Abstract
High-through put methods for analyzing genome structure and function are having a large impact in song-bird neurobiology. Methods include genome sequencing and annotation, comparative genomics, DNA microarrays and transcriptomics, and the development of a brain atlas of gene expression. Key emerging findings include the identification of complex transcriptional programs active during singing, the robust brain expression of non-coding RNAs, evidence of profound variations in gene expression across brain regions, and the identification of molecular specializations within song production and learning circuits. Current challenges include the statistical analysis of large datasets, effective genome curations, the efficient localization of gene expression changes to specific neuronal circuits and cells, and the dissection of behavioral and environmental factors that influence brain gene expression. The field requires efficient methods for comparisons with organisms like chicken, which offer important anatomical, functional and behavioral contrasts. As sequencing costs plummet, opportunities emerge for comparative approaches that may help reveal evolutionary transitions contributing to vocal learning, social behavior and other properties that make songbirds such compelling research subjects.
Keywords: Songbirds, Zebra finch, Gene expression, Vocal learning, Genomics, Transcriptomics, Microarrays, High-throughput, Birdsong, Networks, Avian models
1. Introduction
Deciphering the molecular and genetic basis of learned behaviors is one of the central challenges in neurobiology. Through the early pioneering efforts of neurobiologists like Gabriel Horn, birds were shown to be highly informative model organisms with regards to uncovering plastic changes in the brain that may underlie learning and memory, especially in the context of visual imprinting (e.g., Horn et al., 2001; reviewed in Horn, 2004). These early studies in birds contributed substantially to the broadly accepted notion that the laying down of long-lasting memories requires specific and localized biochemical and molecular changes in the brain. Avian studies have particularly benefitted from a relatively simpler brain organization than in mammals, with telencephalic pallial-areas having a nuclear and thus less heterogeneous spatial distribution than the mammalian cortex (Reiner et al., 2004a; Jarvis et al., 2005). This structural organization considerably facilitates anatomical, molecular and physiological studies of the avian brain. Nonetheless, the avian telencephalon shares with the mammalian brain the occurrence of specialized areas involved in sensory and perceptual processing, motor control, multi-sensory and sensorimotor integration, and various aspects of learning and cognition (Reiner et al., 2004a,b; Jarvis et al., 2005, 2013a). The analogs of thalamo-recipient cortical layers, long descending projections to sub-cortical targets, and intricate cortical-like microcircuitry with abundant inhibitory interneurons have also been identified in birds. These similarities point to conserved aspects of the functional brain organization of birds and mammals, and support the notion that insights on brain function and behavior gained from avian studies can be highly informative with regards to mammals, including humans.
A remarkable example is the study of songbirds, one of the few groups of organisms where juveniles are capable of learning their vocalizations by imitating a model, typically the song of an adult male, which is usually referred to as a tutor (Marler and Peters, 1977; Nottebohm, 1972). Songbird learning bears remarkable similarities to human speech learning, including the early production of immature babbling-like vocalizations (subsong), the requirement for auditory feedback during the sensorimotor vocal learning phase, and the occurrence of regional variations or dialects in the adult repertoire, characterizing a cultural transmission of vocal patterns (Doupe and Kuhl, 1999). Furthermore, vocal production and learning depend upon a set of discrete interconnected brain areas collectively known as the song control system (Nottebohm and Arnold, 1976), which includes nuclei in cortical-like areas, basal ganglia, and thalamus (Fig. 1). Different parts of this circuitry are involved in the production and/or learning of complex vocal patterns, as demonstrated very elegantly in zebra finches (authoritative reviews on the song system in Zeigler and Marler, 2004, 2008). Output projections from this circuitry allow for cortical-like areas to exert descending control over vocal and respiratory brain-stem areas, a connectivity feature that has only been seen in animals that have evolved vocal learning (Jarvis, 2004). This vocal control circuitry operates in concert with auditory processing centers, thought to be important for the perceptual processing and auditory memorization of song (Chew et al., 1995; Mello and Clayton, 1994; Bolhuis et al., 2001), which is also an essential step in songbird learning (London and Clayton, 2008). Besides vocal learning per se, studies of the vocal control system in finches and other song-bird species have contributed substantially to our understanding of brain sex dimorphisms, the effects of sex steroid on the brain and behavior, and neurogenesis and neuronal replacement in adult-hood, among several other contributions (reviewed in Zeigler and Marler, 2004, 2008).
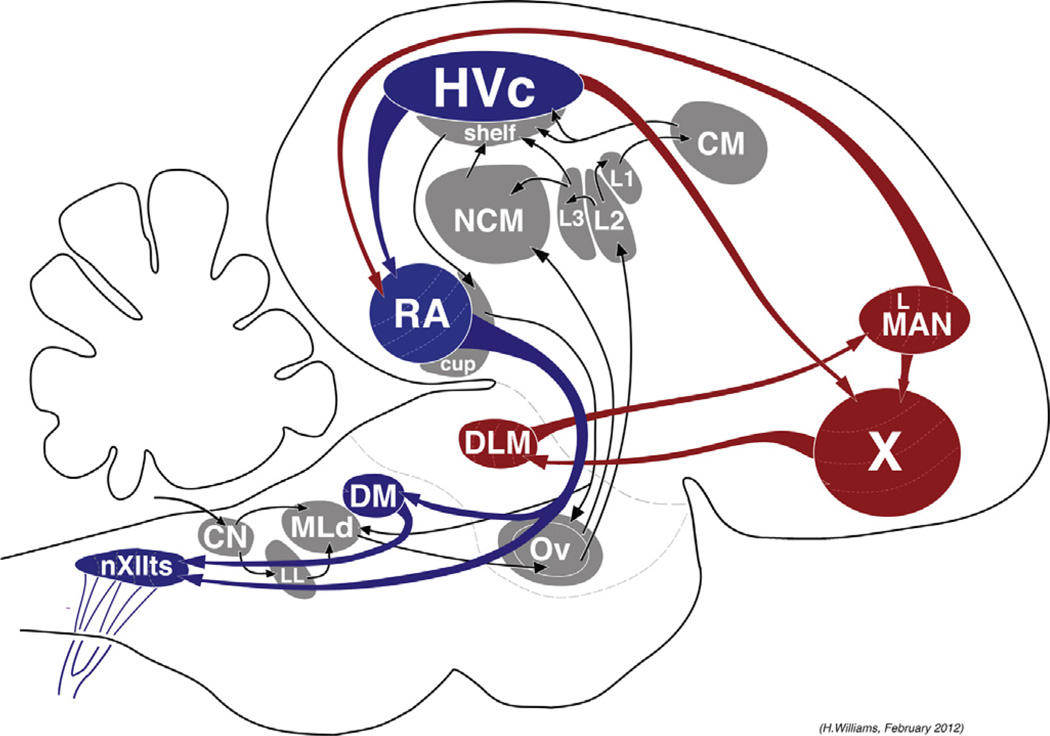
Fig. 1.
Simplified schematic diagram showing major elements of the song control system (sagittal view). Auditory input pathways are shown in gray: the experience of hearing song activates gene expression in many of these areas. The primary motor output pathway is in blue: the act of singing activates gene expression in these areas. The anterior forebrain pathway (AFP) is shown in red: this pathway is necessary for song learning and plasticity, and gene activity in its component nuclei varies with context of singing. Some additional elements (e.g., respiratory control pathways) are not shown but may be seen in a more comprehensive version of this figure produced and maintained online by Heather Williams (http://web.williams.edu/Biology/Faculty_Staff/hwilliams/Finches/circuits.html). Common abbreviations for the individual nuclei are used here; a full account of the circuitry is given in Reiner et al. (2004a). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
The detailed knowledge available on the anatomical and functional organization of the song system has set the stage for defining the molecular properties of its component auditory and vocal control pathways, and for identifying molecular and genetic correlates of learning. Early efforts benefitted greatly from the analysis of a few activity-inducible genes (a.k.a. immediate early genes, IEGs), including ZENK (a.k.a. zif-268, egr1, ngfia and krox-24) and c-fos (Mello and Clayton, 1994; Mello et al., 1992; Kimpo and Doupe, 1997 and reviewed in Clayton, 1997, 2000; Mello, 2002). More recently, new methods for broad scale high-throughput analyses have drastically changed the landscape of songbird research. Here we review the progression of this research from its early focus on IEGs to the current emphasis on high-throughput approaches. We consider both the insights gained and the challenges still to be met in future research.
2. Lessons/insights from single activity inducible genes
IEGs are rapidly and transiently induced in activated neuronal cells, and their study, which traditionally uses methods focusing on single genes, has been very useful for mapping brain activation (Mello et al., 1992; Kimpo and Doupe, 1997; Velho et al.,2005; Park and Clayton, 2002; Wada et al., 1521; Stripling et al.,2001; Chaudhuri and Cynader, 1993; Curran and Morgan, 1985; Morgan et al., 1987; McCabe and Horn, 1994; Bailey et al., 2002; Gentner et al., 2001; Phillmore et al., 2003). Importantly, animals are allowed to behave freely, thus minimizing issues related to stress or restraint. Furthermore, as one can map the entire brain through serial sections, this approach is unbiased, and can lead to novel and sometimes surprising findings. The method also allows for a direct assessment of activated cell populations, as well as the determination of their phenotype through multiple labelling or by combining gene expression with other approached such as tract-tracing. Importantly, IEGs like ZENK and c-fos encode transcription factors that can modify programs of gene expression in the activated cells (Clayton, 1997, 2000; Curran and Morgan, 1985; Morgan et al., 1987). Thus, mapping with IEGs like ZENK potentially reveals areas that are undergoing activity-induced neuroplasticity changes.
Due to a combination of all the features above, IEG expression analysis can contribute many important insights. In songbirds, analysis of the IEG ZENK has been instrumental for the identification and functional studies of brain structures that are activated in the context of vocal communication (Mello, 2002). Of particular interest, the act of hearing song was found to predominantly activate the caudomedial nidopallium (NCM) and mesopallium (CMM), structures subsequently shown to be part of the avian central auditory processing pathways (Mello and Clayton, 1994; Vates et al., 1996; Mello et al., 1998). In contrast, the act of singing activates IEG expression in the primary nuclei of the vocal control circuit (Kimpo and Doupe, 1997; Jarvis and Nottebohm, 1997; Jin and Clayton, 1997). This distinction, considered surprising at the time, clearly establishes distinct subsystems in the brain for perceiving vs. producing complex vocal signals. There is an interesting parallel in chicken, namely the demonstration that the IEG c-fos is induced in the intermediate medial mesopallium (IMM, named IMHV before implementation of the newer avian brain nomenclature; Reiner et al., 2004a) in the context of imprinting (McCabe and Horn, 1994), which contributed to solidifying the notion that this structure undergoes critical changes during the imprinting memory formation (Horn, 2004). Thus, findings in different avian species illustrate well the insights that can be gained from the expression analysis of single or a few genes in terms of localizing brain activation and possible sites of memory formation.
The early ZENK studies in finches triggered a broad interest in using IEG mapping to investigate the role that modulatory influences including experience, sex and hormonal status, developmental stages and others factors exert on song perceptual processing and memorization (Stripling et al., 2001; Mello et al., 2004; Maney et al., 2006; Bailey and Wade, 2005; Jeong et al., 2011). Other studies revealed components of the signal transduction machinery involved in song-induced IEG expression in NCM and CMM, in particular MAP kinases (Velho et al., 2005; Cheng and Clayton, 2004; Dong and Clayton, 2008). Manipulations of this pathway then provided highly compelling evidence that molecular events occurring in NCM are important for tutor song learning (London and Clayton, 2008). These findings helped cement a role for NCM in the auditory tutor song memorization, a role that had been suggested by correlative analysis of ZENK expression and strength of tutor song imitation (Bolhuis et al., 2000, 2001; Gobes and Bolhuis, 2007), as well as from electrophysiological evidence that NCM is the site of storage of learning dependent memory traces of the tutor song (Phan et al., 2006). Another important advance was the demonstration that synapsins are regulated by song in NCM and are a very likely downstream target of IEGs like ZENK (Velho and Mello, 2008). Recent evidence strongly points to noradrenergic transmission as a key regulator that triggers the gene induction cascade (Velho et al., 2012). Thus, neuronal stimulation by song leads to a cascade of molecular events, from signal transduction to early and downstream target gene activation, possibly resulting in long-lasting synaptic changes (Clayton, 2000; Velho et al., 2005; Moorman et al., 2012).
ZENK expression mapping has also provided novel and highly significant insights into the functional organization of vocal production pathways. For example, areas within the anterior forebrain pathway of the song system (Fig. 1), traditionally considered important for vocal learning, were shown to also be activated during singing in adulthood (Jarvis and Nottebohm, 1997; Jin and Clayton, 1997). This finding paralleled electrophysiological data demonstrating firing by neuronal cells within the basal ganglia parts of the vocal circuitry during singing in adults (Hessler and Doupe, 1999). These studies thus provided a much more accurate definition of brain areas that are activated during the active production of song in adults, and evidence that plasticity-related neuronal pathways remain actively involved in vocal control, even after the end of the critical period for vocal learning. Furthermore, activation of this anterior pathway was shown to be context dependent, being highest in contexts related to the practice of learned song, when vocal variability is high (Jarvis et al., 1998). These data have contributed significantly to elucidating a role of the basal ganglia in vocal plasticity and learning.
3. From single genes to pathways and networks
Altogether, studies of single or a few IEGs provided significant novel insights into vocal communication and learning in birds, but there has always been a strong suspicion that these early activity-regulated genes were just the tip of an iceberg. For example, many more genes could be temporally co-regulated with ZENK and other known IEGs, whereas yet other genes could be down-stream targets of early transcription factors, eventually leading to changes in neuronal cellular properties (Clayton, 2000; Velho et al., 2005; Lovell and Mello, 2011). Similarly, early studies of individual molecular markers (e.g. Clayton, 1997) provided only a very partial picture of gene regulation within the song control system. This all changed with the availability of several new molecular and genomic resources, in particular the construction of large libraries of annotated brain derived cDNAs/ESTs from the zebra finch (Wada et al., 1521; Replogle et al., 2008; Li et al., 2007), and more recently the closely related Bengalese finch (Kato and Okanoya, 2010), a species with a more elaborate syntax than the zebra finch. These libraries were deeply sequenced using automated procedures, and blast searches of NCBI databases were used to identify and annotate most of the genes and transcripts expressed in the zebra finch brain. The availability of these resources greatly facilitated the direct assessment of the brain expression of a vastly more comprehensive number of genes in a wide variety of songbird species, experimental paradigms, behavioral contexts, and laboratories.
As an example, the ESTIMA collection (Replogle et al., 2008) has enabled a systematic analysis utilizing in situ hybridization to determine the distribution of specific transcripts throughout the brain (e.g. Lovell et al., 2008, 2013; London and Clayton, 2010; Jarvis et al., 2013b). These efforts in turn culminated in the construction of a molecular atlas of the zebra finch brain (ZEBrA; www.zebrafinchatlas.org), an ongoing and expanding effort to map the distribution of a large set of transcripts on serial brain sections, in registration with a histological atlas (Karten et al., 2013). This large-scale approach in a songbird species is allowing the neurochemical characterization of areas of interest (e.g. nuclei of the song control system), the identification of molecular signatures for various brain areas, the identification of subregional molecular specializations within the major avian brain subdivisions, and the identification of unsuspected molecular relationships between different areas. The utility and broad use of the mouse brain atlas from the Allen Institute, with its many applications and adepts, attests to the importance of generating a comparable dataset in birds (Ng et al., 2009; Lein et al., 2007).
Under the NIH-supported Songbird Neurogenomics Initiative (SoNG; Replogle et al., 2008; Drnevich et al., 2012), the ESTIMA set of cDNA clones was also used to advance a broad collaborative effort to apply microarray technologies across a range of research questions involving songbirds. A set of non-redundant clones (~18,000) was selected for the construction of ~1000 glass slide microarrays, and 16 different research groups were recruited to participate via a standardized central pipeline for data generation and analysis. Initial tests demonstrated the feasibility of using zebra finch microarrays to monitor gene expression changes in several different songbird species and experimental paradigms (Replogle et al., 2008), taking advantage of the diversity of songbirds to explore a broad range of factors affecting brain gene regulation (e.g., regulatory modules in Fig. 2).
Fig. 2.
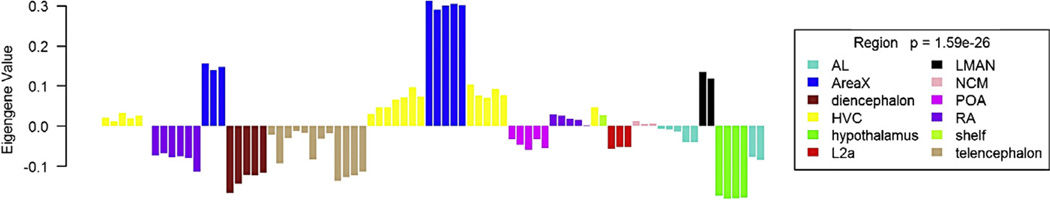
Normalized relative expression level for genes in Module 91 from the Songbird Neurogenomics Initiative meta-analysis (Drnevich et al., 2012). The figure illustrates an integration of data from 15 different experiments using a common microarray analysis pipeline. The WGCNA algorithm was used to sort ~18,000 probes into 95 modules based on co-expression across the 80 different treatment groups in the collected data. Each bar represents one treatment group from one experiment (i.e., one specific combination of laboratory, species, brain region, age, sex and experiential exposure). The bars are colored here according to tissue of origin, which helps visualize an emergent pattern in the data; the eigengenes consist of the sum of the vectors related to the expression levels of all genes within a given experiment. Of note, genes in Module 91 vary strongly according to brain region (p-value shown for main effect of region), with especially strong expression in Area X and also in lMAN, two nuclei of the anterior forebrain pathway in the song vocal control system (Fig. 1). Elevated expression is also seen in HVC, part of the song output control pathway (Fig. 1) whereas expression is notably low in the diencephalon and the shelf underlying HVC. There are 46 probes in this module. Gene ontology analysis revealed significant enrichment (FDR 0.019) for the term “dopamine metabolic process” in this gene set (Drnevich et al., 2012).
One of the studies under the SoNG Initiative performed a comprehensive accounting of the genes that are regulated by song exposure in higher auditory areas. Early studies using double labeling approaches revealed that several inducible genes (ZENK, c-fos, arc) are indeed co-regulated in the same cells (Velho et al., 2005). However, microarray analysis revealed that several hundred RNAs changed their expression in auditory areas within minutes after song stimulation onset, and an even larger number showed changes a day after repeated exposures had created a demonstrable memory of the song (Dong et al., 2009). Genes that increased in expression immediately after song playback tended to encode proteins involved in transcriptional control and RNA processing, whereas later changes occurred in genes involved in energetics and macromolecular synthesis. Interestingly, as many RNAs decreased as increased in response to song, with the most rapid changes involving ion channel genes and non-coding RNAs. An important focus of ongoing research is to identify mechanisms that coordinate these large-scale experience-dependent shifts in brain gene expression. MicroRNAs are of particular interest as they have now been shown to respond to song playbacks (Gunaratne et al., 2011) and to singing activity (Shi et al., 2013), and they can regulate the expression of multiple target mRNAs through post-transcriptional mechanisms. Of note, the singing-induced response is context dependent, pointing to an influence of social factors on the brain expression of microRNAs, an important novel insight contributed by songbird studies.
Other microarray studies under SoNG turned up evidence for large-scale changes in brain gene expression in response to other environmental, experiential and social factors. To cite a few examples, Mukai et al. (2009) found that the experience of a simulated territorial intrusion (effectively a song playback) caused changes in gene expression in the hypothalamus of song sparrows. The response differed in the breeding (spring) and nonbreeding (autumn) seasons, with 88 cDNAs showing significant interactions between season and the simulated intrusion. In both starlings and white-crowned sparrows, changes in photoperiod triggered complex gene response patterns in major telencephalic nuclei and hypothalamus (Thompson et al., 2012; Stevenson et al., 2012). The range of genes involved in these responses is wide, encompassing functions in thyroid hormone action and neuroplasticity, neurogenesis and angiogenesis, electrophysiology and epigenetic processes. Importantly, all studies under SoNG were coordinated, with samples processed and analyzed by a central laboratory, which facilitated a subsequent large scale meta-analysis to assess the relative contribution of a range of factors on brain gene expression (Drnevich et al., 2012). This analysis found that almost all genes in the brain vary their expression according to brain region, age, species and sex, with brain region acting as a dominating factor in differential gene expression (Fig. 2).
Microarray studies also influenced the molecular and neurochemical characterization of the song system. Early studies using in situ hybridization or immunohistochemistry (e.g., Clayton, 1997) revealed that hormone receptors, peptides, calcium binding and metabolic proteins are differentially expressed in different nuclei of the song system. However, these studies had to rely on tedious cloning efforts to obtain individual finch probes, due to poor cross-hybridization of mammalian probes with avian tissues, or the use of antibodies raised against epitopes from other species, often with no knowledge of the degree of sequence conservation or species cross-reactivity. The use of microarrays exponentially accelerated the rate of discovery. For example, a study using the SoNG microarrays revealed that several hundred genes are differentially expressed in nucleus HVC compared to the adjacent areas (dorsal nidopallium), representing molecular specializations of this song nucleus (Lovell et al., 2008). Many sets of markers represent novel molecular features of the song system related to axon guidance, cell death and proliferation, cell excitability, and others. Other efforts using different microarray platforms uncovered further evidence of differential gene expression in HVC of zebra finches (Li et al., 2007; Gunaratne et al., 2011) and Bengalese finch (Kato and Okanoya, 2010), greatly expanding the range of identified song nuclei markers. More recently, studies using an oligonucleotide array based on the complete zebra finch genome annotation have shown further evidence of complex variation in gene expression within and across brain regions involved in song control (Hilliard et al., 2012a,b). Overall, these findings helped solidify the notion that song nuclei possess molecular features that distinguish them from the rest of the brain, suggesting that pathways and processes within the song system are subject to distinct regulatory mechanisms. Together with the ZEBrA atlas, these studies have led to a greater understanding of the molecular makeup of song nuclei and identified several likely candidate regulators of the distinct properties of the song system.
Technological advances such as microarrays, which allow for monitoring the coordinated expression of vast cohorts of genes, have begun to bring us closer to a more realistic view of how the brain changes in response to a variety of modulatory factors. Collectively, results from the microarray era reveal that gene expression in the brain is anything but a simple “housekeeping” activity, and that neural tissue is exceedingly diverse in macromolecular makeup. Gene expression pattern analysis is exquisitely sensitive to detecting variation in physiological state, developmental history, and the animal’s immediate behavioral context and experience (Drnevich et al., 2012; Hilliard et al., 2012a). A major challenge moving forward will be to find ways to understand how distinct gene expression networks emerge in different brain systems, and to relate variations in specific gene expression to variations in neurological and behavioral function. Some initial progress has been made in this direction by applying new statistical tools for identifying sets of genes that are expressed in similar patterns across complex data sets (e.g., Weighted Gene Coefficient Network Analysis; WGCNA; Drnevich et al., 2012; Hilliard et al., 2012b) and for integrating predicted regulatory relationships (e.g. predicted target genes for regulated transcription factors) into dynamic models of gene expression changes (Warren et al., 2010). Achieving real progress with these approaches, however, may require very large and dense data sets to account for the high cellular heterogeneity of neural tissue. As attested by the outcomes of collaborative microarray efforts, these are challenges that are best met with an open approach, where different labs collaborate to generate resources, protocols and analytical methods of common interest, potentiating the rate of discovery and helping the field move forward fast.
The more recent use of transcriptome analysis by direct sequencing of RNA (RNAseq) is another important innovation that heralds yet new waves of discovery in the field. In this case, tissue RNAs are extracted and used for the construction of libraries, which are then sequenced with next generation technologies. In other organisms, applying advanced analytical tools for transcript counting has emerged as quantitatively more accurate than the kind of data provided by microarrays (e.g. Belgard et al., 2011, 2013). Besides being more sensitive to gene expression changes, this approach is also less biased as it does not depend on having a pre-established set of probes, thus overcoming some important limitations of microarrays. With steadily declining sequencing costs, this approach is becoming increasingly attractive to a broad range of researchers. Moreover, an important hurdle for its use has been removed with the recent availability of avian genomes (see below). As of this writing, published examples of RNAseq technology in songbird research include a contribution to the initial genome annotation (Warren et al., 2010), the characterization of song-responsive microRNAs (Gunaratne et al., 2011), a profiling of gene expression in two zebra finch cell lines (Balakrishnan et al., 2012), and the assembly of brain transcriptomes from songbird species included the violet eared waxbill (Balakrishnan et al., 2013), and three sparrow species (Balakrishnan et al., 2014). While still in its early days for the songbird research field, this kind of analysis holds great promise for further unraveling complex patterns of gene regulation in a tissue like the brain, and provides hope that we are approximating a more complete understanding of the molecular underpinnings of learning and memory. Overall, the challenge of efficiently handling the vast amounts of transcriptomics information and yet be able to extract functional correlates in a synthetic and comprehensible manner, for example by identifying the regulatory nodes in complex regulated networks, becomes increasingly more acute.
4. Avian genomes
A fundamental advance that occurred in parallel with the emergence of resources for large scale gene expression analysis was the sequencing and assembly of avian genomes, first the chicken (Gallusgallus; Hillier et al., 2004) followed by the zebra finch (Taeniopygiagutatta; Warren et al., 2010). The availability of these genomes, followed by annotation and ongoing curation efforts, provided for a first opportunity to define the complete set of genes in any avian species. This advance had several important implications for song-bird researchers.
For instance, applying predictive algorithms (e.g., Ensembl) to these genome sequences and/or mapping onto them the already well characterized orthologous genes from species like mouse or humans, greatly facilitated the characterization of the structure of genes of key interest, including the identification of exon/intron boundaries and of candidate promoter regions. The latter is then instrumental for defining gene regulatory elements (i.e. promoters). Another example is that the genome greatly expanded the usefulness of resources like the ESTIMA collection of cDNAs and microarrays, by allowing the mapping of numerous ESTs representing the 3′UTR region of transcripts which otherwise could not be identified through cross-species alignments with other vertebrate species due to low sequence conservation. The precise mapping of these previously unplaced transcripts in turn provided evidence for the brain expression of an even larger number of specific genes, as well as a larger set of useful molecular probes with defined specificity for expression analysis efforts like the ZEBrA atlas. Another aspect of practical importance was the availability of coding sequences that provide information on the conservation of specific epitopes that are used in other species for generating antibodies, including commercially available ones. The new avian genomic data have allowed researchers to make much more informed decisions with regards to the suitability of cross-reacting antibodies for use in avian tissue, and/or the need to develop antibodies against avian antigens for adequate tissue reactivity and specificity.
High quality genomes (i.e. genomes that have been sequenced with high redundancy to resolve ambiguous regions, and whose assemblies have few and/or short unsequenced gaps) also serve as references for rooting the complex data derived from transcriptome analysis (e.g. RNAseq), and open the door to tools for storing, accessing, and analyzing such data. Importantly, automated ab initio algorithms tend to predict the 5′and 3′non-coding regions poorly, and thus are usually not effective in the precise identification of transcription start sites, which is crucial for accurately defining promoter regions. In this regard, transcript mapping provides complementary evidence that confirms and often extends the output of gene prediction algorithms, improving genome annotations, as attested by the mapping of high quality zebra finch brainc DNA/EST databases with good representation of 5′UTR regions (Wada et al., 1521). However, these improved cDNA libraries do not contain all brain-expressed genes, and not all 5′UTR regions are represented. In contrast, transcriptome analysis using RNAseq provides a much more in-depth coverage of transcripts and is quickly becoming an invaluable source of information for analysis of gene expression patterns and for helping establish gene structure with precision. The application of effective long read technology to transcriptome sequencing (e.g. PacBio, Sharon et al., 2013) will likely have a major further impact on these issues, especially through better resolution of GC-rich regions and more accurate identification of transcript variants.
Defining the complete sets of genes present in avian genomes has also allowed for the precise identification of the set of 1-to-1 orthologous gene sets that are present in birds in comparison with other vertebrate organisms, as well as across different avian species that are representative of different lineages. A precise definition of orthologous gene sets is a fundamental requirement for studies of genome evolution trying to correlate the occurrence of novel and/or unique genomic features with the emergence of species- or lineage-specific phenotypic traits. Analysis of the chicken genome indeed revealed genomic correlates of some avian traits, such as the absence of vomeronasal receptors, keratins, casein and enamel genes that are associated with characteristic mammalian traits such as the vomeronasal organ, hair, milk production and lactation, and teeth. In contrast, some genes expanded in birds, including genes with SRCR domains, and a large family of olfactory receptors, potentially rewriting the dogma that birds have a poor sense of smell (Hillier et al., 2004). In large part these were corroborated by the zebra finch genome. Comparison of chicken and zebra finch, on the other hand, has allowed the identification of genomic features that appear as songbird specific, such as management of sex chromosome gene expression, accelerated evolution of ion transport genes, large expansions of specific gene families, including kinases (e.g. PAK3), PHF7, and MHC genes implicated in a range of regulatory processes (Lovell et al., 2013; Warren et al., 2010; Nam et al., 2010). While these features could potentially relate to the occurrence of characteristic songbird traits like the presence of vocal learning and associated brain structures, which are absent in species like the chicken, it is important to realize that these finches and chicken are phylogenetically very distant and differ in several other respects, most notably an altricial form of post-natal development in songbirds, as opposed to the precocious development typical of galliformes. Further comparative genomics involving species that provide a more complete coverage of avian radiations will be needed for establishing stronger correlations between genome features and lineage specific phenotypic traits. The increasing availability of a number of other avian genomes, including species like turkey, budgerigar and Darwin’s finch, already in NCBI, and the upcoming genomes from other sources (http://phybirds.genomics.org.cn/) holds great promise in this regard.
While the genomes are a tremendously useful resource, several significant challenges still remain. For example, the genome sequences are not complete, and contain numerous gaps that often disrupt the continuity of genes and interfere with the efficacy of gene predictive algorithms such as those used by Ensembl. As a result, numerous genes might appear to be incomplete or resemble pseudogenes, leading to erroneous annotations. In some severe cases of sequence disruption the gene fails to be predicted altogether, even though it is present at the correct syntenic location, as most effectively shown by local BLAT alignments. A related common issue is that many sequence gaps tend to occur in domains that correspond to the promoter region, likely due to the characteristic high GC content that render such regions particularly difficult to fully sequence. This problem considerably complicates the task of isolating and characterizing gene regulatory domains.
Another significant issue with the current avian genomes is the relatively frequent occurrence of apparent tandem gene duplications (Warren et al., 2010). These often have suspicious features, including the fact that one or both copies are often flanked by gaps and their percent identity is very high (often >98%), suggesting that they are allelic copy variants rather than real duplications with sequence divergence (Mello, unpublished). It seems likely that this kind of artifact is due to difficulties in correctly assembling regions containing repetitive sequences, especially when local sequence quality and/or coverage is low. Related problems include difficulties of assembling the small and highly repetitive W sex chromosome characteristic of birds, and poor mapping of genes and genome scaffolds to the numerous small microchromosomes that are also characteristic of birds. For example, the initial assembly of the zebra finch genome only defines chromosomes 1–27, with the remaining microchromosomes presumably represented by unassigned genome contigs (“chrUN”). Due to a combination of these and other factors, including the difficulties in correctly assembling complex heterozygous loci, gene prediction efforts using automated procedures are still fairly incomplete, compromising the ability to conduct accurate quantitative functional transcriptome analysis and comparative studies of genome evolution in birds.
Some of the approaches that are being taken to address these concerns include further sequencing with next generation technologies, gap-filling algorithms and novel assembly strategies. This approach has been successfully applied to improve the quality of the chicken genome (Galgal4; Warren, unpublished), and is now being applied to the zebra finch genome (Mello, Warren, unpublished). Also crucial are the efforts to manually curate the lists of orthologs present in avian genomes, especially focusing on candidate species-specific novel genomic features. As a representative example, optimized curation and gene alignment efforts have been applied to potassium channel genes, which are of critical importance for regulating neuronal excitability (Gutman et al., 2005; Wei et al., 2005). The effort of curating this gene family in the zebra finch genome corrected several annotation errors due to gene misidentifications, detected previously unidentified orthologs and paralogs, and generated more accurate data with regards to specific members undergoing high selective pressure in the avian lineage. Furthermore, several family members were found to be differentially expressed in the song system, in some cases representing remarkable molecular markers of song nuclei. These findings support the notion that selective gene regulation likely helps modulate basic neuronal properties within the vocal circuitry, and suggest that the evolution of avian vocal learning systems involved modified regulation of the brain expression of specific ion channel genes (Lovell et al., 2013). Systematically applying this comprehensive approach to other gene families involved in developmental circuitry assembly, neuronal physiology and behavioral regulation will likely bring novel fundamental insights into mechanisms that regulate the physiology and evolution of vocal learning and associated pathways in songbirds. Due to the large scale of the endeavor, conducting these efforts under a collaborative and open network is most likely to lead to rapid advances in this field.
Acknowledgments
We thank Peter Lovell for critical reading, suggestions and editorial assistance. C.V.M. received funding from the NIH/NIGMS (R24-GM092842).
References
- Bailey DJ, Wade J. FOS and ZENK responses in 45-day-old zebra finches vary with auditory stimulus and brain region, but not sex. Behav. Brain Res. 2005;162:108–115. doi: 10.1016/j.bbr.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Bailey DJ, Rosebush JC, Wade J. The hippocampus and caudomedial neostriatum show selective responsiveness to conspecific song in the female zebra finch. J. Neurobiol. 2002;52:43–51. doi: 10.1002/neu.10070. [DOI] [PubMed] [Google Scholar]
- Balakrishnan CN, Lin YC, London SE, Clayton DF. RNA-seq transcriptome analysis of male and female zebra finch cell lines. Genomics. 2012;100:363–369. doi: 10.1016/j.ygeno.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan CN, Chapus C, Brewer MS, Clayton DF. Brain transcriptome of the violet-eared waxbill Uraeginthus granatina and recent evolution in the songbird genome. Open Biol. 2013;3:130063. doi: 10.1098/rsob.130063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan CN, Mukai M, Gonser RA, Wingfield JC, London SE, et al. Brain transcriptome sequencing and assembly of three songbird model systems for the study of social behavior. PeerJ. 2014;2:e396. doi: 10.7717/peerj.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Montiel JF, Wang WZ, Garcia-Moreno F, Margulies EH, et al. Adult pallium transcriptomes surprise in not reflecting predicted homologies across diverse chicken and mouse pallial sectors. Proc. Natl. Acad. Sci. U. S. A. 2013;110:13150–13155. doi: 10.1073/pnas.1307444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis JJ, Zijlstra GG, den Boer-Visser AM, Van Der Zee EA. Localized neuronal activation in the zebra finch brain is related to the strength of song learning. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2282–2285. doi: 10.1073/pnas.030539097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis JJ, Hetebrij E, Den Boer-Visser AM, De Groot JH, Zijlstra GG. Localized immediate early gene expression related to the strength of song learning in socially reared zebra finches. Eur. J. Neurosci. 2001;13:2165–2170. doi: 10.1046/j.0953-816x.2001.01588.x. [DOI] [PubMed] [Google Scholar]
- Chaudhuri A, Cynader MS. Activity-dependent expression of the transcription factor Zif268 reveals ocular dominance columns in monkey visual cortex. Brain Res. 1993;605:349–353. doi: 10.1016/0006-8993(93)91765-k. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Clayton DF. Activation and habituation of extracellular signal-regulated kinase phosphorylation in zebra finch auditory forebrain during song presentation. J. Neurosci. 2004;24:7503–7513. doi: 10.1523/JNEUROSCI.1405-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew SJ, Mello C, Nottebohm F, Jarvis E, Vicario DS. Decrements in auditory responses to a repeated conspecific song are long-lasting and require two periods of protein synthesis in the songbird forebrain. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3406–3410. doi: 10.1073/pnas.92.8.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DF. Role of gene regulation in song circuit development and song learning. J. Neurobiol. 1997;33:549–571. [PubMed] [Google Scholar]
- Clayton DF. The genomic action potential. Neurobiol. Learn. Mem. 2000;74:185–216. doi: 10.1006/nlme.2000.3967. [DOI] [PubMed] [Google Scholar]
- Curran T, Morgan JI. Super induction of c-fos by nerve growth factor in the presence of peripherally active benzodiazepines. Science. 1985;229:1265–1268. doi: 10.1126/science.4035354. [DOI] [PubMed] [Google Scholar]
- Dong S, Clayton DF. Partial dissociation of molecular and behavioral measures of song habituation in adult zebra finches. Genes Brain Behav. 2008;7:802–809. doi: 10.1111/j.1601-183X.2008.00423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Replogle KL, Hasadsri L, Imai BS, Yau PM, et al. Discrete molecular states in the brain accompany changing responses to a vocal signal. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11364–11369. doi: 10.1073/pnas.0812998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupe AJ, Kuhl PK. Birdsong and human speech: common themes and mechanisms. Annu. Rev. Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- Drnevich J, Replogle KL, Lovell P, Hahn TP, Johnson F, et al. Impact of experience-dependent and -independent factors on gene expression in songbird brain. Proc. Natl. Acad. Sci. U. S. A. 2012;109(Suppl. 2):17245–17252. doi: 10.1073/pnas.1200655109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentner TQ, Hulse SH, Duffy D, Ball GF. Response biases in auditory forebrain regions of female songbirds following exposure to sexually relevant variation in male song. J. Neurobiol. 2001;46:48–58. doi: 10.1002/1097-4695(200101)46:1<48::aid-neu5>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Gobes SM, Bolhuis JJ. Birdsong memory: a neural dissociation between song recognition and production. Curr. Biol. 2007;17:789–793. doi: 10.1016/j.cub.2007.03.059. [DOI] [PubMed] [Google Scholar]
- Gunaratne PH, Lin YC, Benham AL, Drnevich J, Coarfa C, et al. Song exposure regulates known and novel microRNAs in the zebra finch auditory forebrain. BMC Genomics. 2011;12:277. doi: 10.1186/1471-2164-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman GA, Chandy KG, Grissmer S, Lazdunski M, McKinnon D, et al. International union of pharmacology. LIII. Nomenclature and molecular relationships of voltage-gated potassium channels. Pharmacol. Rev. 2005;57:473–508. doi: 10.1124/pr.57.4.10. [DOI] [PubMed] [Google Scholar]
- Hessler NA, Doupe AJ. Singing-related neural activity in a dorsal forebrain-basal ganglia circuit of adult zebra finches. J. Neurosci. 1999;19:10461–10481. doi: 10.1523/JNEUROSCI.19-23-10461.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard AT, Miller JE, Horvath S, White SA. Distinct neurogenomic states in basal ganglia subregions relate differently to singing behavior in songbirds. PLoS Comput. Biol. 2012a;8:e1002773. doi: 10.1371/journal.pcbi.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard AT, Miller JE, Fraley ER, Horvath S, White SA. Molecular microcircuitry underlies functional specification in a basal ganglia circuit dedicated to vocal learning. Neuron. 2012b;73:537–552. doi: 10.1016/j.neuron.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Miller W, Birney E, Warren WC Consortium atmot, C.G. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Horn G. Pathways of the past: the imprint of memory. Nat. Rev. Neurosci. 2004;5:108–120. doi: 10.1038/nrn1324. [DOI] [PubMed] [Google Scholar]
- Horn G, Nicol AU, Brown MW. Tracking memory’s trace. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5282–5287. doi: 10.1073/pnas.091094798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED. Learned birdsong and the neurobiology of human language. Ann. N. Y. Acad. Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Nottebohm F. Motor-driven gene expression. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4097–4102. doi: 10.1073/pnas.94.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F. For whom the bird sings: context-dependent gene expression. Neuron. 1998;21:775–788. doi: 10.1016/s0896-6273(00)80594-2. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Gunturkun O, Bruce L, Csillag A, Karten H, et al. Avian brains and a new understanding of vertebrate brain evolution. Nat. Rev. Neurosci. 2005;6:151–159. doi: 10.1038/nrn1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Yu J, Rivas MV, Horita H, Feenders G, et al. Global view of the functional molecular organization of the avian cerebrum: mirror images and functional columns. J. Comp. Neurol. 2013a;521:3614–3665. doi: 10.1002/cne.23404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Yu J, Rivas MV, Horita H, Feenders G, et al. A global view of the functional molecular organization of the avian cerebrum: mirror images and functional columns. J. Comp. Neurol. 2013b doi: 10.1002/cne.23404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JK, Burrows K, Tremere LA, Pinaud R. Neurochemical organization and experience-dependent activation of estrogen-associated circuits in the songbird auditory forebrain. Eur. J. Neurosci. 2011;34:283–291. doi: 10.1111/j.1460-9568.2011.07743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Clayton DF. Localized changes in immediate-early gene regulation during sensory and motor learning in zebra finches. Neuron. 1997;19:1049–1059. doi: 10.1016/s0896-6273(00)80396-7. [DOI] [PubMed] [Google Scholar]
- Karten HJ, Brzozowska-Prechtl A, Lovell PV, Tang DD, Mello CV, et al. Digital atlas of the zebra finch (Taeniopygia guttata) brain: a high resolution photo atlas. J. Comp. Neurol. 2013 doi: 10.1002/cne.23443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Okanoya K. Molecular characterization of the song control nucleus HVC in Bengalese finch brain. Brain Res. 2010;1360:56–76. doi: 10.1016/j.brainres.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Kimpo RR, Doupe AJ. FOS is induced by singing in distinct neuronal populations in a motor network. Neuron. 1997;18:315–325. doi: 10.1016/s0896-6273(00)80271-8. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li X, Wang XJ, Tannenhauser J, Podell S, Mukherjee P, et al. Genomic resources for songbird research and their use in characterizing gene expression during brain development. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6834–6839. doi: 10.1073/pnas.0701619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London SE, Clayton DF. Functional identification of sensory mechanisms required for developmental song learning. Nat. Neurosci. 2008;11:579–586. doi: 10.1038/nn.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London SE, Clayton DF. Genomic and neural analysis of the estradiol-synthetic pathway in the zebra finch. BMC Neurosci. 2010;11:46. doi: 10.1186/1471-2202-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Mello CV. Brain expression and song regulation of cholecystokinin gene in the zebra finch (Taeniopygia guttata) J. Comp. Neurol. 2011 doi: 10.1002/cne.22513. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Clayton DF, Replogle KL, Mello CV. Birdsong transcriptomics: neurochemical specializations of the oscine song system. PLoS ONE. 2008;3:e3440. doi: 10.1371/journal.pone.0003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Carleton JB, Mello CV. Genomics analysis of potassium channel genes in songbirds reveals molecular specializations of brain circuits for the maintenance and production of learned vocalizations. BMC Genomics. 2013;14:470. doi: 10.1186/1471-2164-14-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney DL, Cho E, Goode CT. Estrogen-dependent selectivity of genomic responses to birdsong. Eur. J. Neurosci. 2006;23:1523–1529. doi: 10.1111/j.1460-9568.2006.04673.x. [DOI] [PubMed] [Google Scholar]
- Marler P, Peters S. Selective vocal learning in a sparrow. Science. 1977;198:519–521. doi: 10.1126/science.198.4316.519. [DOI] [PubMed] [Google Scholar]
- McCabe BJ, Horn G. Learning-related changes in Fos-like immunoreactivity in the chick forebrain after imprinting. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11417–11421. doi: 10.1073/pnas.91.24.11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV. Mapping vocal communication pathways in birds with inducible gene expression. J. Comp. Physiol. A: Neuroethol. Sens. Neural. Behav. Physiol. 2002;188:943–959. doi: 10.1007/s00359-002-0347-1. [DOI] [PubMed] [Google Scholar]
- Mello CV, Clayton DF. Song-induced ZENK gene expression in auditory pathways of songbird brain and its relation to the song control system. J. Neurosci. 1994;14:6652–6666. doi: 10.1523/JNEUROSCI.14-11-06652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV, Vicario DS, Clayton DF. Song presentation induces gene expression in the songbird forebrain. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV, Vates GE, Okuhata S, Nottebohm F. Descending auditory pathways in the adult male zebra finch (Taeniopygia guttata) J. Comp. Neurol. 1998;395:137–160. [PubMed] [Google Scholar]
- Mello CV, Velho TA, Pinaud R. Song-induced gene expression: a window on song auditory processing and perception. Ann. N. Y. Acad. Sci. 2004;1016:263–281. doi: 10.1196/annals.1298.021. [DOI] [PubMed] [Google Scholar]
- Moorman S, Gobes SM, Kuijpers M, Kerkhofs A, Zandbergen MA, et al. Human-like brain hemispheric dominance in birdsong learning. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12782–12787. doi: 10.1073/pnas.1207207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987;237:192–197. doi: 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- Mukai M, Replogle K, Drnevich J, Wang G, Wacker D, et al. Seasonal differences of gene expression profiles in song sparrow (Melospiza melodia) hypothalamus in relation to territorial aggression. PLoS ONE. 2009;4:e8182. doi: 10.1371/journal.pone.0008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K, Mugal C, Nabholz B, Schielzeth H, Wolf JB, et al. Molecular evolution of genes in avian genomes. Genome Biol. 2010;11:R68. doi: 10.1186/gb-2010-11-6-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Bernard A, Lau C, Overly CC, Dong HW, et al. An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 2009;12:356–362. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- Nottebohm F. The origins of vocal learning. Am. Nat. 1972;106:116–140. [Google Scholar]
- Nottebohm F, Arnold AP. Sexual dimorphism in vocal control areas of the songbird brain. Science. 1976;194:211–213. doi: 10.1126/science.959852. [DOI] [PubMed] [Google Scholar]
- Park KH, Clayton DF. Influence of restraint and acute isolation on the selectivity of the adult zebra finch zenk gene response to acoustic stimuli. Behav. Brain Res. 2002;136:185–191. doi: 10.1016/s0166-4328(02)00129-8. [DOI] [PubMed] [Google Scholar]
- Phan ML, Pytte CL, Vicario DS. Early auditory experience generates long-lasting memories that may subserve vocal learning in songbirds. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1088–1093. doi: 10.1073/pnas.0510136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillmore LS, Bloomfield LL, Weisman RG. Effects of songs and calls on ZENK expression in the auditory telencephalon of field- and isolate-reared black capped chickadees. Behav. Brain Res. 2003;147:125–134. doi: 10.1016/s0166-4328(03)00155-4. [DOI] [PubMed] [Google Scholar]
- Reiner A, Perkel DJ, Mello CV, Jarvis ED. Songbirds and the revised avian brain nomenclature. Ann. N. Y. Acad. Sci. 2004a;1016:77–108. doi: 10.1196/annals.1298.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, et al. The avian brain nomenclature forum: terminology for a new century in comparative neuroanatomy. J. Comp. Neurol. 2004b;473:E1–E6. doi: 10.1002/cne.20119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle KL, Arnold AP, Ball GF, Band M, Bensch S, et al. The Songbird Neurogenomics (SoNG) Initiative: community-based tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 2013;31:1009–1014. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Luo G, Fu L, Fang Z, Wang X, et al. miR-9 and miR-140-5p target FoxP2 and are regulated as a function of the social context of singing behaviour in zebra finches. J. Neurosci. 2013;33:16510–16521. doi: 10.1523/JNEUROSCI.0838-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TJ, Replogle K, Drnevich J, Clayton DF, Ball GF. High through-put analysis reveals dissociable gene expression profiles in two independent neural systems involved in the regulation of social behavior. BMC Neurosci. 2012;13:126. doi: 10.1186/1471-2202-13-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stripling R, Kruse AA, Clayton DF. Development of song responses in the zebra finch caudomedial neostriatum: role of genomic and electrophysiological activities. J. Neurobiol. 2001;48:163–180. doi: 10.1002/neu.1049. [DOI] [PubMed] [Google Scholar]
- Thompson CK, Meitzen J, Replogle K, Drnevich J, Lent KL, et al. Seasonal changes in patterns of gene expression in avian song control brain regions. PLOS ONE. 2012;7:e35119. doi: 10.1371/journal.pone.0035119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vates GE, Broome BM, Mello CV, Nottebohm F. Auditory pathways of caudal telencephalon and their relation to the song system of adult male zebra finches. J. Comp. Neurol. 1996;366:613–642. doi: 10.1002/(SICI)1096-9861(19960318)366:4<613::AID-CNE5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Velho TA, Mello CV. Synapsins are late activity-induced genes regulated by birdsong. J. Neurosci. 2008;28:11871–11882. doi: 10.1523/JNEUROSCI.2307-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velho TA, Pinaud R, Rodrigues PV, Mello CV. Co-induction of activity-dependent genes in songbirds. Eur. J. Neurosci. 2005;22:1667–1678. doi: 10.1111/j.1460-9568.2005.04369.x. [DOI] [PubMed] [Google Scholar]
- Velho TA, Lu K, Ribeiro S, Pinaud R, Vicario D, et al. Noradrenergic control of gene expression and long-term neuronal adaptation evoked by learned vocalizations in songbirds. PLOS ONE. 2012;7:e36276. doi: 10.1371/journal.pone.0036276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Howard JT, McConnell P, Whitney O, Lints T, et al. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15212–15217. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei AD, Gutman GA, Aldrich R, Chandy KG, Grissmer S, et al. International union of pharmacology. LII. Nomenclature and molecular relationships of calcium-activated potassium channels. Pharmacol. Rev. 2005;57:463–472. doi: 10.1124/pr.57.4.9. [DOI] [PubMed] [Google Scholar]
- Zeigler HP, Marler P, editors. Ann. N. Y. Acad. Sci. 2004. Behavioral Neurobiology of Birdsong. [Google Scholar]
- Zeigler HP, Marler P, editors. Neuroscience of Birdsong. Cambridge Univ. Press; 2008. [Google Scholar]