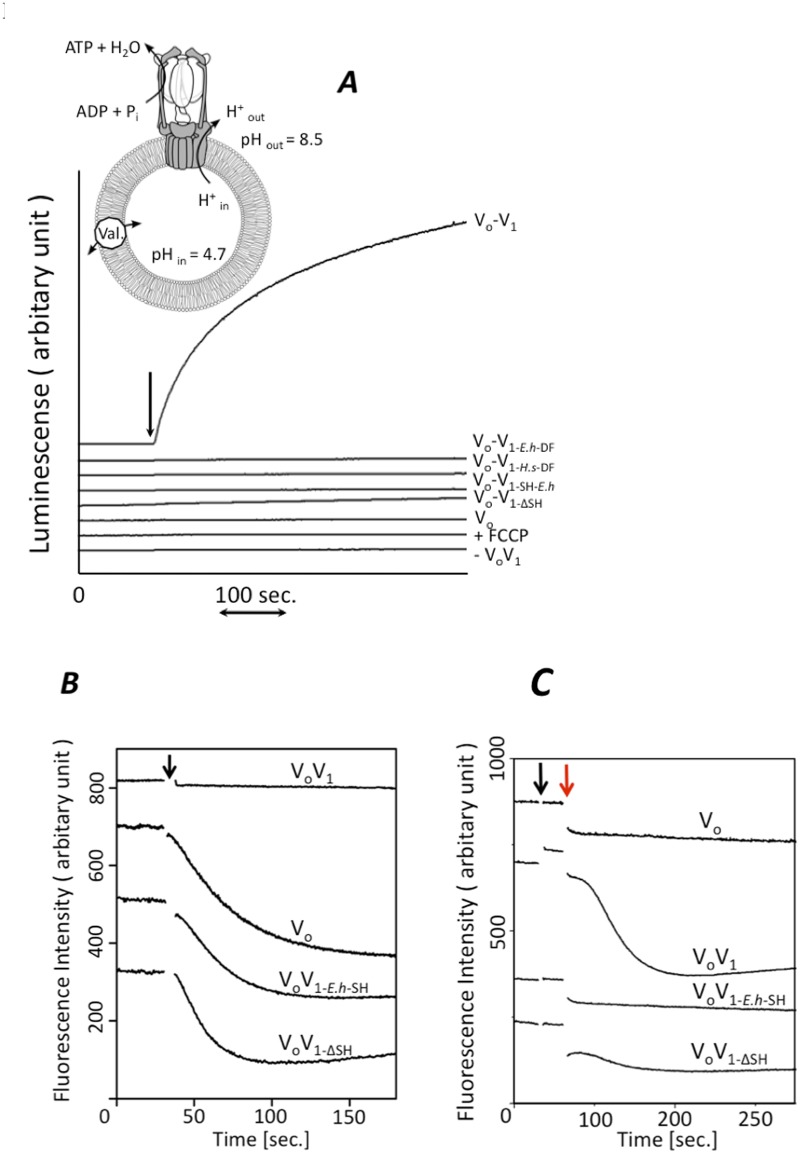
To further investigate the role of the short helix of the V1-D subunit in energy coupling between V1 and Vo, ATP synthesis activity of the reconstituted VoV1 were assessed. The reconstituted VoV1 constructs were purified by gel permeation chromatography and reconstituted into liposomes by freeze-thaw methods. Proton motive force was generated across the membranes of the reconstituted liposomes by acid-base transition (ref. [23], see inset in Fig. 4A). As shown in Fig. 4A, wild type reconstituted VoV1 showed continuous ATP synthesis. In contrast, the reconstituted VoV1ΔSH showed no ATP synthesis activity, indicating that lack of the short helix in VoV1 causes an intra molecular uncoupling between V1 and Vo. In addition, the reconstituted VoV1 including the exogenous V1-DF (VoV1-DF-H.s, VoV1-DF-E.h) or the short helix (VoV1-SH-E.h) showed no ATP synthesis activity.
Fig 4. Function of the short helix of V1-D subunit for energy coupling.
A, Analysis of ATP synthesis by the reconstituted complexes. inset; schematic model of the experimental system [16]. The reconstituted VoV1 was incorporated into liposomes, then energized by an acid base transition procedure described in the Materials and Methods. Each line shows the raw data for ATP synthesis by each reconstituted complex at a Δ pH of 3.8. The reactions were initiated by addition of acidified proteoliposomes into the base buffer, as indicated by the arrow. Final concentrations were 2 μg/ml of VoV1 and 1 mM ADP. The synthesized ATP was monitored by the luciferin-luciferase assay [18]. Analysis of proton channel (B) and proton pump activity (C) coupled with ATP hydrolysis by proteoliposomes. K+-loaded proteoliposomes containing the reconstituted complexes were prepared as described in the Materials and Methods. Proton influx was initiated by addition of 20 ng of valinomycin at the time indicated by the black arrow (B) and proton pump was initiated by addition of ATP-Mg at finally 1mM followed by the addition of valinomycin at the time indicated by the red arrow (C). The ACMA fluorescence emission (480nm) was recorded at 25°C.
Next, proton channel and proton pump activity of the reconstituted VoV1 were measured to investigate the energy coupling efficiency between the mutated V1 and Vo in the complexes. To facilitate ATP hydrolysis activity measurements, a mutated V1 incorporating the TSSA substitutions (A-S232A/A-T235S) to overcome ADP inhibition was used [5]. The wild-type VoV1 did not show proton channel activity but did show proton pump activity (Fig. 4B, C). These results indicate that the wild-type VoV1 is tightly coupled. In contrast, mutated VoV1 including the exogenous short helix or lacking the short helix did not show proton pump activity (Fig. 4C). In contrast, the mutated VoV1s had proton pump activity almost identical to the proton channel activity of Vo (Fig. 4B), indicating that the mutated VoV1s were completely uncoupled. These results strongly suggest that a proper match between the short helix in V1-D subunit and Vo-C subunit is essential for tight energy coupling between V1 and Vo.
Discussion
In this study, we have directly demonstrated a low binding affinity between V1-DF and Vo-C by both FRET and reconstitution experiments (Fig. 2). This is consistent with our previous result indicating that the two EG peripheral stalks are the major mediators of association of V1 with Vo [16]. This low binding affinity between V1-DF and Vo-C is relevant to reversible dissociation/association of V1 from Vo in eukaryotic VoV1 [26, 27]. However such low binding affinity is unfavorable for energy coupling between V1 and Vo; for ATP synthesis (ΔG = ~55 kJ/mol, [16]), the torque generated in the Vo rotor ring needs to be transmitted to V1-DF via Vo-C. Thus, an ingenious structure, that is both detachable and sticky, is required at the boundary surface between V1-DF and Vo-C. The EM density map of T. thermophilus VoV1 [11] provided a clue to unraveling the molecular basis of these seemingly contradictory properties. The short helix of V1-DF apparently lies in the cavity of Vo-C in Vo, suggesting that it may play an important role in association of V1-DF and Vo-C.
Here, we have investigated the role of the short helix in V1-D subunit on VoV1 assembly and energy coupling between V1 and Vo. Not only the chimeric V1 containing the exogenous V1-DF and the short helix, but also V1 lacking the short helix could reconstitute the complex with Vo (Fig. 3), indicating that the short helix is not a key factor for VoV1 complex formation. However, these mutant VoV1s showed neither ATP synthesis or proton pump activities (Fig. 4). As shown in Fig. 1C, amino acid residues in the short helix of V1-D are not highly conserved among species (T. thermophilus, E. hirae, H. sapiens). The different surface shape of the exogenous short helix compared to the endogenous one likely generates repulsion between V1-DF and Vo-C. The intact fitting of the short helix into the recess of Vo-C appears to be required for tight energy coupling between V1 and Vo.
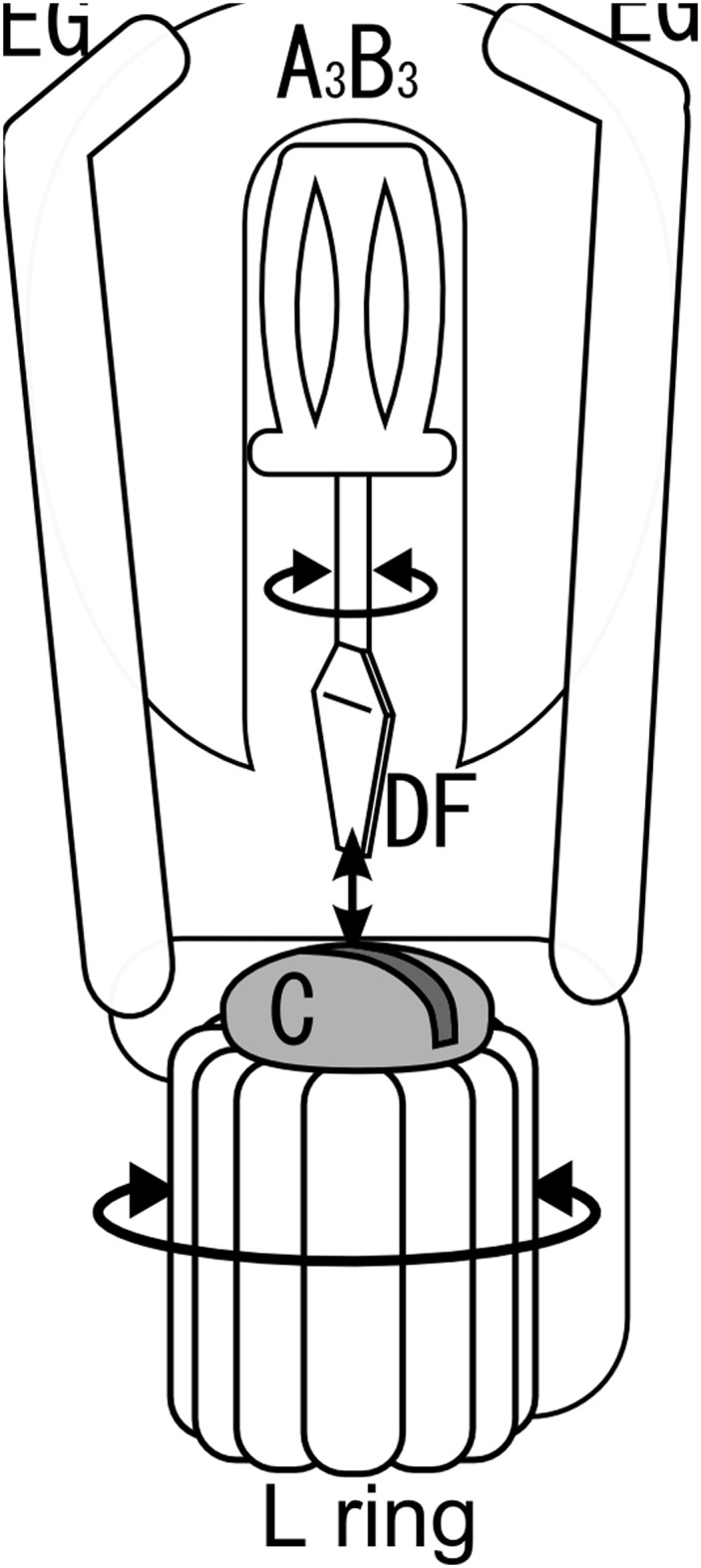
Here we propose a model to account for a detachable V1-DF/Vo-C boundary surface, which can transmit torque. In this model, the relationship between V1-DF and Vo-C is analogous to that between a slotted screwdriver and a head of slotted screw (Fig. 5). In the VoV1, two EG peripheral stalks push the short helix of V1-D into the socket of Vo-C. Thus, the short helix of V1-D binds into the socket of Vo-C, forming a sufficiently close interaction for transmission of torque from the rotating V1-DF to Vo-C. The rigid interaction between the V1-DF and Vo-C would be abolished by a loss of interaction between the two EG peripheral stalks and A3B3, consistent with the results of reconstitution experiments (Fig. 2) and EM analysis of sequential disassembly of the VoV1 [28]. FoF1 does not contain a counterpart of Vo-C so that the F1 shaft composed of the γ subunit directly attaches to the c6–10 rotor ring [12]. Thus, the F1-c6–10 stator-less complex is easily isolated from FoF1.
Fig 5. A schematic diagram for a slotted screwdriver and a head of slotted screw in VoV1 [16].
Supporting Information
The solid lines show fit with the Michaelis-Menten equation.
(TIF)
Acknowledgments
We thank Dr. Bernadette Byrne for critical reading of the manuscript, Dr Takeshi Murata for the gift of pCemtp18.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan No. 24370059 and 26650039 to KY. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007;8: 917–929 [DOI] [PubMed] [Google Scholar]
- 2. Mulkidjanian AY, Makarova KS, Galperin MY, Koonin EV. Inventing the dynamo machine: the evolution of the F-type and V-type ATPases. Nat. Rev. Microbiol. 2007;5: 892–899. [DOI] [PubMed] [Google Scholar]
- 3. Yokoyama K, Imamura H. Rotation, structure, and classification of prokaryotic V-ATPase. J. Bioenerg. Biomembr. 2005;37: 405–410 [DOI] [PubMed] [Google Scholar]
- 4. Muench SP, Trinick J, Harrison MA. Structural divergence of the rotary ATPase. Q. Rev. Biophys. 2011;44: 311–356 10.1017/S0033583510000338 [DOI] [PubMed] [Google Scholar]
- 5. Nakano M, Imamura H, Toei M, Tamakoshi M, Yoshida M, Yokoyama K. ATP hydrolysis and synthesis of a rotary motor V-ATPase from Thermus thermophilus . J. Biol. Chem. 2008;283: 20789–20896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kakinuma Y, Yamato I, Murata T. Structure and function of vacuolar Na+-translocating ATPase in Enterococcus hirae . J. Bioenerg. Biomembr. 1999;31: 7–14 [DOI] [PubMed] [Google Scholar]
- 7. Yoshida M, Muneyuki E, Hisabori T. ATP synthase—a marvellouse rotary engine of the cell. Nat. Rev. Mol. Biol. 2001;2: 669–677 [DOI] [PubMed] [Google Scholar]
- 8. Kishikawa J, Ibuki T, Nakamura S, Nakanishi A, Minamino T, Miyata T, et al. Common evolutionary origin for the rotor domain ATPase and flagellar protein export apparatus. PLoS. ONE. 2013;8: e64695 10.1371/journal.pone.0064695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jefferies KC, Cipriano DJ, Forgac M. Function, structure and regulation of the vacuolar H+-ATPases. Arch Biochem Biophys. 2008;476: 33–42. 10.1016/j.abb.2008.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nagamatsu Y, Takeda K, Kuranaga T, Numoto N, Miki K. Origin of asymmetry at the intersubunit interfaces of V1-ATPase from Thermus thermophilus . J. Mol. Biol. 2013; 425: 2699–2708 10.1016/j.jmb.2013.04.022 [DOI] [PubMed] [Google Scholar]
- 11. Lau WC, Rubinstein JL. Subnenometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature 2011;481: 214–218 [DOI] [PubMed] [Google Scholar]
- 12. Stock D, Leslie AG, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science 1999;286: 1700–1705 [DOI] [PubMed] [Google Scholar]
- 13. Iwata M, Imamura H, Stambouli E, Ikeda C, Tamakoshi M, Nagata K, et al. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 2004;101: 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stewart AG, Lee LK, Donohoe M, Chaston JJ, Stock D. The dynamic stator stalk of rotary ATPase. Nature Commun. 2012;3:687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kitagawa N, Mazon H, Heck AJ, Wilkens S. Stoichiometry of the peripheral stalk subunits E and G of yeast V1-ATPase determined by mass spectrometry. J. Biol. Chem. 2008;283: 3329–37 [DOI] [PubMed] [Google Scholar]
- 16. Kishikawa J, Yokoyama K. Reconstitution of Vacuolar type rotary H+-ATPase/synthase from Thermus thermophilus . J. Biol. Chem. 2012;287: 24597–24603 10.1074/jbc.M112.367813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saijo S, Arai S, Hossain KM, Yamato I, Suzuki K, Kakinuma Y, et al. Crystal structure of the central axis DF complex of the prokaryotic V-ATPase. Proc. Natl. Acad. Sci. U.S.A. 2011;108: 19955–19960 10.1073/pnas.1108810108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tamakoshi M, Uchida M, Tanabe K, Fukuyama S, Yamagishi A, Oshima T. A new Thermus-Escherichia coli shuttle integration vector system. J. Bacteriol. 1997;179: 4811–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yokoyama K, Nagata K, Imamura H, Ohkuma S, Yoshida M, Tamakoshi M. Subunit arrangement in V-ATPase from Thermus thermophilus . J. Biol. Chem. 2003;278: 42686–42691 [DOI] [PubMed] [Google Scholar]
- 20. Imamura H, Ikeda C, Yoshida M, Yokoyama K. The F subunit of Thermus thermophilus V1-ATPase promotes ATPase activity but is not necessary for rotation. J. Biol. Chem. 2004;279: 18085–18090 [DOI] [PubMed] [Google Scholar]
- 21. Kishikawa J, Nakanishi A, Furuike S, Tamakoshi M, Yokoyama K. Molecular basis of ADP inhibition of vacuolar (V)-type ATPase/synthase. J. Biol. Chem. 2014;289: 403–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murata T, Takase K, Yamato I, Igarashi K, Kakinuma Y. Properties of the VoV1Na+-translocating ATPase from Enterococcus hirae and its Vo moiety. J. Biochem. 1999;125: 414–421 [DOI] [PubMed] [Google Scholar]
- 23. Toei M, Gerle C, Nakano M, Tani K, Gyobu N, Tamakoshi M, et al. Dodecamer rotor ring defines H+/ATP ratio for ATP synthesis of prokaryotic V-ATPase from Thermus thermophilus . Proc. Natl. Acad. Sci. U.S.A. 2007;104: 20256–20261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takeda M, Suno-Ikeda C, Shimabukuro K, Yoshida M, Yokoyama K. Mechanism of inhibition of the V-type Molecular Motor by Tributyltin Chloride. Biophy. J. 2009;96: 1210–1217 10.1016/j.bpj.2008.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soga N, Kinoshita K Jr., Yoshida M, Suzuki T. Kinetic equivalence of transmembrane pH and electrical potential differences in ATP synthesis. J. Biol. Chem. 2012;287: 9633–9639 10.1074/jbc.M111.335356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parra KJ, Kane PM. Reversible association between the V1 and Vo domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol. Cell Biol. 1998;18: 7064–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kane PM. Targeting Reversible Disassembly as a Mechanism of Controlling V-ATPase Activity. Curr. Protein Pept. Sci. 2012;13: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tani K, Arthur CP, Tamakoshi M, Yokoyama K, Mitsuoka K, Fujiyoshi Y, et al. Visualization of two distinct states of disassembly in the bacterial V-ATPase from Thermus thermophilus . Microscopy 2013;62: 467–474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The solid lines show fit with the Michaelis-Menten equation.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.