Abstract
Autism spectrum disorder (ASD) is a severe developmental disorder of the central nervous system characterized by impairments in social interaction, communication, and range of interests and behaviors. The syndrome's prevalence is estimated to be as high as 1 in 150 American children yet its etiology remains largely unknown. Examination of observed cytogenetic variants in individuals with ASD may identify genes involved in its pathogenesis. As part of a multidisciplinary study, an apparently balanced de novo translocation between chromosomes 2 and 9 [46,XY,t(2;9)(p13;p24)] was identified in a subject with pervasive developmental disorder not otherwise specified (PDD-NOS), and no distinctive dysmorphic features. Molecular characterization of the rearrangement revealed direct interruption of the RAB11 family interacting protein 5 (RAB11FIP5) gene. RAB11FIP5 is a Rab effector involved in protein trafficking from apical recycling endosomes to the apical plasma membrane. It is ubiquitously expressed and reported to contribute to both neurotransmitter release and neurotransmitter uptake at the synaptic junction. Detailed analysis of the rearrangement breakpoints suggests that the reciprocal translocation may have formed secondary to incorrect repair of double strand breaks (DSBs) by nonhomologous end-joining (NHEJ).
Keywords: autism, translocation, genetic, RAB11 family interacting protein 5 (class I), human, double-stranded DNA break, type I DNA topoisomerase
INTRODUCTION
Autism spectrum disorder (ASD) [Online Mendelian Inheritance in Man (OMIM) 209850] is a severe developmental disorder of the central nervous system. It is characterized by impairments in three behavioral areas: social interaction; verbal and non-verbal communication; and range of interests, activities and patterns of behavior [American Psychological Association, 1994]. The syndrome is divided into five DSM-IV subtypes: Autistic Disorder, Asperger's Disorder, Disintegrative Disorder, Pervasive Developmental Disorder Not Otherwise Specified (PDD-NOS) and Rett Disorder [American Psychological Association, 1994]. With the exception of Rett Disorder, diagnosis and classification of ASD depends on the pattern of observed behaviors; reliable neurobiological or genetic markers do not exist for most cases of the disorder [Bienvenu and Chelly, 2006]. Given the prevalence of the disorder (as high as 1 in 150 American children), a clearer understanding of etiology is necessary for diagnostic and therapeutic purposes [Centers for Disease Control and Prevention, 2007].
One approach to candidate gene identification involves molecular analysis of observed cytogenetic variants in affected individuals. This strategy has facilitated positional cloning of disease genes in several disorders, including complex and late onset diseases such as schizophrenia and diabetes [Bache et al., 2006]. We describe an apparently balanced de novo translocation between chromosomes 2 and 9 [46,XY,t(2;9)(p13;p24)] in a subject with Pervasive Developmental Disorder Not Otherwise Specified (PDD-NOS). Trans-locations may affect the expression of genes through a variety of mechanisms including, most directly, interruption of specific genes, implicating such genes strongly in disease etiology [Abrams and Cotter, 2004].
MATERIALS AND METHODS
Cytogenetic Analysis
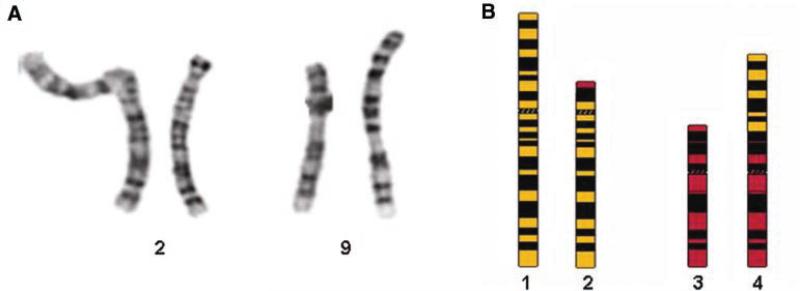
Ninety-two individuals with ASD and their biological parents were recruited into a genetic study of the syndrome [see Roohi et al., in press for study details]. Initial evaluations included cytogenetic analysis of each subject. Twenty trypsin-Giemsa banded metaphase spreads from two culture vessels were analyzed for chromosome number and structure in the proband described in this report. All cells had a normal chromosome number of 46 with one X chromosome and one Y chromosome. In addition all cells had an apparently balanced reciprocal translocation between part of the short arm of one chromosome 2 homolog and part of the short arm of one chromosome 9 homolog [i.e., t(2;9)(p13;p24)]. All other chromosomes were structurally normal at a resolution of 650 chromosome bands (Fig. 1). Parental chromosome analyses revealed that the translocation was de novo. Paternity was confirmed across 16 genetic markers by The Genetic Testing Laboratories, Inc. (www.gtldna.com).
Fig. 1.
De novo chromosomal rearrangement in an autistic male. A: Partial Karyotype of a 46,XY,t(2;9)(p13;p24) individual. B: Ideogram of the Proband's normal chromosome 2, normal chromosome 9 and the two derivative chromosomes. (1) normal chromosome 2; (2) chromosome 2 derivative; (3) normal chromosome 9; (4) chromosome 9 derivative.
Flow Karyotype and Fluorescent In Situ Hybridization (FISH)
The chromosome 9 derivative was isolated as previously described [Dumas et al., 2007]. Purity of the flow sort was confirmed with FISH as previously described [Dumas et al., 2007]. The chromosome 2 derivative co-migrated with normal chromosomes and could not be collected individually.
Bacterial Artificial Chromosome (BAC) Microarray Analysis
The subject's chromosome 9 derivative and chromosomes 2 and 9 from a normal donor were Phi29 amplified (Genomiphi V2 kit, GE Healthcare, Cat# 25-6600-31) and fluorescently labeled with BioPrime Array CGH Genomic Labeling Module (Invitrogen, Cat# 18095-12) (chromosome 9 derivative, Cy3/chromosome 2+chromosome 9, Cy5). Unincorporated nucleotides were removed with G50 column purification (GE Healthcare Cat# 27-5330-01). Fifty microliters of each labeling reaction was combined with 100 μg of Cot-1 and 500 μg of yeast tRNA. The material was ethanol precipitated, resuspended in 34.5 μl of TE, and then heated for 100°C for 10 min. 45.5 μl of 1.6× SlideHyb Buffer #3 (Ambion, Cat# AM8863) (concentrated with a speedvac) was added and the sample incubated at 37°C for 1 hr. Hybridization was performed onto a tiling path BAC array as previously described [Christian et al., in press]. As part of a larger study, BAC microarray analysis was also performed on the subject's genomic DNA [Christian et al., in press].
Oligonucleotide Microarray Analysis
One microgram of the subject's phi29 amplified chromosome 9 derivative and 1 μg each of phi29 amplified chromosomes 2 and 9 was fluorescently labeled with Cy3 or Cy5 as previously described [Selzer et al., 2005]. Hybridization was performed onto a custom 385,000 oligonucleotide NimbleGen fine tiling array spanning chr2: 73,030,173–73,429,557 and chr9: 1–5,000,000. Probes were selected from repeat-masked sequence at an average spacing of 7bp. Cy3/Cy5 [Chr.9 Der./(Chr2 + Chr9)] fluorescent ratios were calculated for each spot after image processing with NimbleScan version 2.3 software (NimbleGen Systems, Inc., Madison, WI). Rearrangement breakpoints were determined by automated segmentation analysis of data sets and the test versus reference log 2 ratios averaged at window sizes corresponding to 1× and 10× the median probe spacing. Data was visualized with SignalMap version 1.9.0.03 software (NimbleGen Systems, Inc.).
Polymerase Chain Reaction (PCR)
PCR was performed to amplify the coding regions of Rab11FIP5 and the derivative chromosome junction fragments in the proband. Amplification of exons was performed as part of an ongoing Hi-Res Melt study of RAB11FIP5 in a cohort of individuals with ASD. In addition to Hi-Res Melt analysis, the proband's products were sequenced directly. PCR conditions are described in Table S1. PCR of the chromosome 9 derivative junction fragment was carried out in a 50 ml reaction mixture containing 50 ng DNA, 0.2 mM dNTP, 1 μM of each primer, 1.25 U HotMaster Taq polymerase, and 1× HotMaster buffer (Fisher, Cat # FP2200310). Reactions were held at 95°C for 2 min, followed by 35 cycles of denaturation at 95°C for 30 sec, annealing at 63°C for 30 sec, and extension at 68°C for 30 sec, followed by a final extension at 68°C for 10 min. Products were visualized on an HDA-GT12 Genetic Analyzer. PCR primers unique to the chromosome 9 derivative were designed using the oligonucleotide array data (chr2FTGAAGGTGTCTGCAGCTTTC, chr9R-TGAAGTCTCTGTCCTGATAATGG).
To identify the chromosome 2 derivative breakpoints, inverse PCR was performed as previously described [Sambrook and Russell, 2001]. Genomic DNA from the proband was digested with DpnII and ligations performed at 0.1, 0.5, and 1 ng/μl concentrations. Primers were designed using the chromosome 9 derivative breakpoints and the ligation products amplified with the reaction conditions described above. Ligation products were amplified with DpnII_chr9FGCACATCTGGAAATCATTGATC and DpnII_chr9R-CCTCAGAAGTCCTCTCTTTTGG or DpnII_chr2F-TGTAGGCCCTTCTGGAGTGT and DpnII_chr2R-GATGCAGATGAGAGGCT-G. PCR primers unique to the chromosome 2 derivative were designed after successful amplification of the junction fragment by inverse PCR (chr9F-TGGAAGAAGCAATGGGGTTT and chr2R-AAGCGTGAGTCGGAGGAGT).
Junction DNA Analysis
GC-content profiles were drawn in draw_chromosome.gc.pl [Paces et al., 2004]. Repetitive elements were identified using the UCSC Genome Browser tracks and RepeatMasker, and Tandem-Repeat Finder [Benson and Dong, 1999; Jurka 2000; Smit et al., 2007]. The EMBOSS program EINVERTED was applied to search the translocation breakpoints for palindromes [Rice et al., 2000]. Regions of sequence similarity were identified with NCBI Blast 2 Sequences (http://www.ncbi.nlm. nih.gov/blast/bl2seq/wblast2.cgi) [Tatusova and Madden, 1999]. Z-DNA was predicted by ZHUNT (http://gac-web.cgrb.oregonstate.edu/zdna) [Ho et al., 1986]. The dreg program from The University of California at Merced (http://ccb.ucmerced.edu/cgi-bin/app/emboss/index/dreg) was used to identify sequence motifs associated with mutations such as deletions and translocations.
RESULTS
Clinical Findings
The proband was evaluated through this study at 10 years 4 months of age with a diagnosis of PDD-NOS. He was born full-term with normal growth parameters after an uncomplicated prenatal course to non-consanguineous, reportedly non-autistic, Caucasian parents. Family history was significant for one maternal first cousin and one paternal first cousin with autism. Parental concern first arose at the age of 18 months due to poor verbal communication, repetitive behaviors and minimal eye contact. His first single words were noted at the age of 7 months but phrase speech was absent until 4 years of age. He was diagnosed with hypotonia at 3 years of age and brain MRI and EEG were unremarkable at that time. Seizures, self-injurious behaviors, regression, auditory and visual deficits were not present. IQ was reported as 80. He received speech, physical therapy and alternative schooling.
At 10 years of age he was described as increasingly social but lacking some ability to understand abstract concepts with persistence of occasional repetitive behaviors. His height, weight and head circumference were at 3%, 97%, and 15%, respectively. He lacked significant dysmorphic features aside from mild hypertelorism and bilateral 5th finger clinodactyly.
Molecular Analysis
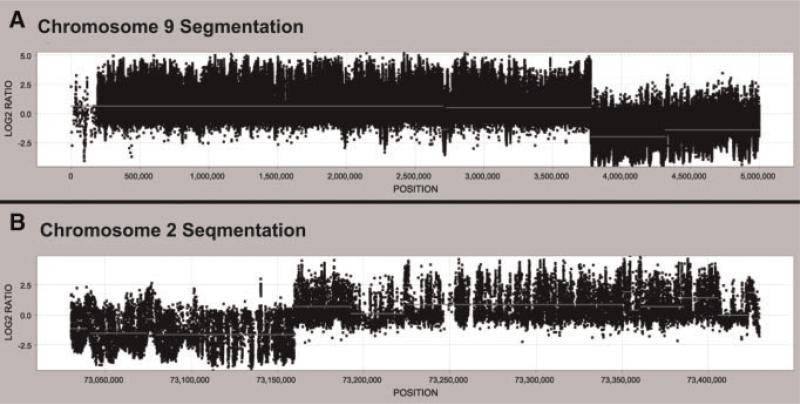
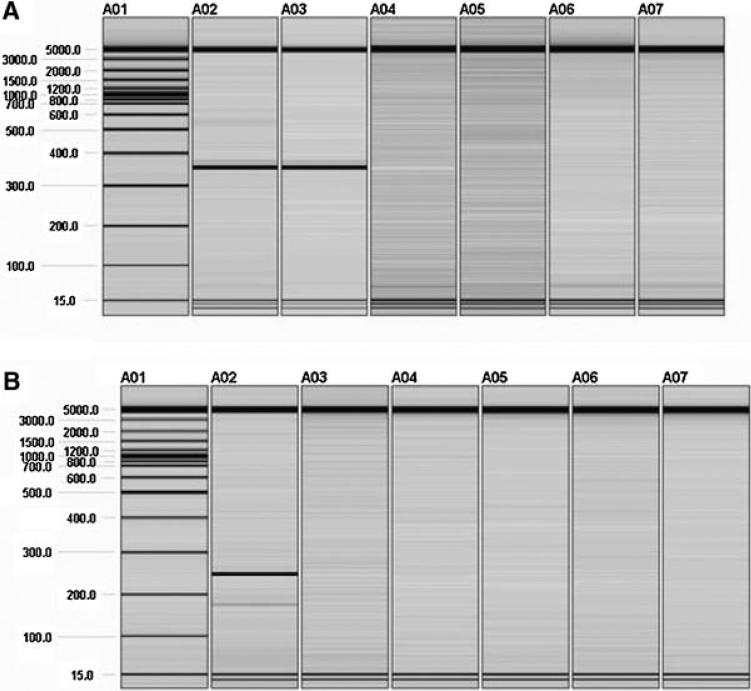
Cytogenetic analysis identified an apparently balanced de novo translocation between chromosomes 2 and 9 [46,XY,t(2;9)(p13;p24)] in an individual with ASD (Fig. 1). Whole genome BAC microarray analysis of the proband's genomic DNA failed to detect any copy number variations at a resolution of ~150 kb (data not shown). To investigate the rearrangement further, chromosome flow sorting was performed and the chromosome 9 derivative successfully isolated (data not shown). This derivative chromosome, normal chromosome 2, and normal chromosome 9 were Phi29 amplified and hybridized onto a tiling path BAC array, roughly mapping the translocation breakpoints to between clones RP11-91O12 and RP11-87C16 on chromosome 2 and clones RP11-17L15 and RP11-632L5 on chromosome 9 (data not shown). The rearrangement was characterized in greater detail with a custom fine tiling oligonucleotide array designed with probes spanning the approximate translocation breakpoints (Fig. 2). The oligonucleotide array data was used to design PCR primers unique to the chromosome 9 derivative (chr2F and chr9R). These primers amplified a 384 bp product in the proband and the proband's flow sorted chromosome 9 derivative. No product was seen in the proband's mother or father or in a normal control genomic DNA sample (Fig. 3A). Sequencing of the junction fragment allowed delineation of the rearrangement boundaries; the 384bp product mapped to chr2: 73,159,733–73,159,877 and chr9: 3,775,575–3,775,783 (Table S2).
Fig. 2.
Oligonucleotide fine tiling array. A custom NimbleGen fine tiling array spanning positions chr2: 73,030,173–73,429,557 and chr9: 1–5,000,000 was used to map the rearrangement breakpoints. The Cy3/Cy5 log 2 ratios [Chr.9 Der./(Chr2 + Chr9)] deviate significantly from 0 at chr2: 73,159,900 and ~chr9: 3,775,600, indicating the approximate rearrangement breakpoints.
Fig. 3.
Junction fragment PCR. A: Amplification with primers unique to the chromosome 9 derivative. A 384 bp product was amplified in the proband (A02) and the proband's flow sorted chromosome 9 derivative (A03). No product was amplified in the proband's mother (A04), father (A05), a normal control (A06) or water (A07). B: Amplification with primers unique to the chromosome 2 derivative. A 248 bp product was amplified in the proband (A02). No product was amplified in the proband's flow sorted chromosome 9 derivative (A03), the proband's mother (A04), the proband's father (A05), a normal control (A06) or water (A07).
Primers were designed to amplify the chromosome 2 derivative junction fragment using the rearrangement breakpoints identified above. However, PCR failed to generate a product in the proband, suggesting the rearrangement could be unbalanced. A small deletion, duplication, or insertion may escape detection by microarray analysis or karyotype. Inverse PCR and sequencing were performed to investigate the chromosome 2 derivative further. Primers were designed assuming at least one of the breakpoints identified for the chromosome 9 derivative was the same in the other derivative chromosome. Amplification with DpnII_chr9F and DpnII_chr9R yielded a 350 bp derivative chromosome fragment (data not shown). Primers were subsequently designed to amplify the chromosome 2 derivative junction fragment from genomic DNA (chr9F and chr2R) (Fig. 3B). PCR performed with these primers amplified a 248 bp product in with the proband's genomic DNA but not with DNA from the flow sorted chromosome 9 derivative or genomic DNA from the proband's mother, the proband's father, or a normal control. Sequencing mapped the derivative 2 product to chr9: 3,775,403–3,775,566 and chr2: 73,160,921–73,161,005 (Table S2). RAB11FIP5 is the only gene directly interrupted by the rearrangement. Exonic sequencing of RAB11FIP5 failed to identify any other mutations in the proband (data not shown).
Junction fragment analysis included examination of the rearrangement breakpoint regions for repetitive elements with RepeatMasker and Tandem-Repeat Finder. Only the chromosome 9 breakpoints were within or near a repetitive element. Specifically, these breakpoints fell within a MSTD element (a long terminal repeat). Alignment of the trans-location junctions with Blast 2 Sequences found no sequence similarity. Using draw_chromosome.gc.pl, the GC-content profiles for 4 Mb, 100 kb, and 10 kb around the translocation breakpoints were examined with sliding windows of 10,000, 1,000, and 100 bp, respectively. The chromosome 2 breakpoints fell within a region of high GC content (data not shown). In comparison, the chromosome 9 breakpoints were within a region of average GC content (data not shown). ZHUNT failed to predict Z-DNA at or near any of the translocation breakpoints. A search for palindromes with EINVERTED uncovered none. However, examination of the rearrangement breakpoints for motifs associated with mutation found the verterbrate/plant topoisomerase I consensus within 7 bp of all four translocation breakpoints (Fig. 4).
Fig. 4.
Localization of vertebrate/plant topoisomerase I consensus cleavage sites at rearrangement breakpoints. Topoisomerase I recognizes the following motifs: CAT, CTY, GTY, RAT. Sequence present on the derivative chromosome indicated by 8 bp long black bar. Topoisomerase I recognition site indicated by 3 bp long black bar. A: Topoisomerase I recognition site at chromosome 9 derivative breakpoint on chromosome 2. B: Topoisomerase I recognition site at chromosome 9 derivative breakpoint on chromosome 9. C: Topoisomerase I recognition site at chromosome 2 derivative breakpoint on chromosome 2. D: Topoisomerase I recognition site near chromosome 2 derivative breakpoint on chromosome 9.
DISCUSSION
The reciprocal translocation described in our subject affects only one gene directly, Rab11 family interacting protein 5 (RAB11FIP5) on chromosome 2. Along with the Ras GTPase Rab11, RAB11FIP5 is localized to subapical recycling endosomes (ARE) and the apical plasma membrane of polarized epithelial cells where it is involved in vesicle recycling, plasma membrane recycling, and transcytosis [Prekeris et al., 2000; Hales et al., 2001]. In neurons, recycling endosomes participate in the sorting and transport of neurotransmitter receptors. Park et al. found that recycling endosomes containing Rab11, in conjunction with myosin V, trafficked glutamate receptors to postsynaptic sites, contributing to long-term potentiation (LTP) of synaptic strength [Park et al., 2004; Lise et al., 2006]. RAB11FIP5, an effector of Rab11, is possibly involved in this process as well. This protein has been localized to the post synaptic density (PSD), a cellular structure specialized in receiving and transducing synaptic information, indicating it may play a role in postsynaptic signal transduction [Jordan et al., 2004]. In addition, RAB11FIP5 may contribute to the release of neurotransmitters into the synapse. A proteomic analysis of in vivo phosphorylated synaptic proteins identified it as a protein of the presynaptic active zone (a region where vesicles dock, fuse, and release their neurotransmitters into the synaptic cleft) [Collins et al., 2005]. Taken together, these findings suggest RAB11FIP5 is involved in neuro-transmitter signal transduction, possibly participating in neurotransmitter release and uptake [Jordan et al., 2004].
In our proband, the translocation directly interrupts RAB11FIP5, causing a complete loss of function of the gene product from the affected chromosome. No deleterious mutations were detected in the proband's uninterrupted copy of RAB11FIP5, suggesting that haploinsufficiency of RAB11-FIP5 may contribute to subject's ASD. Certain gene functions are inherently dosage-sensitive. These include gene products involved in signal transduction with a function dependent on partial or variable occupancy of a receptor, DNA-binding site, etc.; gene products that compete with each other to determine a developmental or metabolic switch; and gene products that co-operate with each other in interactions with fixed stoichiometry [Fisher and Scambler, 1994; Strachan and Read, 1996]. RAB11FIP5 and Rab11 form a complex with a 1:1 stoichiometry and perhaps haploinsufficiency of either gene may be sufficient to produce a pathologic phenotype such as ASD [Meyers and Prekeris, 2002].
The mechanism of translocation formation is unknown for most rearrangements [Gajecka et al., 2006]. In some cases, a reciprocal translocation may be the result of two random double-strand breaks (DSBs) followed by ligation between these broken chromosomes or the result of errors in programmed DSB repair mechanisms [Gajecka et al., 2006]. Two general mechanisms mediate DNA repair: homologous recombination (HR) and nonhomologous end-joining (NHEJ). Little is known about how these repair pathways contribute to a rearrangement but NHEJ is often suspected as the mechanism when a translocation occurs between regions of little or no homology and there is an additional genomic alteration at the breakpoint junction such as a small deletion, duplication, or insertion [Gajecka et al., 2006].
To uncover potential factors contributing to the trans-location, the rearrangement breakpoints were investigated in detail. No sequence homology, including repetitive elements, was identified between the translocation breakpoints. In addition, detailed molecular analysis of the derivative chromosomes detected two small deletions, 1,044 bp on chromo-some 2 (chr2: 73,159,878–73,160,920) and 8bp on chromosome 9 (chr9:3,775,567–3,775,574). Taken together, this data suggested NHEJ and improper repair of a DSB were involved in the creation of the translocation. DSBs may occur in regions prone to the formation of non-B DNA conformations, such as Z DNA. DNA features like a high GC-content or palindromes may promote such conformations but analysis of the junction fragment regions found neither [Gajecka et al., 2006]. Examination of the translocation breakpoints for sequence motifs associated with site-specific recombination, mutation, cleavage, and gene rearrangement identified the verterbrate/plant topoisomerase I consensus sequence at, or near, all four breakpoints [Abeysinghe et al., 2003]. Incorrect repair of DNA topoisomerase I mediated DSBs may have been involved in the generation of the translocation. DNA topoisomerase I creates transient single-strand breaks while relaxing supercoils in DNA. These single-strand breaks can be converted into DSBs upon collision with replication forks [Adachi et al., 2004]. Repair of topoisomerase I induced DSBs by NHEJ, instead of HR, may create mutations, including reciprocal translocations [Adachi et al., 2004]. This mechanism is perhaps responsible for the formation of the reciprocal translocation observed in our subject.
The subject's family history is positive for ASD but there are no affected siblings. Furthermore, the presence of autism in both maternal and paternal relatives suggests multiple mechanisms in this family (the parents are nonconsanguineous). Because of the high prevalence of ASD in the general population, it is likely that in some families, different individuals with autism may have different underlying causes. Several siblings, from multiplex families, have already been described with different causes of ASD [Szatmari et al., 2007; Alarcon et al., 2008]. Despite the reasoning stated above, it is still possible that the disruption of RAB11FIP5 in our proband may be coincidental and not the direct cause of his PDD-NOS. However, we believe that disruption of RAB11FIP5 may contribute to ASD in our proband, for the following reasons. The gene's sequence is highly conserved and RAB11FIP5 plays an important role in the recycling endosomes. To date, no structural variation in this region has been described in normal individuals. In addition, RAB11FIP5's possible roles at both the presynaptic and postsynaptic junctions suggests it may be a strong candidate gene for the disorder. Several diseases have been linked to mutations or altered functioning of Rab proteins or Rab effectors, including mental retardation [Bienvenu et al., 1998; Stein et al., 2003; Strutz-Seebohm et al., 2006]. RAB11FIP5 may also be involved in these disorders, as it has been shown that, generally, the pathology is the same whether it is the effector or the GTPase is itself that is not functioning [Stein et al., 2003]. Our future plans include examining RAB11FIP5 in large cohorts of normal individuals and individuals affected with ASD.
ACKNOWLEDGMENTS
This work was supported in part by the Cody Center for Autism and Developmental Disabilities, funding from the SBUMC GCRC MO1RR10710 and a Clinical Research Scholar grant to DHT through the Stony Brook University School of Medicine. This research was supported in part by the Intramural Research Program of the NIH, NCI, CCR, NCI-Frederick.
Footnotes
Please cite this article as follows: Roohi J, Tegay DH, Pomeroy JC, Burkett S, Stone G, Stanyon R, Hatchwell E. 2008. A De Novo Apparently Balanced Translocation [46,XY,t(2;9)(p13;p24)] Interrupting RAB11FIP5 Identifies a Potential Candidate Gene for Autism Spectrum Disorder. Am J Med Genet Part B 147B:411–417.
REFERENCES
- Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum Mutat. 2003;22(3):229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- Abrams L, Cotter PD. Prenatal diagnosis of de novo X;autosome translocations. Clin Genet. 2004;65(5):423–428. doi: 10.1111/j.0009-9163.2004.00255.x. [DOI] [PubMed] [Google Scholar]
- Adachi N, So S, Koyama H. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J Biol Chem. 2004;279(36):37343–37348. doi: 10.1074/jbc.M313910200. [DOI] [PubMed] [Google Scholar]
- Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychological Association . Diagnostic and Statistical Manual of Mental Disorders. American Psychological Association; Washington, DC: 1994. [Google Scholar]
- Bache I, Hjorth M, Bugge M, Holstebroe S, Hilden J, Schmidt L, Brondum-Nielsen K, Bruun-Petersen G, Jensen PK, Lundsteen C, et al. Systematic re-examination of carriers of balanced reciprocal translocations: A strategy to search for candidate regions for common and complex diseases. Eur J Hum Genet. 2006;14(4):410–417. doi: 10.1038/sj.ejhg.5201592. [DOI] [PubMed] [Google Scholar]
- Benson G, Dong L. Reconstructing the duplication history of a tandem repeat. Proc Int Conf Intell Syst Mol Biol. 1999:44–53. [PubMed] [Google Scholar]
- Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: When DNA methylation goes unrecognized. Nat Rev Genet. 2006;7(6):415–426. doi: 10.1038/nrg1878. [DOI] [PubMed] [Google Scholar]
- Bienvenu T, des Portes V, Saint Martin A, McDonell N, Billuart P, Carrie A, Vinet MC, Couvert P, Toniolo D, Ropers HH, et al. Non-specific X-linked semidominant mental retardation by mutations in a Rab GDP-dissociation inhibitor. Hum Mol Genet. 1998;7(8):1311–1315. doi: 10.1093/hmg/7.8.1311. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Autism Information Center. 2007.
- Christian S, Sudi J, Kumar RA, Liu S, KaraMohamed S, Badner JA, Matsui S, Conroy J, McQuaid D, Gergel J, Hatchwell E, Conrad Gilliam T, Gershon ES, Nowak NJ, Dobyns WB, Cook EH., Jr Novel submicroscopic chromosomal abnormalities detected in Autism Spectrum Disorder. Biological Psychiatry. doi: 10.1016/j.biopsych.2008.01.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, Yu L, Coba MP, Husi H, Campuzano I, Blackstock WP, Choudhary JS, Grant SG. Proteomic analysis of in vivo phosphorylated synaptic proteins. J Biol Chem. 2005;280(7):5972–5982. doi: 10.1074/jbc.M411220200. [DOI] [PubMed] [Google Scholar]
- Dumas F, Stanyon R, Sineo L, Stone G, Bigoni F. Phylogenomics of species from four genera of New World monkeys by flow sorting and reciprocal chromosome painting. BMC Evol Biol. 2007;7(Suppl 2):S11. doi: 10.1186/1471-2148-7-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher E, Scambler P. Human haploinsufficiency—One for sorrow, two for joy. Nat Genet. 1994;7(1):5–7. doi: 10.1038/ng0594-5. [DOI] [PubMed] [Google Scholar]
- Gajecka M, Pavlicek A, Glotzbach CD, Ballif BC, Jarmuz M, Jurka J, Shaffer LG. Identification of sequence motifs at the breakpoint junctions in three t(1;9)(p36.3;q34) and delineation of mechanisms involved in generating balanced translocations. Hum Genet. 2006;120(4):519–526. doi: 10.1007/s00439-006-0222-1. [DOI] [PubMed] [Google Scholar]
- Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EK, Lapierre LA, Goldenring JR. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem. 2001;276(42):39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- Ho PS, Ellison MJ, Quigley GJ, Rich A. A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. EMBO J. 1986;5(10):2737–2744. doi: 10.1002/j.1460-2075.1986.tb04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics. 2004;3(9):857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- Jurka J. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 2000;16(9):418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Lise MF, Wong TP, Trinh A, Hines RM, Liu L, Kang R, Hines DJ, Lu J, Goldenring JR, Wang YT, et al. Involvement of myosin Vb in glutamate receptor trafficking. J Biol Chem. 2006;281(6):3669–3678. doi: 10.1074/jbc.M511725200. [DOI] [PubMed] [Google Scholar]
- Meyers JM, Prekeris R. Formation of mutually exclusive Rab11 complexes with members of the family of Rab11-interacting proteins regulates Rab11 endocytic targeting and function. J Biol Chem. 2002;277(50):49003–49010. doi: 10.1074/jbc.M205728200. [DOI] [PubMed] [Google Scholar]
- Paces J, Zika R, Paces V, Pavlicek A, Clay O, Bernardi G. Representing GC variation along eukaryotic chromosomes. Gene. 2004;333:135–141. doi: 10.1016/j.gene.2004.02.041. [DOI] [PubMed] [Google Scholar]
- Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305(5692):1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- Prekeris R, Klumperman J, Scheller RH. A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol Cell. 2000;6(6):1437–1448. doi: 10.1016/s1097-2765(00)00140-4. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Roohi J, Tegay DH, Palmer LE, DeVincent C, Pomeroy JC, Christian SL, Nowak N, Hatchwell E. Disruption of Contactin 4 in 3 Subjects with Autism Spectrum Disorder. Journal of Medical Genetics. doi: 10.1136/jmg.2008.057505. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Selzer RR, Richmond TA, Pofahl NJ, Green RD, Eis PS, Nair P, Brothman AR, Stallings RL. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44(3):305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 2007.
- Stein MP, Dong J, Wandinger-Ness A. Rab proteins and endocytic trafficking: Potential targets for therapeutic intervention. Adv Drug Deliv Rev. 2003;55(11):1421–1437. doi: 10.1016/j.addr.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Strachan T, Read AP. Human molecular genetics. BIOS Scientific Publishers; Wiley-Liss; Oxford New York: 1996. p. xiv.p. 597. [Google Scholar]
- Strutz-Seebohm N, Korniychuk G, Schwarz R, Baltaev R, Ureche ON, Mack AF, Ma ZL, Hollmann M, Lang F, Seebohm G. Functional significance of the kainate receptor GluR6(M836I) mutation that is linked to autism. Cell Physiol Biochem. 2006;18(4–5):287–294. doi: 10.1159/000097675. [DOI] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174(2):247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]