Abstract
Proteus mirabilis isolates commonly have decreased susceptibility to imipenem. Previously, we found P. mirabilis hfq mutant was more resistant to imipenem and an outer membrane protein (OMP) could be involved. Therefore, we investigated the role of this OMP in carbapenem susceptibility. By SDS-PAGE we found this OMP (named ImpR) was increased in hfq mutant and LC-MS/MS revealed it to be the homologue of Salmonella YbfM, which is a porin for chitobiose and subject to MicM (a small RNA) regulation. We demonstrated that ImpR overexpression resulted in increased carbapenem MICs in the laboratory strain and clinical isolates. Chitobiose induced expression of chb (a chitobiose utilization operon). Real-time RT-PCR and SDS-PAGE were performed to elucidate the relationship of hfq, impR, chb and MicM in P. mirabilis. We found MicM RNA was decreased in hfq mutant and chbBC-intergenic region (chbBC-IGR) overexpression strain (chbIGRov), while impR mRNA was increased in hfq mutant, micM mutant and chbIGRov strain. In addition, mutation of hfq or micM and overexpression of chbBC-IGR increased ImpR protein level. Accordingly, chitobiose made wild-type have higher levels of ImpR protein and are more resistant to carbapenems. Hfq- and MicM-complemented strains restored wild-type MICs. Mutation of both impR and hfq eliminated the increase in carbapenem MICs observed in hfq mutant and ImpR-complementation of hfq/impR double mutant resulted in MICs as hfq mutant, indicating that the ImpR-dependent decreased carbapenem susceptibility of hfq mutant. These indicate MicM was antisense to impR mRNA and was negatively-regulated by chbBC-IGR. Together, overexpression of ImpR contributed to the decreased carbapenem susceptibility in P. mirabilis.
Introduction
Proteus mirabilis is an important pathogen of the urinary tract, especially in patients with indwelling urinary catheters [1]. Because of intrinsic resistance to polymyxin B [2] and production of enzymes, such as extended-spectrum β-lactamases (ESBLs) [3], carbapenemases [4] and AmpC [5], treatment of P. mirabilis infections could be difficult. Carbapenems are often the drugs of last resort for ESBL-producing organisms which are increasingly multi-drug resistant [6,7]. However, the emergence of carbapenem-resistant bacteria (CRB) jeopardizes the use of carbapenems [6,8]. In particular, P. mirabilis is intrinsically less susceptible to imipenem which is active for most enterobacteria [9,10].
Mechanisms of carbapenem resistance include target alterations, production of carbapenemases, efflux pumps and porin deficiency. Reduced expression of penicillin binding proteins (PBP) is associated with carbapenem resistance in Acinetobacter and Proteus [9,11]. A consistent number of acquired carbapenemases have been identified during the past few years, belonging to either metallo-β-lactamases or serine carbapenemases, and genes encoding these enzymes are associated with mobile genetic elements that allow their rapid dissemination in the clinical setting [6–8]. Overexpression of Acinetobacter AdeABC and Pseudomonas CzcCBA efflux pumps has been reported to be implicated in resistance to carbapenems [12,13]. Loss of porins engenders imipenem resistance in Pseudomonas, Klebsiella and Acinetobacter [13–15].
The study of carbapenem resistance in P. mirabilis is still lacking. Neuwirth et al. found reduced imipenem-affinity of PBP2 in two imipenem-insusceptible P. mirabilis isolates [9]. In addition, Tibbetts et al. first reported a carbapenem resistant P. mirabilis caused by the acquisition of bla KPC-2 [4]. P. mirabilis isolates exhibiting decreased susceptibility to imipenem also have been shown to carry a bla VIM-1 or a bla NDM-1 metallo-β-lactamase gene [16,17]. Although there is a report of imipenem resistance in a P. mirabilis strain associated with the loss of a 24 kDa OMP [18], no conclusion was drawn concerning P. mirabilis OMPs and carbapenem susceptibility.
Previously, we found Hfq is a pivotal coordinator for a diversity of regulatory circuits including surface components and virulence in P. mirabilis [19] and hfq mutant exhibited increased susceptibility to many antibiotics except imipenem (data not shown). Hfq is a posttranscriptional regulator that binds small RNAs (sRNAs) and mRNA and facilitates RNA-RNA interaction [20]. Numerous cellular processes, such as stress responses and OMP biogenesis are subject to the control of sRNAs and Hfq [21]. OMP analysis of P. mirabilis hfq mutant revealed a protein band (around 48 kDa) of increased intensity. We investigated the role of this OMP in carbapenem susceptibility and disclosed an Hfq-MicM (a sRNA) mediated process involved in decreased carbapenem susceptibility via upregulation of this OMP, named ImpR. This is a novel report elucidating how a sRNA-regulated OMP contributes to decreased susceptibility to carpapenems in P. mirabilis. The results indicate that an overexpressed OMP, neither a part of an efflux pump nor an OprD-like porin for drug entry, is associated with decreased susceptibility to carbapenems. This study also highlights the importance of sRNAs in drug susceptibility.
Materials and Methods
Bacterial strains, plasmids, reagents and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were routinely cultured at 37°C in Luria-Bertani (LB) broth. The LSW- agar [2] was used to prevent the phenotypic expression of swarming motility. Chitobiose, the β-1, 4-linked disaccharide of N-acetylglucosamine, was prepared using Serratia marcescens chitinase A [22].
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotype or relevant phenotype | Source or reference |
|---|---|---|
| P. mirabilis | ||
| wt | Wild-type N2; Tcr | [2] |
| hfq | wt derivative; hfq-knockout mutant; Kmr | [19] |
| micM | wt derivative; micM-knockout mutant; Kmr | This study |
| impR | wt derivative; impR-knockout mutant; Cmr | This study |
| hfq/impR | wt derivative; hfq/impR-knockout mutant; Kmr Cmr | This study |
| chbIGRov | wt containing pGEM-T Easy-chbBC-IGR; chbBC-IGR overexpressing strain; Ampr | This study |
| ImpRov | wt containing pGEM-T Easy-impR; impR overexpressing strain; Ampr | This study |
| hfqca | hfq mutant containing pGEM-T Easy-hfq; Hfq-complemented strain; Ampr | [19] |
| hfq/impRc | hfq/impR mutant containing pGEM-T Easy-impR; hfq/impR mutant complemented with ImpR; Ampr | This study |
| hfqca/impR | hfq/impR mutant containing pGEM-T Easy-hfq; hfq/impR mutant complemented with Hfq; Ampr | This study |
| micMc | micM mutant containing pGEM-T Easy-micM; MicM-complemented strain; Ampr | This study |
| E. coli | ||
| DH5α | fhuA2 lac(del)U169 phoA glnV44 Φ80' lacZ(del)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | Invitrogen |
| S17–1 λ pir | λ pir lysogen of S17–1 [thi pro hsdR - hsdM + recA RP4 2-Tc::Mu-Km::Tn7 (Tpr Smr)]; permissive host able to transfer suicide plasmids requiring the Pir protein by conjugation to recipient cells | [2] |
| Plasmids | ||
| pGEM-T Easy | High-copy TA cloning vector; Ampr | Promega |
| pUT-Km1 | Suicide plasmid requiring the Pir protein for replication and containing a mini-Tn5 cassette containing Kmr gene | [2] |
| pACYC184 | Low-copy cloning vector, P15A replicon; Cmr Tetr | [2] |
| pGEM-T Easy-chbBC-IGR | pGEM-T Easy containing intact chbBC-IGR sequence; Ampr | This study |
| pGEM-T Easy-impR (pImpR) | pGEM-T Easy containing intact impR sequence including its ribosome binding site only for overexpression or its promoter for complementation; Ampr | This study |
| pACYC184-chb-xylE | chb reporter plasmid, pACYC184 containing intact chb promoter sequence before xylE; Cmr | This study |
| pGEM-T Easy-micM | pGEM-T Easy containing intact micM sequence including its promoter; Ampr | This study |
Construction of P. mirabilis mutants
Sequences flanking micM gene were amplified by PCR using the primer pairs micM-upF/XbaI-micM-upR and XbaI-micM-downF/micM-downR, respectively (Table 2) and cloned into pGEM-T Easy (Promega) to generate pGmicM-up and pGmicM-dn. pGmicM-up was digested with SalI/XbaI, and the micM upstream sequence-containing fragment was ligated to SalI/XbaI-digested pGmicM-dn to produce the pGmicM-updn plasmid, which contains the combined upstream and downstream sequences of micM. A Kmr cassette was inserted in the XbaI-digested pGmicM-updn plasmid to generate pGmicM-updn-Km. pGimpR-updn-Cm was constructed in a similar way except using primer pairs impR-upF/XbaI-impR-upR and XbaI-impR-downF/impR-downR and a Cmr cassette for insertion. The DNA fragment containing the combined upstream and downstream sequence of micM or impR gene disrupted by Kmr or Cmr cassette was cleaved from pGmicM-updn-Km or pGimpR-updn-Cm and ligated into SalI/SphI-cleaved pUT-Km1 to generate pUTmicM-Km and pUTimpR-Cm, respectively. Gene inactivation mutagenesis by homologous recombination and confirmation of mutants with double-crossover events were performed as described previously [2]. The hfq/impR double mutant (Kmr and Cmr) was constructed in a similar way using an existing hfq mutant (Kmr). Those mutants were validated by sequencing and determining carbapenem MICs of mutants and their complemented strains to demonstrate the specificity of deletion.
Table 2. Primers used in this study.
| Primers | Sequence (5’ to 3’) | Description |
|---|---|---|
| micM-upF | GAGATCCACACATTCAATC | For micM knockout |
| XbaI-micM-upR | TCTAGAAAGCCTCTGGTATCTAAAG | |
| XbaI-micM-downF | TCTAGACCTCTTAACGATAGAATAGC | For micM knockout |
| micM-downR | AGGCTGAAATGTATTCACC | |
| impR-upF | CTGGTGCTGAAGAGGATTTC | For impR knockout |
| XbaI-impR-upR | TCTAGATGCCTGGTTTAGCATGTTG | |
| XbaI-impR-downF | TCTAGAGTGATCATGCCATTTACG | For impR knockout |
| impR-downR | AAATAGACCACACTACGGG | |
| chbBC-IGR-overF | GGCCAAGAAGCGGATGTCG | For chbBC-IGR overexpression |
| chbBC-IGR-overR | TACCGTCATAAAGGCGGCG | |
| impR-overF | AGTCAAAGTCAAAGAGGAAAC | For impR overexpression |
| impR-overR | AATACACTTTATCCTTATTG | |
| micMc-F | TGATTCTACCATAGAACCATTTCC | For micM complementation |
| micMc-R | TGATATCGCCATTGAAAC | |
| chbre-F | GCATGCTGTAACGCGAAACAATGC | Amplification of chb promoter for reporter assay |
| chbre-R | CTGCAGAACCCTCTCAAATTGGCC | |
| impRrt-F | GAAAATGTCTTATGCTGAAG | For impR real-time RT-PCR |
| impRrt-R | CGTTAAAGGTAGAGTGGTG | |
| chbBC-IGRrt-F | ATATGGGAAAGTGGATGGATTAGG | For chbBC-IGR real-time RT-PCR |
| chbBC-IGRrt-R | ACCCTTAAAATGCCGCTAATTG | |
| micMrt-F | AAGAGGGCGGAGTGATGA | For micM real-time RT-PCR |
| micMrt-R | CGGCCAGTCAAAGAGGAATT | |
| gyrBrt-F | GACCCGTACGCTAAACAAC | Internal control for real-time RT-PCR |
| gyrBrt-R | AGAAATAACCGCAATCAGG |
For complementation of micM mutant, the fragment containing full-length micM was amplified by PCR using the primer pair, micMc-F/micMc-R, and cloned into pGEM-T Easy (Promega) to generate the plasmid, pmicM-com. The pmicM-com was transformed into the micM mutant to generate the MicM-complemented strain. The ImpR-complemented plasmid (pimpR-com) was constructed in a same way using the primer pair, impR-upF/impR-overR, to amplify the fragment containing full-length impR. pimpR-com and pGEM-T Easy-hfq [19] were transformed into hfq/impR double mutant, respectively, to generate the ImpR- and Hfq-complemented strains (hfq/impRc and hfqca/impR).
Construction of impR and chbBC-IGR overexpressing strains
Full-length genes of impR and chbBC-IGR were amplified by PCR and cloned into pGEM-T Easy to generate pGimpR and pGchbBC-IGR, respectively. impR and chbBC-IGR are thus driven by the lac promoter in the pGEM-T Easy plasmid. The primers used in this study are listed in Table 2. pGimpR, and pGchbBC-IGR were then transformed into the wild-type P. mirabilis to generate the impR and chbBC-IGR overexpression strains. To study the effect of impR overexpression on the carbapenem susceptibility of clinical P. mirabilis isolates, pGimpR was also transformed into the clinical isolates.
Minimum inhibitory concentration assay
The carbapenem MICs were determined by the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (M07-A9) [23].
Outer membrane protein analysis
Analysis of OMPs was carried out by SDS-PAGE. OMPs were prepared from bacteria grown overnight in LB according to the protocol described previously [24]. The OMP obtained was quantified by the Bio-Rad protein assay and adjusted to the same concentration before SDS-PAGE. We identified the OMP band that was significantly increased in hfq mutant, micM mutant, ImpRov (impR-overexpressing strain), and chbIGRov (chbBC-IGR-overexpressing strain) relative to wild-type and also this band of wild-type in the presence of chitobiose by liquid chromatography-tandem mass spectrometry (LC–MS/MS) using a hybrid dual-cell quadrupole linear ion trap (LTQ-Orbitrap Velos, Thermofisher Scientific) at Medical Center, College of Medicine, National Taiwan University.
Reporter assay
The promoter region of the putative chb operon was amplified by the primer pair, chbre-F/chbre-R, (Table 2) and cloned into pGEM-T Easy to generate pGchbp. pGchbp was cut by SphI/PstI and the promoter-containing fragment was ligated with the xylE containg pACYC184 to construct the chb-xylE reporter plasmid. The overnight cultures of the wild-type transformed with the reporter plasmid (chb-xylE) were diluted 100 fold in the same medium with or without chitobiose and the XylE activity was measured as described previously [2] at time points indicated after incubation at 37°C.
Real-time reverse transcription PCR (real-time RT-PCR)
Overnight LB cultures of wild-type and its derived strains (hfq, impR, micM, hfq/impR and chbIGRov) were diluted in LB broth to an optical density at 600 nm of 0.1, and grown overnight at 37°C adding chitobiose or not. Total RNA was extracted and real time RT-PCR was performed as described [2] to monitor the RNA levels of impR, micM and chbBC-IGR using primer pairs listed in Table 2. The RNA levels were normalized against the housekeeping gene, gyrB.
Nucleotide sequence accession numbers
The nucleotide sequences of P. mirabilis N2 impR and micM genes have been deposited in GenBank under accession no. KM006423 and KM006424, respectively.
Results
Identification of ImpR, an OMP increased in P. mirabilis hfq mutant, contributing to decreased carbapenem susceptibility
Previously, we found P. mirabilis hfq mutant exhibited increased resistance to imipenem. SDS-PAGE analysis revealed an OMP about 48 kDa was increased in hfq mutant (Fig. 1). We confirmed that imipenem MIC of hfq mutant was increased 4-fold compared to the wild-type (Table 3) and identified the OMP (named ImpR) as YbfM protein (an OMP for chitobiose utilization) homologue of Salmonella [25] by LC-MS/MS. Real-time RT-PCR also showed a much higher impR mRNA level in hfq mutant relative to the wild-type which is almost silent (Fig. 2A). To further investigate the role of ImpR in imipenem susceptibility, we constructed the ImpRov strain (Table 1) by transforming the ImpR-overexpressing plasmid (pImpR) to the wild-type and found the ImpRov strain had a 4-fold higher imipenem MIC level than the vector only control (Table 3). SDS-PAGE analysis revealed a band (about 48 kDa) of increased intensity in the ImpRov strain (Fig. 1) and the band was identified as Salmonella YbfM homologue by LC-MS/MS.
Fig 1. The SDS-PAGE profile of OMPs from overnight cultures of wild-type, its derivatives and wild-type treated with chitobiose.
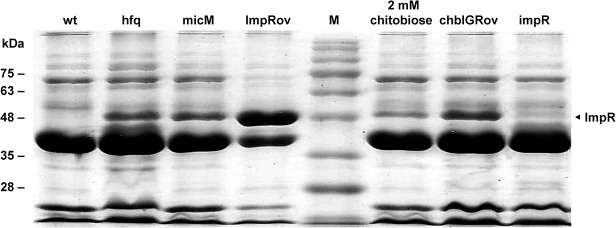
The representative result from three independent experiments is shown. The arrow indicates the band of ImpR. M, marker; wt, wild-type; hfq, hfq mutant; micM, micM mutant; ImpRov, ImpR-overexpressing strain; chbIGRov, chbBC-IGR-overexpressing strain; impR, impR mutant.
Table 3. MICs of imipenem, meropenem and ertapenem for P. mirabilis N2 and its derived strains.
| MIC (μg/ml) | |||
|---|---|---|---|
| Strain | imipenem | meropenem | ertapenem |
| wt | 1 | 0.06 | 0.015 |
| hfq | 4 | 0.24 | 0.06 |
| hfq-vector | 4 | 0.24 | 0.06 |
| hfqca | 1 | 0.06 | 0.015 |
| wt-vector | 1 | 0.06 | 0.015 |
| ImpRov | 4 | 0.24 | 0.06 |
| wt-chitobiose* | 4 | 0.24 | 0.03 |
| impR | 1 | 0.06 | 0.015 |
| impRc | 4 | 0.24 | 0.06 |
| hfq/impR | 1 | 0.06 | 0.015 |
| hfq/impR-vector | 1 | 0.06 | 0.015 |
| hfq/impRc | 4 | 0.24 | 0.06 |
| hfqca/impR | 1 | 0.06 | 0.015 |
| micM | 4 | 0.24 | 0.06 |
| micM-vectormicMc | 41 | 0.240.06 | 0.060.015 |
| chbIGRov | 4 | 0.24 | 0.03 |
*, 2 mM; wt, wild-type; hfq, hfq mutant; hfqca, Hfq-complemented strain; ImpRov, ImpR-overexpressing strain; impR, impR mutant; impRc, impR mutant complemented with ImpR; hfq/impR, hfq/impR double mutant; hfq/impRc, hfq/impR complemented with ImpR; hfqca/impR, hfq/impR complemented with Hfq; micM, micM mutant; micMc, MicM-complemented strain; chbIGRov, chbBC intergenic region-overexpressing strain.
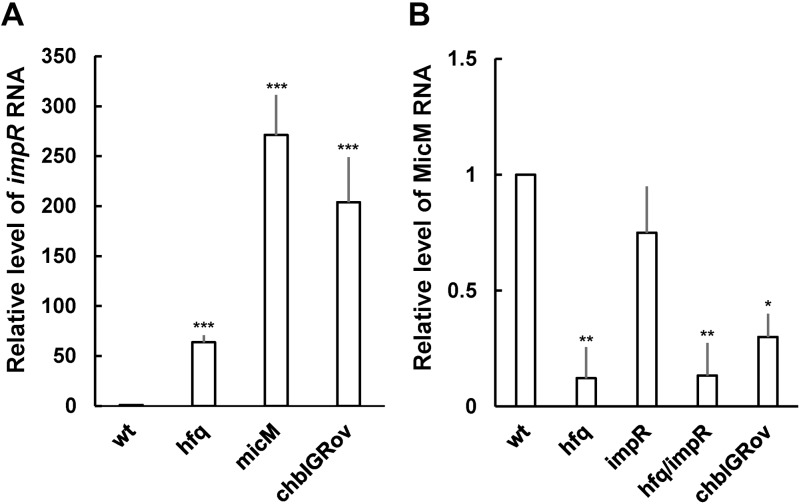
Fig 2. The RNA levels of impR (A) and MicM (B) in the wild-type P. mirabilis and its derived strains.
The relative RNA levels of impR in the wild-type, mutants of hfq and micM, and chbIGRov were quantified by real-time RT-PCR. The RNA was prepared using overnight bacterial cultures. The relative RNA levels of MicM in the wild-type, mutants of hfq, impR and hfq/impR, and chbIGRov were also determined in the same way. The expression level for the wild-type cells was set at 1. The data represent the averages of three independent experiments with standard deviations. Significant difference from the wild-type was indicated with the asterisk (*, P<0.05; **, P<0.01; ***, P<0.001 by Student’s t-test analysis). wt, wild-type; hfq, hfq mutant; impR, impR mutant; hfq/impR, hfq and impR double mutant; chbIGRov, chbBC-IGR overexpressing strain; micM, micM mutant.
The impR gene was identified at nt 583221 to 584615 in the genome of P. mirabilis strain HI4320. It is located in the glnS-impR-PMI0542 cluster as shown in Fig. 3A. The ImpR protein consists of 464 amino acids and shares 64% sequence identity and 77% similarity with its homologue, YbfM of Salmonella. Using primers annealing to conserved sequences, we cloned and sequenced the fragment containing impR and upstream of impR in P. mirabilis N2. The amino acids of N2 ImpR were 100% and 99% identical to those of P. mirabilis HI4320/P. mirabilis BB2000 and P. mirabilis WGLW4, respectively.
Fig 3. Genomic location of impR (A) and micM (B) in P. mirabilis.
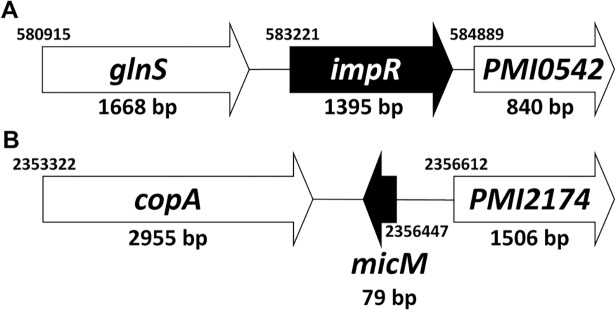
The number indicates the start nucleotide number of each gene in the genome and the size (bp) of each gene is indicated.
Searching for a small RNA regulating expression of impR
It has been known that YbfM is a porin required for uptake of chitobiose [25–27]. In the absence of chitobiose (inducer), ybfM is kept silent by the action of a constitutively-made sRNA, MicM, which pairs with the 5’ UTR of ybfM mRNA [25–27]. Silencing is relieved in the presence of inducer due to accumulation of an RNA that pairs with MicM, thus promoting MicM degradation. The anti-MicM RNA is from an intergenic region (between chbB and chbC, called chbBC-IGR) of the chb operon (chbBCARFG), which contains genes for chitobiose utilization and whose transcription is activated in the presence of chitobiose [28]. We first searched P. mirabilis MicM homologue in the website, http://bac-srna.org/BSRD/index.jsp#, and located it in genome of P. mirabilis strain HI4320. Although the ybaK-micM-ybaP region is well conserved in many enterobacteria, we found P. mirabilis MicM is between copA gene and PMI2174 (Fig. 3B). We also found the existence of chb operon homologue in P. mirabilis strain HI4320. We cloned and sequenced the fragment containing micM and upstream of micM in P. mirabilis N2. The nucleotide sequences of micM were 100% identical to those of P. mirabilis HI4320.
Sequence analysis revealed ‘5’UGAAAAAUUCCUCUUUGACUGG’ could be the site for MicM to bind with impR mRNA and the anti-MicM region of the chb mRNA.
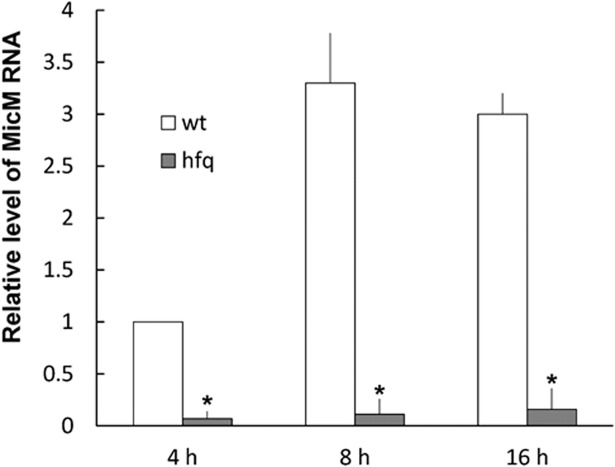
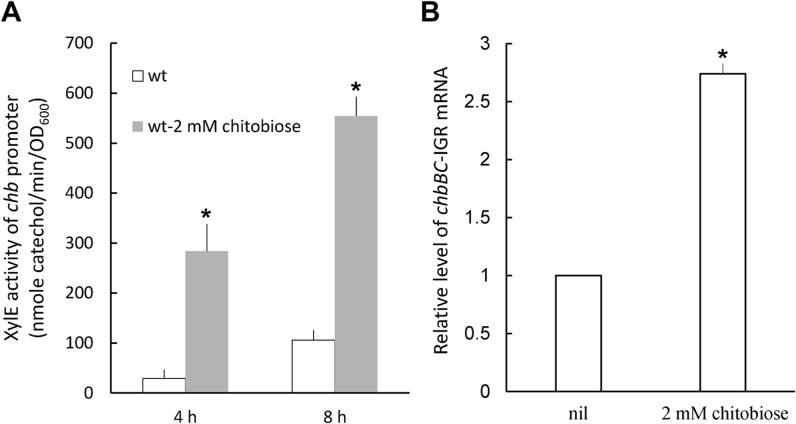
Knowing the existence of MicM and chb operon in P. mirabilis, we first performed real-time RT-PCR to assess the MicM level in wild-type and hfq mutant. Fig. 4 indicated MicM was constitutively expressed and subjected to the positive control of Hfq. Constitutive expression of MicM could explain the very low level of ImpR in wild-type (Fig. 2). In addition, Fig. 5 showed that chitobiose can induce chb promoter activity and consequently lead to the increased chb mRNA (chbBC-IGR) level.
Fig 4. The expression of MicM in the wild-type and hfq mutant.
The RNA amount of MicM in the wild-type or hfq mutant was quantified by real-time RT-PCR. Overnight bacterial cultures were diluted to OD600 of 0.1 and incubated for 4 h, 8 h and 16 h before total RNA was prepared. The expression level for the wild-type cells at 4 h was set at 1. The data represent the averages of three independent experiments with standard deviations. Significant difference from the wild-type at different time point was indicated with the asterisk (*, P<0.01 by Student’s t-test analysis). wt, wild-type; hfq, hfq mutant.
Fig 5. (A) Chitobiose induced chb promoter activity.
The xylE activity in the chb-xylE reporter plasmid-transformed wild-type P. mirabilis in the presence or absence of 2 mM chitobiose was measured after induction for 4 h and 8 h. (B) Chitobiose induced expression of chb mRNA. The RNA of the wild-type was prepared using overnight bacterial cultures and relative chb mRNA amount was quantified by real-time RT-PCR using primers for chbBC-IGR. The expression level of control cells without chitobiose (nil) was set at 1. The data represent the averages of three independent experiments with standard deviations. Significant difference from the no chitobiose control at different time point was indicated with the asterisk (*, P<0.01 by Student’s t-test analysis). wt, wild-type.
Characterization of MicM-mediated regulation of impR
To know if the regulation of Hfq-dependent MicM by chb mRNA and impR by MicM exists in P. mirabilis, we constructed single mutants of impR and MicM, hfq/impR double mutant and chbBC-IGR overexpressing strain (chbIGRov). Real-time RT-PCR was performed to clarify the relationship of chb, MicM, hfq and impR. Fig. 2B showed impR mutation didn’t affect MicM level, whereas hfq/impR double mutation led to a significantly decreased level of MicM. The result confirmed MicM is positively regulated by Hfq. In addition, chbIGRov strain exhibited a significantly decreased MicM level. Moreover, either overexpression of chbBC-IGR or mutation in hfq or micM resulted in a high level of impR mRNA. OMP analysis also revealed that chbBC-IGR overexpression strain (chbIGRov), micM mutant and hfq mutant all had an increased level of ImpR protein (Fig. 1). In accordance with the finding that chitobiose can induce expression of chb operon (Fig. 5), ImpR protein was increased in wild-type treated with chitobiose (Fig. 1). Together, these results indicated antisense regulation of impR mRNA by MicM, Hfq-dependent expression of MicM and negative regulation of MicM by chb operon in response to chitobiose.
Significance of MicM-mediated regulation of impR in carbapenem susceptibility of P. mirabilis
MIC assay was conducted to clarify the significance of MicM-mediated regulation of impR in carbapenem susceptibility of P. mirabilis. Firstly, carbapenem MICs were determined in wild-type, derived mutants and the complemented strains to validate that mutation of hfq, impR or micM was involved in carbapenem susceptibility. We noticed that impR mutant had wild-type MIC (Table 3). Hfq- and MicM-complemented strain restored wild-type carbapenem MICs. hfq/impR double mutant complemented with ImpR had the same carbapenem MICs as hfq mutant and exhibited wild-type MICs when complemented with Hfq (Table 3). The finding that MICs of impR single and hfq/impR double mutants were the same as wild-type (Table 3) indicated the low expression of impR in wild-type and the ImpR-dependent carbapenem-resistance for hfq mutant. Besides, chitobiose (the inducer for chb operon) and overexpression of chbBC-IGR caused a 4-fold increase in imipenem MICs compared to the wild-type (Table 3). Accordingly, micM mutant had a 4-fold increase in imipenem MICs (Table 3). It is worth noting that mutation of micM or hfq, overexpression of ImpR or chbBC-IGR, and chitobiose also increased meropenem and ertapenem MICs to 4 and 2–4 fold, respectively (Table 3).
To further demonstrate the significance of ImpR in carbapenem susceptibility of P. mirabilis, we introduced the ImpR-overexpressing plasmid into clinical isolates of P. mirabilis and MICs were determined. Table 4 showed that clinical isolates bearing the ImpR-overexpressing plasmid exhibited 4–8 fold increase in imipenem and meropenem MICs relative to the vector control. We also found that MICs of ertapenem were increased 2–4 fold (Table 4).
Table 4. MICs of imipenem, meropenem, and ertapenem for P. mirabilis clinical isolates transformed with the ImpR-overexpressing plasmid.
| MIC (μg/ml) | ||||
|---|---|---|---|---|
| Clinical isolate | imipenem | meropenem | ertapenem | |
| 1 | vector | 1 | 0.06 | 0.015 |
| pImpR | 4 | 0.25 | 0.06 | |
| 2 | vector | 1 | 0.06 | 0.03 |
| pImpR | 4 | 0.25 | 0.06 | |
| 3 | vector | 1 | 0.06 | 0.06 |
| pImpR | 4 | 0.25 | 0.25 | |
| 4 | vector | 2 | 0.06 | 0.03 |
| pImpR | 8 | 0.25 | 0.12 | |
| 5 | vector | 1 | 0.12 | 0.06 |
| pImpR | 4 | 0.48 | 0.12 | |
| 6 | vector | 1 | 0.03 | 0.03 |
| pImpR | 8 | 0.25 | 0.12 | |
Discussion
It is imperative to investigate carbapenem resistance to escape the public health crisis caused by CRB. Although P. mirabilis is still susceptible to meropenem and ertapenem, the bacterium is known to be intrinsically less susceptible to imipenem [9,10]. Previously, we accidentally found that decreased imipenem susceptibility of P. mirabilis hfq mutant was associated with an OMP. In this study, we described the role of the OMP (ImpR) in decreased imipenem susceptibility, also meropenem and ertapenem. The phenotype was mediated through an Hfq-regulated sRNA, MicM, in a process involving chb operon in response to chitobiose. Several lines of evidence support the notion. First, increased ImpR, either by mutation of hfq or micM (MicM is antisense to impR mRNA) or overexpression of ImpR rendered carbapenem MICs to increase 4-fold (Table 3). Second, levels of impR mRNA and ImpR protein were increased but MicM RNAs were decreased in hfq mutant (Figs. 1 and 2), consistent with that Hfq-dependent MicM is antisense to impR mRNA. Third, chitobiose induced expression of ImpR protein (Fig. 1) and chb mRNAs (Fig. 5), antisense to MicM. chbIGRov strain displayed a decrease in MicM RNA level (Fig. 2) but an increase in expression of impR (Figs. 1 and 2). Both the presence of chitobiose and overexpression of chbBC-IGR resulted in increased carbapenem MICs (Table 3). Fourth, hfq/impR double mutation abolished the increased carbapenem MICs of hfq mutant (Table 3), indicating decreased carbapenem suaceptibility in hfq mutant depends on the ImpR protein. Fifth, the ImpR- overexpressing plasmid made the clinical isolates more resistant than its parent (Table 4).
Bioinformatic analysis revealed ImpR could be a porin of the OprD superfamily, such as E. coli Chip, Salmonella YbfM and Pseudomonas OprD [13,15,25,26], instead of OMPs of the RND efflux pumps, such as TolC [31]. A high glycine content and absence of cysteine residues also indicated ImpR was typical of a gram-negative bacterial porin [32]. In Pseudomonas, OprD loss usually causes a 4 to16-fold increase in MICs of carbapenems [13]. We found P. mirabilis ImpR is probably involved in the uptake of chitobiose because both impR mRNA (Fig. 2) and protein levels (Fig. 1) were induced by overexpression of chbBC-IGR (i.e. the presence of chitobiose). In addition to probably serving as a porin as Chip or YbfM for growing on chitobiose [25,27,33], ImpR also contributed to carbapenem susceptibility when overexpressed. Until now the role of Chip and YbfM in drug susceptibility has not been reported and we found the ImpR-overexpressing plasmid failed to affect carbapenem susceptibility in E. coli (data not shown), suggesting the uniqueness of ImpR overexpression in P. mirabilis. Mutation of impR did not affect carbapenem MICs and ImpR overexpression resulted in an increase in carbapenem MICs instead of a decrease of MICs in overexpression of the porin for carbapenem entry in other bacteria [13–15]. Low expression of ImpR resulting from antisense action of constitutively-expressed MicM in wild-type may explain why impR mutant has wild-type carbapenem MICs. A pump inhibitor, carbonyl cyanide m-chlorophenyl hydrazone, has no effect on carbapenem MICs of ImpRov (data not shown), indicating ImpR not a part of proton-motive pumps.
Our unpublished data showed that RpoE was up-regulated on ImpR overproduction. In this regard, RpoE overexpression has been shown to cause remarkable resistance to the β-lactam antibiotics [30] that cause cell envelope stress by inhibiting peptidoglycan biosynthesis. Accordingly, stress responses have been linked to the development of antimicrobial resistance in Gram-negative bacteria [34]. Oxidative stress is also an end product of antimicrobial exposure. Therefore, the oxidative stress response is expected to promote resistance to antimicrobials [34]. It is possible that the RpoE regulon involved in combating with either envelope or oxidative stresses [29] may contribute to the decreased carbapenem susceptibility in P. mirabilis. Further studies are needed to disclose the mystery.
It has been reported that more attention should be devoted to the mechanisms of low-level resistance in microorganisms, as they can serve as stepping stones to develop high level, clinically relevant resistance [35]. In this study, we found ImpR contributes to decreased susceptibilities (low-level resistance) of carbapenems. What is the importance of ImpR in carbapenem susceptibility in the real world? First, cAMP has been shown to inhibit expression of hfq [36]. In this regard, low level of glucose in urine, a condition of high cAMP level, may lead to repression of Hfq, thus increased ImpR and subsequently decreased carbapenem susceptibility. Second, the presence of chitobiose in urine may also increase the level of ImpR. Chitobiose, the β-1,4-linked disaccharide of N-acetylglucosamine, is the major degradation product of chitin, which constitutes the second-most abundant organic polymer in nature after cellulose [33]. Third, we have found several imipenem-resistant clinical isolates of P. mirabilis whose expression of ImpR is higher than the susceptible ones. In addition, we can not rule out the alterations in the regulatory elements of impR, hfq, micM and chbBC-IGR in the natural environment, which may lead to upregulation of ImpR and thus decreased carbapenem susceptibility. With regard to intrinsic resistance of carbapenems in P. mirabilis, we have found an OMP mutant exhibited increased (8 fold) susceptibility to imipenem. Villar et al. also found decreased expression of an OMP in P. mirabilis was involved in increased susceptibility to imipenem and meropenem [10]. Characterization of the OMP has been underway.
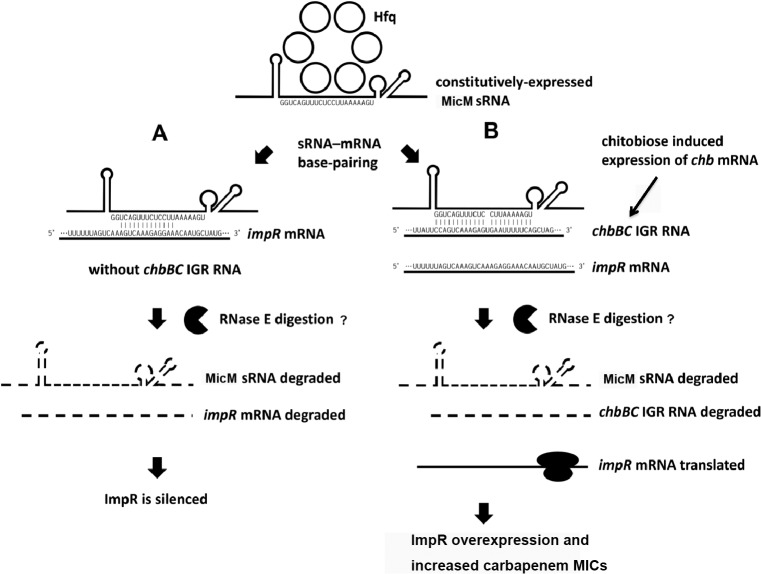
In this work, for the first time, we described the role of an OMP (ImpR) in decreased carbapenem susceptibility and its regulation by a sRNA (MicM) involving the chb operon in P. mirabilis (summarized in Fig. 6). These data suggest that upregulation of ImpR can make P. mirabilis become more resistant to carbapenem treatment, in contrast to down-regulation of OprD in P. aeruginosa [13,15]. The decreased carbapenem susceptibility incurred by increased ImpR has implications in carbapenem therapy against urosepsis caused by P. mirabilis. Clinicians should keep in mind about this acquired low-level resistance of carbapenems in the clinical setting.
Fig 6. Model for ImpR regulation by MicM sRNA.
(A) In the absence of chitobiose, the expression of the ImpR porin is silenced at the post-transcriptional level by pairing of MicM sRNA with the 5’ UTR of impR mRNA, promoting cleavage of the impR mRNA by a ribonuclease (RNase E?). At the same time the chb operon is transcriptionally repressed. (B) In the presence of chitobiose, an inducer to activate transcription of the chb operon, processing of the chb transcript releases chbBC IGR RNA. This RNA base-pairs with MicM making it susceptible to the action of a ribonuclease (RNase E?). The drop in MicM levels relieves impR repression leading to a burst of ImpR translation. ImpR assembles in the outer membrane resulting in increased carbapenem MICs. MicM is an Hfq-dependent and constitutively-expressed sRNA.
Acknowledgments
We would like to thank Professor Yaw-Kuen Li (National Chiao Tung University) for the technical support in preparing chitobiose.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by grants from National Science Council (NSC 101-2320-B-002-028) and National Taiwan University Hospital, Taipei, Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jacobsen SM, Stickler DJ, Mobley HL, Shirtliff ME. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis . Clin Microbiol Rev. 2008;21: 26–59. 10.1128/CMR.00019-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jiang SS, Lin TY, Wang WB, Liu MC, Hsueh PR, Liaw SJ. Characterization of UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase mutants of Proteus mirabilis: defectiveness in polymyxin B resistance, swarming, and virulence. Antimicrob Agents Chemother. 2010;54: 2000–2009. 10.1128/AAC.01384-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pagani L, Migliavacca R, Pallecchi L, Matti C, Giacobone E, Amicosante G, et al. Emerging extended-spectrum beta-lactamases in Proteus mirabilis . J Clin Microbiol. 2002;40: 1549–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tibbetts R, Frye JG, Marschall J, Warren D, Dunne W. Detection of KPC-2 in a clinical isolate of Proteus mirabilis and first reported description of carbapenemase resistance caused by a KPC beta-lactamase in P. mirabilis . J Clin Microbiol. 2008;46: 3080–3083. 10.1128/JCM.00979-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tenover FC, Emery SL, Spiegel CA, Bradford PA, Eells S, Endimiani A, et al. Identification of plasmid-mediated AmpC beta-lactamases in Escherichia coli, Klebsiella spp., and Proteus species can potentially improve reporting of cephalosporin susceptibility testing results. J Clin Microbiol. 2009;47: 294–299. 10.1128/JCM.01797-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Overturf GD. Carbapenemases: a brief review for pediatric infectious disease specialists. Pediatr Infect Dis J. 2010;29: 68–70. 10.1097/INF.0b013e3181c9c118 [DOI] [PubMed] [Google Scholar]
- 7. Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20: 440–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perez F, Van Duin D. Carbapenem-resistant Enterobacteriaceae: a menace to our most vulnerable patients. Cleve Clin J Med. 2013;80: 225–233. 10.3949/ccjm.80a.12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neuwirth C, Siebor E, Duez JM, Pechinot A, Kazmierczak A. Imipenem resistance in clinical isolates of Proteus mirabilis associated with alterations in penicillin-binding proteins. J Antimicrob Chemother. 1995;36: 335–342. [DOI] [PubMed] [Google Scholar]
- 10. Villar HE, Danel F, Livermore DM. Permeability to carbapenems of Proteus mirabilis mutants selected for resistance to imipenem or other beta-lactams. J Antimicrob Chemother. 1997;40: 365–370. [DOI] [PubMed] [Google Scholar]
- 11. Fernandez-Cuenca F, Martinez-Martinez L, Conejo MC, Ayala JA, Perea EJ, Pascual A. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii . J Antimicrob Chemother. 2003;51: 565–574. [DOI] [PubMed] [Google Scholar]
- 12. Abbott I, Cerqueira GM, Bhuiyan S, Peleg AY. Carbapenem resistance in Acinetobacter baumannii: laboratory challenges, mechanistic insights and therapeutic strategies. Expert Rev Anti Infect Ther. 2013;11: 395–409. 10.1586/eri.13.21 [DOI] [PubMed] [Google Scholar]
- 13. Fournier D, Richardot C, Muller E, Robert-Nicoud M, Llanes C, Plésiat P, et al. Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa . J Antimicrob Chemother. 2013;68: 1772–1780. 10.1093/jac/dkt098 [DOI] [PubMed] [Google Scholar]
- 14. Doumith M, Ellington MJ, Livermore DM, Woodford N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother. 2009;63: 659–667. 10.1093/jac/dkp029 [DOI] [PubMed] [Google Scholar]
- 15. Fernandez L, Hancock RE. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012;25: 661–681. 10.1128/CMR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vourli S, Tsorlini H, Katsifa H, Polemis M, Tzouvelekis LS, Kontodimou A, et al. Emergence of Proteus mirabilis carrying the blaVIM-1 metallo-β-lactamase gene. Clin Microbiol Infect. 2006;12: 691–694. [DOI] [PubMed] [Google Scholar]
- 17. Williamson DA, Sidjabat HE, Freeman JT, Roberts SA, Silvey A, Woodhouse R, et al. Identification and molecular characterisation of New Delhi metallo-β-lactamase-1 (NDM-1)- and NDM-6-producing Enterobacteriaceae from New Zealand hospitals. Int J Antimicrob Agents. 2012;39: 529–533. 10.1016/j.ijantimicag.2012.02.017 [DOI] [PubMed] [Google Scholar]
- 18. Mehtar S, Tsakris A, Pitt TL. Imipenem resistance in Proteus mirabilis . J Antimicrob Chemother. 1991;28: 612–615. [DOI] [PubMed] [Google Scholar]
- 19. Wang MC, Chien HF, Tsai YL, Liu MC, Liaw SJ. The RNA chaperone Hfq is involved in stress tolerance and virulence in uropathogenic Proteus mirabilis . PLoS One. 2014;9: e85626 10.1371/journal.pone.0085626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288: 7996–8003. 10.1074/jbc.R112.441386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13: 24–33. 10.1016/j.mib.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 22. Wu YJ, Cheng CY, Li YK. Cloning and expression of chitinase A from Serratia marcescens for large-scale preparation of N,N-diacetyl chitobiose. J Chin Chem Soc. 2009;56: 688–695. [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M07-A9, Wayne, PA; 2012.
- 24. Fukuoka T, Masuda N, Takenouchi T, Sekine N, Iijima M, Ohya S. Increase in susceptibility of Pseudomonas aeruginosa to carbapenem antibiotics in low-amino-acid media. Antimicrob Agents Chemother. 1991;35: 529–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23: 2004–2015. 10.1101/gad.541609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rasmussen AA, Johansen J, Nielsen JS, Overgaard M, Kallipolitis B, Valentin-Hansen P. A conserved small RNA promotes silencing of the outer membrane protein YbfM. Mol Microbiol. 2009;72: 566–577. 10.1111/j.1365-2958.2009.06688.x [DOI] [PubMed] [Google Scholar]
- 27. Vogel J. An RNA trap helps bacteria get the most out of chitosugars. Mol Microbiol. 2009;73: 737–741. 10.1111/j.1365-2958.2009.06806.x [DOI] [PubMed] [Google Scholar]
- 28. Verma SC, Mahadevan S. The chbG gene of the chitobiose (chb) operon of Escherichia coli encodes a chitooligosaccharide deacetylase. J Bacteriol. 2012;194: 4959–4971. 10.1128/JB.00533-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rowley G, Spector M, Kormanec J, Roberts M. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol. 2006;4: 383–394. [DOI] [PubMed] [Google Scholar]
- 30. Gupta N, Kumar S, Mishra MN, Tripathi AK. A constitutively expressed pair of rpoE2-chrR2 in Azospirillum brasilense Sp7 is required for survival under antibiotic and oxidative stress. Microbiology. 2013;159: 205–218. 10.1099/mic.0.061937-0 [DOI] [PubMed] [Google Scholar]
- 31. Huang YW, Hu RM, Yang TC. Role of the pcm-tolCsm operon in the multidrug resistance of Stenotrophomonas maltophilia . J Antimicrob Chemother. 2013;68: 1987–1993. 10.1093/jac/dkt148 [DOI] [PubMed] [Google Scholar]
- 32. del Mar Tomas M, Beceiro A, Perez A, Velasco D, Moure R, Villanueva R, et al. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii . Antimicrob Agents Chemother. 2005;49: 5172–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Overgaard M, Johansen J, Moller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Mol Microbiol. 2009;73: 790–800. 10.1111/j.1365-2958.2009.06807.x [DOI] [PubMed] [Google Scholar]
- 34. Poole K. Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol. 2012;20: 227–234. 10.1016/j.tim.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 35. Baquero F. Low-level antibacterial resistance: a gateway to clinical resistance. Drug Resist Updat. 2001;4: 93–105. [DOI] [PubMed] [Google Scholar]
- 36. Lin HH, Hsu CC, Yang CD, Ju YW, Chen YP, Tseng CP. Negative effect of glucose on ompA mRNA stability: a potential role of cyclic AMP in the repression of hfq in Escherichia coli . J Bacteriol. 2011;193: 5833–5840. 10.1128/JB.05359-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.