Abstract
Chromatin modifying enzymes are predominantly nuclear; however, these factors are also localized to the cytoplasm and very little is known about their role in this compartment. In this report, we reveal a non-chromatin linked role for the lysine-specific demethylase KDM4A. We demonstrate that KDM4A interacts with the translation initiation complex and impacts the distribution of translation initiation factors within polysome fractions. Furthermore, KDM4A depletion reduced protein synthesis and enhanced the protein synthesis suppression observed with mTOR inhibitors, which paralleled an increased sensitivity to these drugs. Lastly, we demonstrate that JIB-04, a JmjC demethylases inhibitor, suppresses translation initiation and enhances mTOR inhibitor sensitivity. These data highlight an unexpected cytoplasmic role for KDM4A in regulating protein synthesis and suggest novel potential therapeutic applications for this class of enzyme.
Keywords: KDM, lysine demethylase, mTOR, DNA methylation/epigenetics, combination chemotherapy, cellular responses to anticancer drugs
INTRODUCTION
While sites of lysine methylation are emerging for non-histone substrates, little is known about how the enzymes responsible for the addition and removal of methylation, KMTs (lysine methyltransferases) and KDMs (lysine demethylases), respectively, are impacting the associated targets or their downstream processes. Most studies are focused on how these enzymes are targeting nuclear proteins even though there are important roles outside the nucleus (1–3). Therefore, studying the functions of KMTs and KDMs in the cytoplasmic compartment could reveal unexpected roles, which will likely identify relationships between these enzymes and signaling pathways (1). Understanding the nuclear and cytoplasmic roles for these chromatin-modifying enzymes is important since they are commonly altered in cancer (4), impact numerous diseases (5), and are emerging as important therapeutic targets.
In the present study, we describe a new and unexpected cytoplasmic role for the lysine demethylase KDM4A. KDM4A is a JmjC domain-containing enzyme that demethylates H3K9me3, H3K36me3 and H1.4K26me3 (6). Previous studies have documented roles for KDM4A in modulating DNA replication, site-specific copy number regulation and gene expression (7–9). These functions relate to the nuclear role of KDM4A; however, KDM4A is also localized to the cytoplasm, suggesting non-chromatin mediated functions. We report that KDM4A interacts with the translation initiation factors and is present in the initiating fractions of polysome profiles. Consistent with these observations, KDM4A depletion altered the distribution of translation initiation factors in polysome profiles, reduced protein synthesis and enhanced the cell sensitivity and effect of mTOR inhibitors. Lastly, we demonstrate that chemical inhibition with JIB-04, a JmjC demethylase inhibitor, potently inhibits translation initiation, reduces overall translation and enhances mTOR inhibitor sensitivity. Taken together, the findings in this report reveal an unexpected direct interaction between KDM4A and components of the translation machinery and highlight KDM4A as well as other JmjC proteins as potentially new targets in combined cancer therapy.
RESULTS
KDM4A interacts with the translation initiation machinery
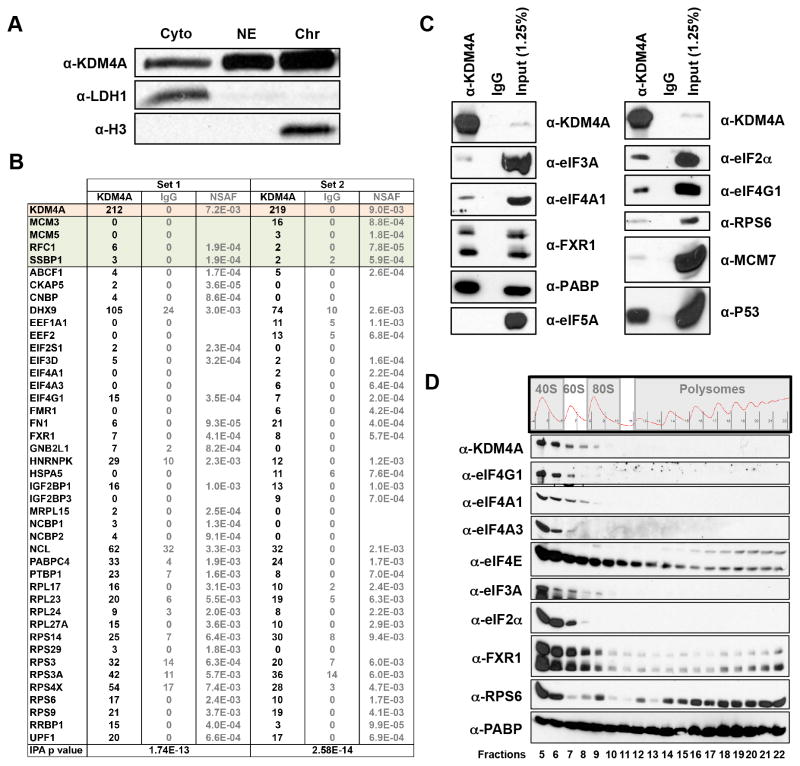
The lysine demethylase KDM4A is located in both the nucleus and cytoplasm (Figure 1A). For this reason, we performed mass spectrometry analysis on endogenously immunoprecipitated KDM4A from whole cell extracts to identify interacting proteins from both compartments. The interacting proteins from two independent immunoprecipitations were analyzed by Ingenuity Pathway Analysis (IPA), which revealed a significant enrichment for proteins involved in translation (p=1.74E-13 and 2.58E-14, respectively; Figure 1B). We further confirmed these interactions with a separate KDM4A antibody by conducting co-immunoprecipitations (Figure 1C). Some of these associations were equal to, or more enriched than previously confirmed KDM4A interactors [e.g., compare eIF2α to MCM-7 or P53 (8, 10)]. We further strengthened these observations by isolating fractions from polysome profiles and western blotting for KDM4A and associated proteins as well as for positive controls for the collections. KDM4A was enriched in the translation initiating fractions: predominantly in the 40S fractions and to a lesser extent in the 60S fractions (Figure 1D; fractions 5–9). These data suggest that KDM4A could directly impact translation.
Figure 1. KDM4A interacts with the translation initiation machinery.
(A) KDM4A is present in the cytoplasm and the nucleus. RPE cells were fractionated and western blotted for KDM4A and control proteins for the fractionations. (B) KDM4A interacts with the translation initiation machinery. Endogenous KDM4A was immunoprecipitated with a rabbit polyclonal antibody from HEK 293T cells and analyzed by mass spectrometry. The upper part of the table (in color; (8)) represents the peptides and NSAF values for KDM4A and previously confirmed interactors. The bottom part of the table (in white) represents the proteins present in the IPA “translation” category. (C) KDM4A interacts with the translation initiation machinery. These interactions were confirmed by western blotting Fab immunoprecipitated KDM4A from HEK 293T cells. (D) KDM4A sediments in the 40S and 60S polysome fractions. HEK 293T lysates were separated on sucrose gradient before fractions from polysome profiles were collected and immunoblotted with the indicated antibodies.
KDM4A impacts initiation factor distribution in 40/60/80S fractions
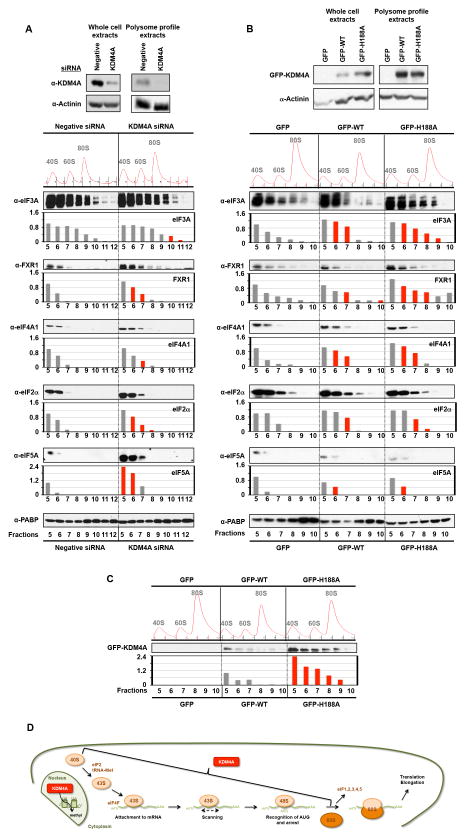
In order to understand the impact that KDM4A could have on the translation machinery, we siRNA depleted KDM4A and assessed whether the protein levels of translation-related factors were altered using multiplexed quantitative mass spectrometry-based proteomics (11). We compared the proteome of unaltered and KDM4A depleted cells in biological duplicates. The protein levels of more than 8,200 individual proteins were quantified. We did not observe changes in levels for translation-related factors. In fact, proteins that are up-regulated (19 proteins greater that 1.7-fold) and down-regulated (four proteins decreased by 1.7-fold; Figure S1A) did not have a significant enrichment in any particular category by IPA analysis (p<0.05). We observed only four proteins that were down regulated by more than 1.7-fold upon KDM4A depletion, including KDM4A (Figure S1A). Consistent with our quantitative proteomics data, western blotting confirmed the absence of major differences in translation-related protein levels (Figure S1B).
Since KDM4A was present in the initiating fractions within the polysome profile, we assessed the impact KDM4A depletion had on initiation factor levels and distribution within the polysome fractions. KDM4A depletion resulted in the enrichment of initiation factors in certain fractions (i.e., 40S) and/or a redistribution of these factors such that there was an extension into larger fractions (i.e., 60/80S; Figures 2A and S1C). Figure 2A depicts the average change across two independent siRNA treated samples. The red bars highlight fractions with >20% change in protein levels for both independent siRNAs. We consistently observed an increase in eIF3A, FXR1, eIF4A1, eIF2α and eIF5A protein levels in some of their corresponding fractions across two independent KDM4A siRNAs (see representative western blots and red bars in accompanying graphs); while, eIF3A, FXR1 and eIF2α presence is extended in larger fractions (fractions 11, 7 and 7–8, respectively). eIF5A is consistently enriched in fractions 5–6 upon KDM4A depletion, while PABP is stable across all fractions and serves as a normalization control.
Figure 2. KDM4A levels impact the distribution of translation initiation factors.
(A–B) Fractions collected from the polysome profiles [Negative Control (Negative) or KDM4A depleted HEK 293T cells for 48h (panel A); HEK 293T cells overexpressing WT or H188A for 24h (panel B)] were immunoblotted with the indicated antibodies. Representative western blots are shown below the graphs and represent the average of the quantification by ImageJ from two independent experiments. The red bars represent a reproducible increase of at least 20% compared to the control fractions from independent KDM4A siRNA treated cells or KDM4A WT or H188A overexpressing cells (see individual experiments in Figure S1C,D). The upper western blot panels represent whole cell extracts or the input extracts used to generate the polysome profiles. (C) Fractions collected from the polysome profiles from HEK 293T cells overexpressing GFP, WT or H188A for 24h were immunoblotted with anti-GFP and the corresponding graph represents the average from two independent experiments. The y-axis represents the fold change for each fraction relative to the fraction 5 from the corresponding control. For (A), the Negative siRNA is the control, whereas, for (B) the GFP is the control. For panel C, the GFP-WT is the control. The red bars represent a reproducible increase of at least 20% compared to the GFP-WT fractions between two independent experiments. (D) Schematic representing where KDM4A could directly be involved in translation. Also see supplementary Figure S1.
We further investigated the impact that KDM4A had on initiation factor distribution by overexpressing enzymatically active and inactive KDM4A. We compared the distribution of translation factors in cells expressing GFP alone (GFP), catalytically active (GFP-WT) or inactive [GFP-H188A; (7)] KDM4A. For both GFP-WT and GFP-H188A, the distribution of initiation factors was altered. For example, eIF3A, FXR1 and eIF4A1 protein levels were consistently higher in fractions 6–11 (Figures 2B and S1D; red bars, >20% between independent experiments). GFP-H188A appeared to have a more pronounced effect on certain initiation factors (i.e., eIF3A was higher in fractions 8–9 and FXR1 was increased in fractions 6–10). Interestingly, while GFP-WT and GFP-H188A had comparable expression (Figure 2B, upper panel), GFP-H188A exhibited much higher levels in fractions 5–9 (Figure 2C). These data are consistent with the catalytic activity being important for KDM4A distribution within the initiating fractions. Taken together, these data highlight the importance of balancing KDM4A protein levels so that proper distribution of translation initiation factors occur within polysome profiles, which supports a role for KDM4A in initiation complex assembly/disassembly (Figure 2D).
KDM4A depletion reduces protein synthesis
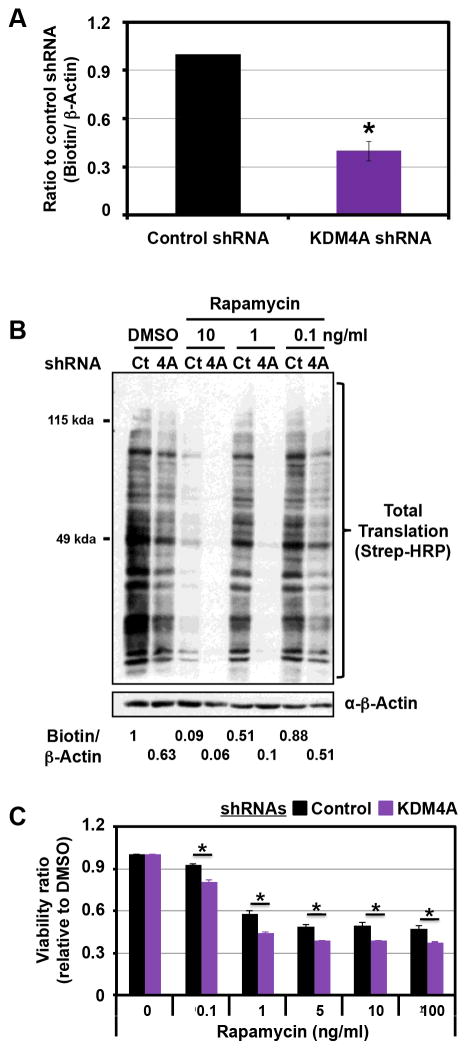
Since KDM4A interacts with translation factors and impacts their distribution, we hypothesized that KDM4A would have a role in protein synthesis. In order to test this possibility, we depleted KDM4A with two different shRNAs and assessed protein synthesis over two hours by measuring the incorporation of the methionine analog AHA (L-Azidohomoalanine) (Figures 3A and S2A). KDM4A depletion significantly reduced the synthesis of nascent proteins (Figure 3A; 40%, p=1E-05).
Figure 3. KDM4A depletion reduces protein synthesis and enhances mTOR inhibitor sensitivity.
(A) HEK 293T cells transfected with KDM4A shRNA present a decrease in overall translation. Forty eight hours after transfection, cells were deprived of cysteine and methionine for 1h and grown in the presence of the nucleotide analog AHA (L-Azidohomoalanine) for 2h. The newly synthesized proteins were labeled with biotin and equal amounts of total protein were immunoblotted with streptavidin-HRP. The graph represents an average of five independent experiments performed with two different KDM4A shRNA. The Y axis represents the ratio of total biotinylated proteins upon KDM4A shRNA to total biotinylated proteins upon control shRNA, which were normalized to β-actin. (B) KDM4A knock-down enhanced the decrease in translation obtained after Rapamycin treatment. Forty eight hours after transfection, HEK 293T cells were treated with the indicated concentration of Rapamycin for 24h and then treated as in (A). The ratio of biotin/β-actin has been calculated with ImageJ and represents the average of two independent experiments. (C) HEK 293T cells depleted for KDM4A are more sensitive to Rapamycin than cells transfected with the control vector. Cells were seeded 24h after the second shRNA transfection and were then treated with the indicated concentrations and associated concentrations 24h later. Forty-eight hours after treatment, samples were analyzed by MTT assay. The assays were normalized to a sample collected and assayed at the treatment time. The Y axis represents the viability ratio relative to DMSO. The average of three independent experiments is represented. All error bars represent the SEM. p values were determined by a two-tailed student’s t test; * represents p<0.05. Also see supplementary Figure S2.
We then assessed whether KDM4A depletion could enhance the inhibition of protein synthesis caused by drugs targeting translation initiation. mTOR is a well-recognized drug target involved in translation initiation (12, 13). Therefore, we depleted KDM4A and assessed AHA incorporation with increasing doses of the mTOR inhibitor Rapamycin. Protein synthesis was further decreased in cells treated with all doses of Rapamycin combined with KDM4A shRNA depletion (Figures 3B and S2B). For example, 0.1ng/ml and 1ng/ml Rapamycin suppressed protein synthesis by ~20% and ~50%, respectively. However, 0.1ng/ml Rapamycin and 1ng/ml coupled to KDM4A depletion resulted in ~50% and 90% reduction in protein synthesis, respectively (Figure 3B, the biotin/β-actin ratio reported at the bottom represents the average of two independent experiments). These observations prompted us to assess the impact of KDM4A depletion on mTOR inhibitor sensitivity in cellular viability assays. As expected, KDM4A depletion coupled to Rapamycin treatment resulted in a statistically significant decrease in cell viability across all drug doses (Figure 3C). Consistent with these results, a polymorphism in KDM4A which results in reduced protein stability also confers sensitivity to mTOR inhibitors (14).
JmjC demethylase inhibition causes translation initiation defects
Previous studies report that KDM4 and KDM5A enzymes are chemical targets for JIB-04, an inhibitor of JmjC demethylases (15). Interestingly, KDM5A is enriched on genes involved in mTOR, p70S6K and EIF2 signaling (16). In fact, KDM5A depletion reduced the expression of ribosomal protein genes [e.g., RPL3, RPL7, RPL24; (16)]. These observations are in contrast with KDM4A, as we did not observe significant alterations in gene expression for genes involved in translation or mTOR signaling, suggesting that these enzymes could crosstalk to translation machinery in different ways (Figure S3A). Consistent with KDM5A regulating mTOR and translation factor genes (16), we observed an increased sensitivity to Rapamycin upon KDM5A depletion (Figures S3B–C) and a reduction in protein synthesis that was comparable to KDM4A depletion (Figure S3D). Since KDM4A was associated with the initiation complex, we also assessed whether KDM5A was present in the polysome fractions. Interestingly, KDM5A was present in the 40S/60S fractions of polysome profiles (Figure S3E). These data suggest that KDM5A could regulate protein synthesis at the level of gene expression and translation complexes. Furthermore, they suggest that multiple KDMs could be involved in regulating protein synthesis and the response to drugs such as Rapamycin. Therefore, we hypothesize that JIB-04 or related compounds could have a significant impact on sensitivity to mTOR inhibitors and protein synthesis by affecting both KDM4A and KDM5A or additional demethylases yet to be linked to protein synthesis.
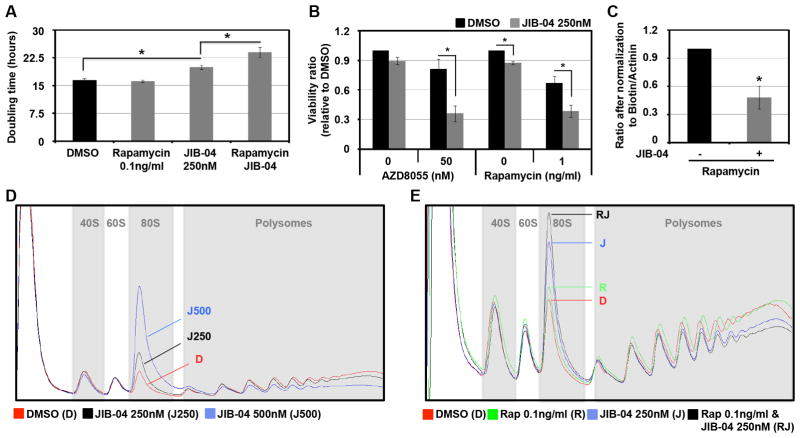
In order to address this hypothesis, cells were co-treated with JIB-04 and Rapamycin or AZD8055 (two different mTOR inhibitors) before proliferation and viability was assessed (Figure 4A,B). JIB-04 treatment enhanced the effect of Rapamycin and AZD8055 on both cell proliferation (Figures 4A and S3F) and viability (Figures 4B). We further demonstrated this increased sensitivity to mTOR inhibitors in additional cancer cell lines (Figure S3G,H). Taken together, these data support the hypothesis that inactivation of KDM activity can be used to sensitize cells to mTOR inhibition. We then tested whether JIB-04 altered overall translation and enhanced the translation defect observed with mTOR inhibitors. As expected, treatment with JIB-04 decreased overall protein synthesis (Figures S3I) and enhanced the translation defect observed upon Rapamycin treatment (Figures 4C and S3I). Consistent with this defect, JIB-04 treatment resulted in a dose-dependent defect in translation initiation (Figure 4D, increase of the 80S peak and decrease in polysomes). Furthermore, JIB-04 enhanced the initiation defect observed with different doses of Rapamycin (Figure 4E and S3J). Taken together, these data highlight the impact that inhibition of KDM4-5 enzymes have on protein synthesis and demonstrate a new and unexpected involvement for JmjC enzymes in translation.
Figure 4. JIB-04 inhibits translation initiation.
(A–B) HEK 293T cells treated with JIB-04 are more sensitive to mTOR inhibitors than cells treated with vehicle. (A) HEK 293T cells were treated with 250nM of JIB-04 and/or 100ng/ml of Rapamycin 24h after seeding. The Y axis represents the doubling time between 5h and 35h after Rapamycin treatment. The average of three independent experiments is represented. (B) HEK 293T cells were treated with the indicated drugs 24h after seeding, and 48h later they analyzed by MTT assay. The Y axis represents the viability ratio relative to DMSO. The average of three independent experiments is represented. (C) JIB-04 enhanced the decrease in translation obtained after Rapamycin treatment. HEK 293T cells were treated with 250nM of JIB-04 and/or 0.1ng/ml of Rapamycin for 24h and then treated as in Figure 3A. The graph represents an average of three independent experiments. The Y axis represents the ratio after normalization to total biotinylated proteins to Actinin. (D) JIB-04 treated cells have a translation initiation defect. HEK 293T cells treated with the indicated concentration of JIB-04 for 24h before being analyzed by polysome profiling. (E) JIB-04 enhanced the translation initiation defect obtained after Rapamycin treatment. HEK 293T cells treated with the 250nM of JIB-04 and/or 0.1ng/ml of Rapamycin for 24h were analyzed by polysome profiling. All error bars represent the SEM. p values were determined by a two-tailed student’s t test; * represents p<0.05. Also see supplementary Figure S3.
DISCUSSION
In the present study, we assess the cytoplasmic role for KDM4A in protein synthesis. Altering the levels of KDM4A changed the distribution of translation initiation factors. This altered distribution could be due to the defective release of initiation factors since there was an accumulation of such factors in 40/60/80S fractions; however, KDM4A could also regulate additional events involved in translation or mRNA processing that could impact initiation and protein synthesis. Our data suggest that KDM4A directly affects initiation and protein synthesis because KDM4A interacts with the translation initiation machinery and is present in the initiating fractions of a polysome profile (i.e., 40S and 60S). Interestingly, there was more catalytically inactive KDM4A in the fractions where KDM4A is present. Therefore, catalytic activity appears to be important for properly regulating KDM4A distribution in these complexes, suggesting that components of the ribosome or ribosome-associated proteins are possible non-histone substrates of KDM4A. However, our data also suggest that there could be a non-enzymatic function since overexpression of both catalytically active and inactive KDM4A impact initiation factors distribution. Therefore, future studies will be focused on understanding the enzymatic and non-enzymatic roles in modulating translation.
Our data demonstrate that reduced KDM4A levels are able to reduce overall protein synthesis without changing cell proliferation. However, the importance of this defect becomes enhanced with chemotherapeutics that target translation. The enhanced effect on translation inhibition correlates with stronger decrease in cell proliferation. These observations are reminiscent of KDM4A depletion resulting in enhanced chemotherapeutic sensitivity to DNA replication drugs, which is another process directly regulated by KDM4A (7). Taken together, these data suggest that inhibition of KDM could potentiate the effect of mTOR inhibitors in the context of various cancer types. mTOR inhibition is a frontline therapy in the treatment of breast cancer and is being tested in ovarian cancer (17–19). This study, albeit limited to a few models, suggests that breast cancer cells [Figure S3B; (20)] and ovarian cancer cells (Figure S3C) might be sensitized by JIB-04 treatment in their response to mTOR inhibition. These data strongly support the combined use of these chemotherapeutics in other cancer cell types.
We also provide evidence that other KDMs such as KDM5A increase mTOR inhibitor sensitivity. Interestingly, Tzatsos et al. demonstrated that KDM5A was enriched at target genes involved in protein synthesis and the mTOR pathway (16). Since we also observe an enrichment of KDM5A in polysome profile fractions this enzyme may impact protein synthesis and mTOR inhibitor sensitivity at multiple steps. Resolving whether one or both aspects are important for the impact of KDM5A on protein synthesis will be important for future studies. Lastly, these studies suggest that other enzymes (e.g., demethylases and methyltransferases) could be involved in pathways that crosstalk to translation or mTOR/PI3K signaling. Since many of these enzymes are altered in cancer (e.g., exhibit mutations, copy number variation or altered expression), they could act as additional biomarkers in treating cancer. Studies evaluating the KDM4A and KDM5A genetic and expression status in relation to treatment efficacy could allow for more refined patient stratification, especially in the context of mTOR inhibitors.
Overall, the data presented in this study highlight the importance of understanding the impact enzymes have inside and outside the nucleus and of identifying the crosstalk between pathways for chemotherapeutics and chromatin regulators. The discovery that KDM4A or KDM5A depletion results in a decrease in protein synthesis suggests there is a benefit in targeting this group collectively. Therefore, the lack of currently available KDM-specific inhibitors could be balanced by the benefit of targeting groups of KDMs, especially in combined therapy. This strategy is currently being applied for HDACs (21). The combined targeting of KDMs and translation could result in an effective therapy with reduced single agent toxicity; while, providing the advantage of reducing the emergence of resistant clones since both KDM4A and KDM5A have been linked to mechanisms that could promote resistance (8, 22). These possibilities are important areas to explore in the future. Lastly, the current study highlights the fact that discovering unappreciated functions for chromatin modifiers can lead to the identification of associated signaling pathways and uncover optimal chemotherapeutic targets for the future.
EXPERIMENTAL PROCEDURES
Cell culture and drug treatments
For tissue culture procedures see (7). Rapamycin (LC Laboratories) and AZD8055 (Selleckchem) were used at indicated concentrations. JIB-04 (Xcessbio) was used at a final concentration of 250nM or 500nM, as annotated. For the translation assays, DMEM depleted of Methionine and Cysteine (Life Technologies 21013-024) was used. HEK 293T cells have been obtained from ATCC, no authentication has been done by the authors.
Plasmids, siRNAs and transfections
Plasmid transfections were performed using X-tremeGENE 9 DNA transfection reagent (Roche) on 6x105 HEK 293T cells plated in 10 cm dishes 20h prior to transfection. The complexes were incubated with the cells in OptiMEM for 4h before being replaced by fresh media. The transfected plasmids were: pMSCV-GFP (7), pMSCV-GFP-KDM4A (7), pMSCV-GFP-KDM4A-H188A (7), pSUPER (7), pSUPER-4C (referred as 4A.2 throughout the figures) (7), pLKO, pLKO-A06 (referred as 4A.6 throughout the figures), pLKO-A10 (referred as 4A.10 throughout the figures). For the MTT assays shRNAs were transfected twice 48h apart. siRNAs transfections were performed using X-tremeGENE siRNA transfection reagent (Roche) or Lipofectamine 3000 (Life Technologies) following vendors instructions. The transfected siRNAs were Silencer Select (Life Technologies) KDM4A (s18636 and s18637), KDM5A (s11834 and s11836), and negative control #1.
Western blot analysis
Western blot analyses were performed according to (7).
Cell Fractionation
Cytoplasmic, nuclear and chromatin fractions were prepared from RPE cells. Cell pellets were washed twice in ice cold PBS and resuspended in ice cold Buffer A (10mM HEPES pH 7.9, 10mM KCl, 0.1M EDTA, 0.5M EGTA) and incubated on ice for 15 minutes. Swollen cells were lysed by addition of NP-40 to 0.8% with 10 seconds of vortex. Lysed cells were centrifuged and the supernatant kept as cytoplasm. The nuclear pellet was resuspended in Buffer C (10mM HEPES pH 7.9, 400mM NaCl, 1mM EDTA, 5mM EGTA), dounced to resuspend and incubated at 4 degrees C for 30 minutes with rotation. Extracts were centrifuged and the supernatant kept as nuclear extract. Chromatin pellets were resuspended in N-Buffer (20mM Trish pH 7.5, 100mM KCl, 2mM MgCl2, 1mM CaCl2, 0.3M Sucrose, 0.1% Triton X-100, 3U per ml micrococcal nuclease. Samples were sonicated for 10 minutes at 70% amplitude in a Q700 cup horn and then incubated at room temperature for 15 minutes for MNase digestion. Reactions were stopped by addition of 5mM EGTA and centrifuged to clear. Supernatant was kept as chromatin extract. For the western-blot analysis a comparable fraction of each compartment was loaded on a gel. The western-blot depicted in Figure 1A was compiled from non-adjacent images from the same exposure of the same blot.
Antibodies
The antibodies used were LDH (Santa Cruz, sc-133123), Histone H3 (Abcam, ab1791) Actinin (Santa Cruz, sc-17829), Streptavidin-HRP (Cell Signaling 3999S), eIF3A (Abcam, ab118357), eIF4A1 (Abcam, ab31217), FXR1 (Abcam, ab129089), PABP (Abcam, ab21060), eIF5A (Abcam, ab32443), eIF2α (Invitrogen, AH00802), eIF4G1 (Abcam, ab2609), P53 (Santa-Cruz, sc-126 X), eIF4A3 (Abcam, ab32485), eIF4E (Cell Signaling, 20675), RPS6 (Abcam, ab58350), eIF3D (Abcam, ab12442), S6K1 (Abcam, ab32529), S6K1pT389 (Abcam, ab2571), RPS6pS235 (Abcam, ab12864), KDM5A (Abcam, ab70892). KDM4A and β-Actin antibodies were described in (23). MCM7 antibody was described in (8). KDM4A immunoprecipitations were performed with KDM4A-P006.
Co-immunoprecipitation
The co-immunoprecipitations experiments were performed as described in (23).
Mass spectrometry analysis on KDM4A immunoprecipitatons
Mass spectrometry analysis were performed as described in (8).
Multiplexed quantitative proteomics
Multiplexed quantitative proteomics using tandem mass tag reagents (TMT, Thermo Scientific) and a synchronous precursor selection based MS3 method (11) on an Orbitrap Fusion mass spectrometer (Thermo Scientific). For a complete methods protocol see the Supplemental Experimental Procedures.
Monitored cell proliferation assay
Seventy-two hours post transfection 1x104 HEK 293T cells were seeded per well of a 96 well plate, and then treated after 24h. Cell proliferation was monitored with an xCELLigence system (Roche) (24).
MTT assays
MTT assays were performed following supplier’s instructions from the Cell Proliferation Kit I (MTT) from Roche. For shRNA experiments, after two subsequent shRNA transfections, cells were seeded in 96wells plates before being treated 24h later. Briefly, 1x104 cells were seeded per well of a 96 well plate and grown for 24h before treatment. Forty-eight hours later, cells were assayed. We determined sensitivity by subtracting the background from the absorbance.
Translation assays
Translation assays were performed following supplier’s instructions from Click-IT Metabolic Labeling Reagents for Proteins (Life Technologies). Cells were incubated in the presence of DMEM without cysteine and methionine for 1h, then grown in presence of 50μM AHA (L-azidohomoalanine, Life Technologies C10102) for 2h, harvested and washed extensively with PBS. Cells were lysed in 1% SDS in 50mM Tris pH8 with 10% Glycerol and sonicated using a bath sonicator (QSonica Q800R) for 30min. The Click-IT reactions were performed following the supplier’s instructions from Click-IT Protein Reaction Buffer Kit (Life Technologies). Briefly, 50μg to 100μg of lysates were used per reaction with 40μM Alkyne-Biotin (Life Technologies B10185), and 10μg were assayed by western blot using a Streptavidin antibody conjugated to HRP.
Polysome profiling
HEK 293T cells were grown to 80% confluence in 10 cm2 plates. Cells were washed and scraped into 100μg/ml cycloheximide/PBS. Cells were pelleted and resuspended in 300–500ul polysome lysis buffer (5mM Tris pH 7.4, 2.4 mM MgCl2, 1.5mM KCl), freshly supplemented with 10μg/ml cyclohexamide, 2uM DTT, 0.5% Triton-X, 0.5% sodium deoxycholate and protease and phosphatase inhibitors. Lysis buffer volume was adjusted for cell number. Lysates were cleared for 10min, 18000xg, 4°C. 250ul of cleared lysates were loaded onto 12ml 10–50% sucrose gradients (prepared in 15mM Tris pH 7.4, 15mM MgCl2, 150mM NaCl) and spun for 2h in a SW40Ti rotor (Beckman Coulter), at 40000rpm at 4°C. Immediately following centrifugation, 0.5ml fractions were collected using a BioComp Gradient Master instrument. For western blot 200μl of each fraction were precipitated by methanol/chloroform extraction and loaded on a gel.
Microarrays analysis
For RNA expression analysis, RNA was extracted using MiRNeasy Mini kit (Qiagen). cDNA synthesis was performed with 2ug of total RNA using the Roche cDNA synthesis System (11 117 831 001, Roche). Hybridization of cDNA to Human Gene Expression 12x135K Arrays (Roche Nimblegen) was performed in triplicate according to manufacturer’s protocol. Arrays were scanned using MS200 scanner (Roche Nimblegen) and extracted using Roche DEVA software. Raw expression array data was RMA-normalized and log2 transformed. Differential expression t-tests were computed using the empirical Bayes shrinkage procedure implemented in the Limma R/Bioconductor package (25). Two-fold up- or downregulated genes with a p-value <0.05 were considered differentially regulated. The accession number for the microarray analysis is GSE63812.
Statistics
All errors bars represent SEM. p values were determined by a two-tailed student’s t test; * represents p<0.05.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
This report documents an unexpected cytoplasmic role for the lysine demethylase KDM4A. We demonstrate that KDM4A interacts with the translation initiation machinery, regulates protein synthesis and, upon co-inhibition with mTOR inhibitors, enhances the translation suppression and cell sensitivity to these therapeutics.
Acknowledgments
Financial support: The studies conducted in this manuscript were funded by the following agencies: American Cancer Society Basic Scholar Grant, MGH Proton Beam Federal Share Grant (CA059267) and NIH R01GM097360 to J.R.W.; NIH (U54 HG006097) to C.H.B.. J.R.W. is the Tepper Family MGH Research Scholar, Leukemia and Lymphoma Scholar as well as the recipient of the American Lung Association Lung Cancer Discovery Award. A post-doctoral fellowship was provided by the Fund for Medical Discovery (C.V.R). C.V.R is the 2014 Skacel Family Marsha Rivkin Center for Ovarian Cancer Research Scholar. This research was supported in part by a grant from the Marsha Rivkin Center for Ovarian Cancer Research. J.C.B. was a Fellow of The Jane Coffin Childs Memorial Fund for Medical Research and is supported by MGH ECOR Tosteson Postdoctoral Fellowship. This investigation has been aided by a grant from The Jane Coffin Childs Memorial Fund for Medical Research. The SGC is a registered charity (no. 1097737) that receives funds from AbbVie, Boehringer Ingelheim, the Canada Foundation for Innovation (CFI), the Canadian Institutes of Health Research (CIHR), Genome Canada, Ontario Genomics Institute Grant OGI-055, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and Wellcome Trust Grant 092809/Z/10/Z. Work in the Kossiakoff lab was funded by SGC and NIH.
We would like to thank Drs. Cailin Joyce and Carl Novina for providing access to the BioComp Gradient Master instrument as well as training. We thank Drs. Mo Motamedi and Elnaz Atabakhsh, and Sweta Mishra as well as Kelly Biette for their comments and suggestions. We thank Tony Kossiakoff, Edyta Marcon, Cheryl Arrowsmith and members of the SGC Antibody team for contributions to generating recombinant KDM4A-P006.
Abbreviation list
- KMT
lysine methyltransferase
- KDM
lysine demethylase
- mTOR
mammalian target of Rapamycin
- PI3K
phosphoinositide kinase 3
- JmjC
jumonji C
- WT
wild type
- AHA
L-azidohomoalanine
- HEK 293T
human embryonic kidney 293T
- HDACs
histone deacetylases
- SEM
standard error of the mean
Footnotes
Conflict of Interest: The authors declare competing financial interests because JRW is a consultant for QSonica.
References
- 1.Mazur PK, Reynoird N, Khatri P, Jansen PW, Wilkinson AW, Liu S, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510:283–7. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–36. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 3.Donlin LT, Andresen C, Just S, Rudensky E, Pappas CT, Kruger M, et al. Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes & development. 2012;26:114–9. doi: 10.1101/gad.177758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Rechem C, Whetstine JR. Examining the impact of gene variants on histone lysine methylation. Biochimica et biophysica acta. 2014;1839:1463–76. doi: 10.1016/j.bbagrm.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–57. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Molecular cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black JC, Allen A, Van Rechem C, Forbes E, Longworth M, Tschop K, et al. Conserved antagonism between JMJD2A/KDM4A and HP1gamma during cell cycle progression. Molecular cell. 2010;40:736–48. doi: 10.1016/j.molcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Black JC, Manning AL, Van Rechem C, Kim J, Ladd B, Cho J, et al. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013;154:541–55. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer research. 2013;73:2936–42. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim TD, Shin S, Berry WL, Oh S, Janknecht R. The JMJD2A demethylase regulates apoptosis and proliferation in colon cancer cells. J Cell Biochem. 2012;113:1368–76. doi: 10.1002/jcb.24009. [DOI] [PubMed] [Google Scholar]
- 11.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8:937–40. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjornsti MA, Houghton PJ. The TOR pathway: a target for cancer therapy. Nature reviews Cancer. 2004;4:335–48. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 13.Populo H, Lopes JM, Soares P. The mTOR Signalling Pathway in Human Cancer. International journal of molecular sciences. 2012;13:1886–918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Rechem C. [Google Scholar]
- 15.Wang L, Chang J, Varghese D, Dellinger M, Kumar S, Best AM, et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nature communications. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, et al. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J Clin Invest. 2013;123:727–39. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciruelos Gil EM. Targeting the PI3K/AKT/mTOR pathway in estrogen receptor-positive breast cancer. Cancer treatment reviews. 2014;40:862–71. doi: 10.1016/j.ctrv.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Jerusalem G, Rorive A, Collignon J. Use of mTOR inhibitors in the treatment of breast cancer: an evaluation of factors that influence patient outcomes. Breast cancer. 2014;6:43–57. doi: 10.2147/BCTT.S38679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mabuchi S, Hisamatsu T, Kimura T. Targeting mTOR signaling pathway in ovarian cancer. Current medicinal chemistry. 2011;18:2960–8. doi: 10.2174/092986711796150450. [DOI] [PubMed] [Google Scholar]
- 20.Roberts PJ, Usary JE, Darr DB, Dillon PM, Pfefferle AD, Whittle MC, et al. Combined PI3K/mTOR and MEK inhibition provides broad antitumor activity in faithful murine cancer models. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:5290–303. doi: 10.1158/1078-0432.CCR-12-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:3970–7. doi: 10.1158/1078-0432.CCR-08-2786. [DOI] [PubMed] [Google Scholar]
- 22.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Rechem C, Black JC, Abbas T, Allen A, Rinehart CA, Yuan GC, et al. The SKP1-Cul1-F-box and Leucine-rich Repeat Protein 4 (SCF-FbxL4) Ubiquitin Ligase Regulates Lysine Demethylase 4A (KDM4A)/Jumonji Domain-containing 2A (JMJD2A) Protein. The Journal of biological chemistry. 2011;286:30462–70. doi: 10.1074/jbc.M111.273508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vistejnova L, Dvorakova J, Hasova M, Muthny T, Velebny V, Soucek K, et al. The comparison of impedance-based method of cell proliferation monitoring with commonly used metabolic-based techniques. Neuro endocrinology letters. 2009;30 (suppl 1):121–7. [PubMed] [Google Scholar]
- 25.Gentleman R, Carey V, Huber W, Dudoit S, Irizarry R. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.