Abstract
Background
Telocytes (TCs) is an interstitial cell with extremely long and thin telopodes (Tps) with thin segments (podomers) and dilations (podoms) to interact with neighboring cells. TCs have been found in different organs, while there is still a lack of TCs-specific biomarkers to distinguish TCs from the other cells.
Results
We compared gene expression profiles of murine pulmonary TCs on days 5 (TC5) and days 10 (TC10) with mesenchymal stem cells (MSCs), fibroblasts (Fbs), alveolar type II cells (ATII), airway basal cells (ABCs), proximal airway cells (PACs), CD8+ T cells from bronchial lymph nodes (T-BL), and CD8+ T cells from lungs (T-LL). The chromosome 17 and 18 genes were extracted for further analysis. The TCs-specific genes and functional networks were identified and analyzed by bioinformatics tools. 16 and 10 of TCs-specific genes were up-regulated and 68 and 22 were down-regulated in chromosome 17 and 18, as compared with other cells respectively. Of them, Mapk14 and Trem2 were up-regulated to indicate the biological function of TCs in immune regulation, and up-regulated MCFD2 and down-regulated E4F1 and PDCD2 had an association with tissue homeostasis for TCs. Over-expressed Dpysl3 may promote TCs self-proliferation and cell-cell network forming.
Conclusions
The differential gene expression in chromosomes 17 and 18 clearly revealed that TCs were the distinctive type of interstitial cells. Our data also indicates that TCs may play a dual role in immune surveillance and immune homoeostasis to keep from immune disorder in acute and chronic pulmonary diseases. TCs also participated in proliferation, differentiation and regeneration.
Reviewers
This article was reviewed by Qing Kay Li and Dragos Cretoiu.
Electronic supplementary material
The online version of this article (doi:10.1186/s13062-015-0042-0) contains supplementary material, which is available to authorized users.
Keywords: Chromosome 17, Chromosome 18, Genes, Lung, Telocytes, Mesenchymal stem cells, Fibroblasts, Alveolar type II cells, Airwayepithelial cells, Lymphocytes
Background
Telocytes (TCs) have been proposed as a new type of interstitial cells with extremely long telopodes (Tps), which were recognized in different organs, such as trachea and lung [1,2], oesophagus [3], intestine [4], liver [5], kidney [6], heart [7,8], skin [9], eyes [10], urinary tract [11], bladder [12], uterus [13], prostate gland [14], and euromuscular spindles [15], as documented on www.telocytes.com. TCs were found to form a three-dimensional network in close contacts with blood vessels, nerve bundles and cells of the local immune system [8,13,16-18]. There is growing evidence to support that TCs have close associations with stem cells and will be a critical player in regeneration medicine [10,19]. TCs may also take part in immune regulation, proliferation, cellular reparation and fibrosis. However, there are no specific biomarkers to distinguish TCs from other tissue-resident cells. The reliable way to identify TCs is dependent on the specific ultrastructural feature - a small nucleated body and extremely long Tps with podomers and podoms by transmission electron microscopy (TEM) [20]. TCs present positive CD34, c-kit and vimentin by immunohistochemistry [19] and other markers like iNOS, caveolin-1, VEGF-D and PDGFRα were also identified to indicate various immunophetypes of TCs existed in different organs [4,21].
Our previous study initially approved the existence of TCs in the trachea and lung tissues of the mouse and human [1,22,23]. We compared genes expression profiles of murine TCs with mesenchymal stem cells (MSCs) and fibroblasts (Fbs) and proteomic profiles of human lung TCs and Fbs to explore TC-specific markers and their biological function [24,25]. MicroRNA signature was also used to differentiate TCs from other interstitial cells [26]. Furthermore, we revealed features and patterns of genes in chromosomes 1 by comparing gene expression profiles of murine pulmonary TCs, MSCs, Fbs, alveolar type II cells (ATII), airway basal cells (ABCs), proximal airway cells (PACs), CD8+ T cells from bronchial lymph nodes (T-BL), and CD8+ T cells from lungs (T-LL) by global analyses [27]. The similar work has been done to detect patterns of TCs-specific genes in chromosome 2 and 3, and showed that TCs played an important role in tissue injury and aging, inhibition of tissue inflammation, tumor promotion, and development of pulmonary fibrosis and other interstitial lung diseases [28]. However, features and patterns of TCs-specific genes in other chromosomes have not been uncovered, and potential function of TCs still remained unclear.
The present study aimed at investigating features and patterns of TCs-specific gene profiles and exploring potential function of TCs by focusing in chromosomes 17 and 18. The significant difference in gene expression profiles of murine pulmonary TCs on days 5 (TC5) and days 10 (TC10) with the other cells, MSCs, Fbs, ATII, ABCs, PACs, T-BL and T-LL were compared to identify TCs-specific genes and the functional networks were identified by bioinformatics tools.
Methods
Cell sampling and data collection
Gene expression profiles of murine pulmonary TCs on days 5 and 10, MSCs and Fbs were obtained from our previous study [24]. TCs were isolated from BALB/c mouse lung tissue and cultured for 5 and 10 days, respectively. MSCs and Fbs were obtained from Sciencell Research Laboratories (M7500-57, Carlsbad, CA, USA) and Chinese Academy of Science (GNM28, Shanghai, China), respectively. ATII, ABCs, PACs, T-BL, T-LL gene expression profiles were obtained from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database (GSE6846, GSE27379, GSE28651) [29]. Alveolar type II cells (GSM157835-GSM157837) were isolated from 8 weeks old sex age matched littermate control mice. CD8+ T-cells from lungs and bronchial lymph nodes (GSM677065) and CD8+ T-cells from bronchial lymph nodes, lymphocytes (GSM677065) derived from mice (CL4). Murine proximal airway duct (GSM709834-GSM709836) and basal cells (GSM709832, GSM709833) were isolated from 8–12 week old C57BL/6 mice.
Data mining and preprocessing
The gene expression profiles of murine lung TCs, MSCs and Fbs from our previous study contained 23861 probes. About 13236 probes and 11532 genes were defined after eliminating the probes without corresponding official symbols. Gene expression of ATII, ABCs, PACs, T-BL, and T-LL from GEO originally included 45,101 probes. 5684 probes without corresponding official symbols were eliminated and 39,417 probes and 21,680 genes were obtained. Data integration and genes detected in all the samples were selected to analysis, of which 11532 genes were elected finally. Totally, 582 genes of the chromosome 17 and 267 genes of the chromosome 18 were focused and furthermore analyzed in the present study.
Identification of differentially expressed genes
Gene expression data were normalized and imported into Agilent GeneSpring GX software (version 11.5.1) for further analysis. Differentially expressed genes between two samples were identified through Fold Change filtering as our previous research [27]. The propensity of functional changes was reflected in different levels of the gene expression in each cell types. The genes in TCs on days 5 and 10, which were up- or down-regulated more than one-fold compared with other cells, were identified as TCs-specific genes in the present study. Up- or down-regulated folds of TCs genes were calculated by comparing with other cells, after the averages of gene expression in cells were obtained from the raw data of multi-databases, as shown in Additional files 1 and 2.
Results
Fifty three genes were up-regulated and 92 down-regulated in chromosome 17 of TC5 as compared with other cells, and 28 genes were up-regulated and 167 down-regulated in TC10. Of genes up or down-regulated at TC5 and TC10, 16 over-expressed genes (i.e. 2900073G15Ri, Ccnd3, Chtf18, Clic1, Fem1a, Fez2, Kifc1, Mapk14, Mcfd2, Mtch1, Pgp, Tbcc, Tubb4, Tubb5, Zfand3, Trem2) were obtained in chromosome 17 of TCs. Among them, Trem2 was up-regulated over five folds in TC5 and TC10, as compared with others. Sixty eight genes were down-regulated in TC5 and TC10, of which C030034I22Rik was expressed in TCs 2–5 folds lower than in other cells (Tables 1, 2, 3 and 4). In chromosome 18, we found 22 up-regulated genes and 31 down-regulated genes in TC5 and 15 up-regulated genes and 57 down-regulated genes in TC10 as compared with other cells. Eleven genes (9430020K01Rik, Bin1, Cdh2, Fech, Txnl4a, Usp14, Yipf5, Dpysl3, Lims2, Tubb6) were up-regulated and 22 genes down-regulated (fold >1). Among them, Dpysl3, Lims2 and Tubb6 were up-regulated 2–5 folds in TC5 and TC10, while Scgb3a2 and Zfp397 was expressed in TCs 2–5 folds lower than other cells (Tables 5, 6, 7 and 8).
Table 1.
Summary of genes expressed preferentially in TCs, as compared with others in chromosome 17
| Compaired pairs/fold up-regulated | >1 | >2 | >5 |
|---|---|---|---|
| TC5 vs. others | 47 | 4 | 2 |
| TC10 vs. others | 22 | 4 | 2 |
| TCs vs. others | 15 | 0 | 1 |
Table 2.
Summary of genes expressed preferentially in TCs, as compared with others in chromosome 17
| Compaired pairs/fold down-regulated | >1 | >2 | >5 |
|---|---|---|---|
| TC5 vs. others | 83 | 9 | 0 |
| TC10 vs. others | 146 | 21 | 0 |
| TCs vs. others | 67 | 1 | 0 |
Table 3.
The up-regulated fold of specific genes in TCs as compared with others in chromosome 17
| Folds(TC5 vs. others/TC10 vs. others) | |||||||
|---|---|---|---|---|---|---|---|
| (A) Genes up-regulated between one- and twofold in TCs as compared with others | |||||||
| Gene symbol | Fibroblast | Stem | ATII | CD8_T_BL | CD8_T_LL | Basal_cell | Duct_cell |
| 2900073G15Rik | 2.90/1.87 | 1.81/1.17 | 2.81/1.81 | 4.89/3.16 | 3.79/2.45 | 3.70/2.45 | 3.45/2.22 |
| Ccnd3 | 2.49/2.33 | 2.37/2.22 | 1.40/1.31 | 1.47/1.38 | 1.73/1.62 | 2.44/1.62 | 5.29/4.96 |
| Chtf18 | 2.41/1.86 | 1.45/1.12 | 4.34/3.35 | 2.24/1.72 | 12.43/9.59 | 5.69/9.59 | 2.39/1.84 |
| Clic1 | 3.64/2.26 | 2.10/1.30 | 2.01/1.25 | 2.37/1.47 | 3.10/1.93 | 2.45/1.93 | 1.94/1.21 |
| Fem1a | 1.99/1.67 | 1.86/1.56 | 2.24/1.87 | 6.71/5.62 | 7.09/5.94 | 1.22/5.94 | 1.31/1.10 |
| Fez2 | 3.14/2.86 | 1.87/1.71 | 9.51/8.69 | 3.84/3.51 | 4.21/3.85 | 3.11/3.85 | 3.49/3.19 |
| Kifc1 | 2.23/2.42 | 1.10/1.19 | 8.53/9.27 | 1.91/2.08 | 1.05/1.14 | 5.49/1.14 | 4.52/4.91 |
| Mapk14 | 1.57/1.02 | 1.71/1.11 | 7.98/5.19 | 3.38/2.20 | 3.96/2.57 | 1.96/2.57 | 2.63/1.71 |
| Mcfd2 | 1.04/1.03 | 1.38/1.37 | 1.24/1.23 | 2.88/2.85 | 3.35/3.31 | 6.02/3.31 | 4.26/4.22 |
| Mtch1 | 2.24/1.63 | 1.66/1.20 | 2.73/1.98 | 88.12/64.07 | 144.31/104.92 | 35.78/104.92 | 27.26/19.82 |
| Pgp | 1.25/1.20 | 1.72/1.65 | 1.50/1.44 | 2.85/2.73 | 2.99/2.86 | 3.01/2.86 | 4.71/4.51 |
| Tbcc | 2.63/2.04 | 1.97/1.53 | 2.34/1.82 | 2.85/2.21 | 2.84/2.20 | 1.81/2.20 | 1.87/1.45 |
| Tubb4 | 4.41/3.16 | 1.77/1.27 | 4.23/3.04 | 13.43/9.63 | 40.35/28.94 | 8.72/28.94 | 2.66/1.90 |
| Tubb5 | 3.95/2.55 | 1.76/1.13 | 5.87/3.79 | 2.81/1.82 | 4.11/2.66 | 4.28/2.66 | 3.33/2.15 |
| Zfand3 | 1.53/1.26 | 1.47/1.21 | 4.12/3.39 | 3.50/2.89 | 5.50/4.53 | 4.37/4.53 | 5.48/4.51 |
| (B) Genes up-regulated between five- and tenfold in TCs as compared with others | |||||||
| Trem2 | 15.11/76.80 | 36.31/184.56 | 30.83/156.68 | 25.98/132.03 | 51.13/259.88 | 30.45/259.88 | 44.38/225.54 |
Table 4.
The down-regulated fold of specific genes in TCs as compared with others in chromosome 17
| Folds(TC5 vs others/TC10 vs others) | |||||||
|---|---|---|---|---|---|---|---|
| (A) Genes up-regulated between one- and twofold in TCs as compared with others | |||||||
| Gene symbol | Fibroblast | Stem | ATII | CD8_T_BL | CD8_T_LL | Basal_cell | Duct_cell |
| 1600014C23Rik | 2.69/3.97 | 1.21/1.78 | 12.47/18.43 | 2.66/3.93 | 2.55/3.77 | 9.29/13.72 | 7.04/10.40 |
| 2310061I04Rik | 1.54/1.80 | 1.27/1.49 | 6.68/7.82 | 3.66/4.29 | 3.06/3.58 | 6.36/7.44 | 6.74/7.89 |
| 4930539E08Rik | 1.40/1.69 | 1.07/1.29 | 5.65/6.83 | 31.25/37.81 | 38.16/46.16 | 32.79/39.68 | 24.54/29.69 |
| 5430411C19Rik | 2.30/2.15 | 1.13/1.05 | 3.40/3.18 | 23.14/21.65 | 15.46/14.47 | 7.61/7.12 | 14.76/13.81 |
| A630033E08Rik | 2.67/3.15 | 1.24/1.46 | 7.91/9.35 | 29.81/35.22 | 41.89/49.49 | 20.02/23.65 | 6.67/7.88 |
| Atl2 | 1.32/1.84 | 2.22/3.10 | 21.29/29.73 | 14.23/19.88 | 13.07/18.25 | 33.45/46.71 | 32.74/45.72 |
| Atp6v1g2 | 1.05/1.44 | 1.72/2.36 | 2.20/3.02 | 1.80/2.47 | 1.61/2.20 | 2.38/3.26 | 3.17/4.34 |
| BC003965 | 2.62/2.97 | 1.70/1.93 | 8.77/9.92 | 21.82/24.69 | 17.18/19.44 | 39.62/44.82 | 25.30/28.62 |
| BC011426 | 1.06/1.32 | 1.25/1.56 | 5.65/7.04 | 15.51/19.30 | 18.28/22.76 | 1.50/1.86 | 3.09/3.85 |
| BC051142 | 1.12/1.60 | 1.16/1.67 | 1.38/1.98 | 2.89/4.15 | 1.45/2.08 | 3.18/4.58 | 2.00/2.87 |
| Cdc5l | 1.79/1.69 | 2.19/2.08 | 171.49/162.64 | 269.71/255.80 | 210.90/200.03 | 271.87/257.85 | 238.68/226.37 |
| Clpp | 1.05/1.06 | 1.63/1.64 | 6.30/6.34 | 4.95/4.97 | 3.45/3.47 | 8.03/8.08 | 7.75/7.79 |
| Cox7a2l | 1.42/2.18 | 1.21/1.87 | 1.41/2.17 | 3.45/5.30 | 2.34/3.60 | 4.80/7.39 | 3.91/6.02 |
| Dpp9 | 1.33/1.63 | 1.44/1.76 | 18.97/23.22 | 8.31/10.17 | 5.90/7.22 | 3.96/4.84 | 2.59/3.17 |
| Dpy30 | 1.38/2.23 | 1.11/1.78 | 6.93/11.15 | 12.89/20.76 | 7.90/12.73 | 16.78/27.02 | 15.21/24.49 |
| E4f1 | 1.60/1.63 | 2.12/2.16 | 2.86/2.92 | 14.41/14.68 | 23.06/23.50 | 3.05/3.11 | 2.74/2.79 |
| Eml4 | 1.08/1.61 | 4.20/6.27 | 1.20/1.79 | 2.43/3.63 | 2.77/4.15 | 2.32/3.47 | 2.45/3.66 |
| Gabbr1 | 1.09/1.34 | 3.45/4.23 | 9.29/11.39 | 29.00/35.58 | 35.59/43.67 | 19.70/24.17 | 12.19/14.96 |
| Glo1 | 3.48/4.60 | 1.46/1.93 | 3.77/4.98 | 3.69/4.87 | 2.29/3.03 | 4.37/5.77 | 6.12/8.08 |
| Gm88 | 1.52/1.02 | 2.10/1.41 | 52.54/35.36 | 11.10/7.47 | 8.02/5.40 | 73.00/49.13 | 27.98/18.83 |
| H2-M3 | 1.86/1.96 | 3.10/3.27 | 4.80/5.07 | 50.87/53.67 | 44.63/47.09 | 10.46/11.04 | 6.11/6.45 |
| Hn1l | 2.75/4.64 | 2.51/4.24 | 6.48/10.95 | 2.83/4.79 | 1.18/2.00 | 4.37/7.39 | 7.69/13.00 |
| Hsf2bp | 4.35/4.89 | 1.34/1.50 | 4.61/5.18 | 4.61/5.19 | 7.26/8.16 | 7.73/8.69 | 14.52/16.33 |
| Khsrp | 1.29/1.72 | 1.16/1.55 | 2.24/2.99 | 3.88/5.18 | 3.32/4.43 | 16.48/22.01 | 13.07/17.45 |
| Mdc1 | 1.52/1.72 | 2.00/2.26 | 3.04/3.45 | 2.69/3.05 | 2.71/3.07 | 4.07/4.60 | 2.42/2.74 |
| Mettl4 | 1.26/1.67 | 1.32/1.75 | 1.34/1.77 | 3.57/4.74 | 3.75/4.98 | 7.81/10.37 | 4.95/6.57 |
| Mrps18a | 2.16/3.03 | 1.25/1.75 | 2.71/3.80 | 1.90/2.67 | 1.52/2.14 | 3.50/4.91 | 3.08/4.32 |
| Msh6 | 1.26/1.82 | 2.07/3.00 | 1.92/2.78 | 3.44/4.99 | 2.42/3.51 | 3.41/4.94 | 2.44/3.53 |
| Ndufv2 | 1.76/2.27 | 1.52/1.96 | 1.42/1.82 | 1.64/2.11 | 1.59/2.05 | 2.38/3.06 | 2.02/2.60 |
| Pdcd2 | 1.57/1.98 | 2.66/3.36 | 1.83/2.31 | 3.34/4.22 | 2.81/3.55 | 2.83/3.58 | 2.09/2.65 |
| Pdpk1 | 1.51/2.36 | 1.27/1.99 | 1.17/1.83 | 3.12/4.88 | 2.17/3.41 | 1.46/2.29 | 1.74/2.72 |
| Phf1 | 1.10/1.17 | 1.92/2.04 | 19.66/20.88 | 6.46/6.86 | 9.04/9.61 | 4.90/5.21 | 3.46/3.67 |
| Pknox1 | 1.27/1.17 | 1.89/1.75 | 21.31/19.71 | 25.53/23.61 | 19.15/17.71 | 25.79/23.86 | 24.24/22.42 |
| Polr1c | 1.49/2.18 | 1.37/2.00 | 9.61/14.04 | 7.75/11.33 | 6.96/10.18 | 11.71/17.11 | 9.91/14.48 |
| Prrt1 | 2.80/2.43 | 1.92/1.67 | 6.39/5.55 | 6.29/5.46 | 11.82/10.27 | 2.39/2.08 | 1.41/1.23 |
| Psors1c2 | 6.73/5.81 | 1.31/1.13 | 12.72/10.98 | 13.83/11.94 | 12.50/10.79 | 8.46/7.30 | 8.08/6.98 |
| Rab5a | 1.08/1.42 | 1.34/1.75 | 4.47/5.86 | 7.74/10.15 | 8.45/11.08 | 3.48/4.56 | 3.64/4.78 |
| Rdbp | 1.22/1.55 | 1.48/1.88 | 1.93/2.45 | 2.51/3.18 | 1.42/1.80 | 4.19/5.32 | 3.61/4.58 |
| Ring1 | 1.28/1.35 | 1.05/1.11 | 59.34/62.76 | 20.66/21.85 | 3.77/3.99 | 92.26/97.58 | 73.81/78.07 |
| Rnps1 | 1.49/1.83 | 1.67/2.05 | 20.20/24.85 | 60.50/74.42 | 53.28/65.54 | 44.57/54.83 | 47.99/59.03 |
| Skiv2l | 1.44/1.33 | 1.55/1.44 | 25.93/24.00 | 24.77/22.93 | 20.72/19.18 | 4.41/4.09 | 3.28/3.04 |
| Slc22a1 | 1.79/2.00 | 1.58/1.76 | 7.12/7.94 | 1.22/1.36 | 1.48/1.65 | 11.28/12.57 | 11.06/12.33 |
| Slc25a23 | 1.03/1.45 | 1.28/1.79 | 44.78/62.78 | 3.84/5.38 | 2.57/3.61 | 40.36/56.58 | 32.46/45.51 |
| Solh | 1.21/1.22 | 1.24/1.24 | 88.56/89.01 | 148.74/149.50 | 153.65/154.43 | 111.96/112.53 | 76.12/76.51 |
| Srd5a2 | 1.05/1.37 | 1.09/1.43 | 2.87/3.76 | 6.38/8.38 | 2.32/3.05 | 5.92/7.78 | 4.08/5.36 |
| Srrm2 | 1.13/1.54 | 1.79/2.42 | 24.93/33.79 | 39.53/53.59 | 62.88/85.24 | 31.91/43.25 | 19.68/26.67 |
| Stk38 | 1.55/1.43 | 1.09/1.00 | 10.55/9.75 | 64.22/59.33 | 56.48/52.17 | 28.77/26.58 | 12.08/11.16 |
| Supt3h | 1.91/1.52 | 2.66/2.12 | 15.28/12.16 | 16.32/12.98 | 12.85/10.22 | 49.56/39.43 | 27.98/22.27 |
| Sytl3 | 1.15/1.28 | 1.06/1.19 | 4.59/5.14 | 4.63/5.18 | 6.65/7.44 | 10.58/11.85 | 5.56/6.23 |
| Taf8 | 1.47/1.50 | 1.87/1.90 | 2.86/2.90 | 3.23/3.28 | 2.51/2.55 | 4.39/4.45 | 2.28/2.31 |
| Tap1 | 3.12/2.15 | 1.91/1.32 | 25.60/17.68 | 173.22/119.61 | 180.68/124.76 | 46.31/31.97 | 36.19/24.99 |
| Tbc1d5 | 1.03/1.16 | 1.53/1.72 | 1.16/1.31 | 1.18/1.33 | 1.32/1.49 | 2.09/2.35 | 1.13/1.27 |
| Tbp | 1.01/1.45 | 2.31/3.33 | 1.11/1.60 | 2.79/4.02 | 1.77/2.56 | 1.87/2.70 | 1.09/1.58 |
| Tcf19 | 1.65/2.21 | 1.26/1.68 | 2.05/2.74 | 3.70/4.95 | 2.53/3.38 | 2.89/3.86 | 2.89/3.87 |
| Telo2 | 1.35/1.69 | 1.69/2.13 | 1.35/1.70 | 1.43/1.80 | 1.35/1.69 | 4.90/6.16 | 3.84/4.83 |
| Thumpd2 | 1.66/1.62 | 1.38/1.35 | 1.52/1.48 | 4.88/4.77 | 4.47/4.37 | 5.39/5.27 | 2.68/2.62 |
| Wdr27 | 1.30/1.15 | 1.28/1.13 | 8.01/7.09 | 2.61/2.31 | 3.82/3.38 | 15.14/13.40 | 1.80/1.60 |
| Wdr43 | 1.08/1.44 | 3.01/4.00 | 1.89/2.52 | 3.40/4.52 | 3.56/4.73 | 4.38/5.83 | 6.09/8.10 |
| Zfp101 | 3.45/4.24 | 1.27/1.56 | 3.57/4.39 | 14.70/18.07 | 15.77/19.38 | 6.56/8.06 | 2.51/3.09 |
| Zfp160 | 1.94/2.10 | 1.67/1.81 | 2.14/2.32 | 14.45/15.68 | 10.13/10.99 | 2.96/3.22 | 2.67/2.90 |
| Zfp161 | 1.70/1.76 | 1.71/1.77 | 5.14/5.32 | 10.17/10.51 | 10.33/10.67 | 14.79/15.28 | 4.90/5.07 |
| Zfp318 | 1.11/1.54 | 2.00/2.77 | 1.53/2.11 | 3.15/4.36 | 2.63/3.65 | 4.30/5.95 | 2.39/3.31 |
| Zfp472 | 1.85/2.12 | 1.17/1.34 | 4.32/4.94 | 45.25/51.74 | 33.07/37.81 | 2.46/2.82 | 1.59/1.82 |
| Zfp54 | 1.24/1.86 | 1.08/1.61 | 1.75/2.63 | 5.39/8.08 | 7.68/11.52 | 1.90/2.85 | 1.47/2.20 |
| Zfp563 | 2.16/2.00 | 1.39/1.29 | 5.71/5.28 | 9.05/8.37 | 4.10/3.79 | 12.25/11.33 | 6.19/5.72 |
| Zfp677 | 1.32/2.15 | 1.02/1.66 | 2.43/3.96 | 12.31/20.09 | 14.11/23.02 | 2.32/3.79 | 1.10/1.80 |
| Znrd1 | 1.29/1.64 | 1.17/1.49 | 1.82/2.31 | 2.95/3.74 | 2.57/3.26 | 1.88/2.39 | 1.61/2.04 |
| (B) Genes down-regulated between two- and fivefold in TCs as compared with others | |||||||
| C030034I22Rik | 5.83/6.02 | 3.09/3.18 | 3.61/3.72 | 26.28/27.11 | 28.59/29.49 | 10.78/11.11 | 5.35/5.52 |
Table 5.
Summary of genes expressed preferentially in TCs, as compared with others in chromosome 18
| Compaired pairs/fold up-regulated | >1 | >2 | >5 |
|---|---|---|---|
| TC5 vs. others | 14 | 8 | 0 |
| TC10 vs. others | 12 | 3 | 0 |
| TCs vs. others | 7 | 3 | 0 |
Table 6.
Summary of genes expressed preferentially in TCs, as compared with others in chromosome 18
| Compaired pairs/fold down-regulated | >1 | >2 | >5 |
|---|---|---|---|
| TC5 vs. others | 28 | 3 | 0 |
| TC10 vs. others | 50 | 6 | 1 |
| TCs vs. others | 20 | 2 | 0 |
Table 7.
The up-regulated fold of specific genes in TCs as compared with others in chromosome 18
| Folds(TC5 vs others/TC10 vs others) | |||||||
|---|---|---|---|---|---|---|---|
| (A) Genes up-regulated between one- and twofold in TCs as compared with others | |||||||
| Gene symbol | Fibroblast | Stem | ATII | CD8_T_BL | CD8_T_LL | Basal_cell | Duct_cell |
| 9430020K01Rik | 1.16/1.02 | 1.85/1.64 | 1.47/1.30 | 24.11/21.33 | 55.70/49.26 | 5.37/4.75 | 6.55/5.79 |
| Bin1 | 4.38/3.55 | 1.57/1.28 | 10.40/8.44 | 13.96/11.33 | 11.95/9.70 | 20.18/16.37 | 20.95/16.99 |
| Cdh2 | 167.95/180.20 | 1.55/1.67 | 86.02/92.30 | 144.75/155.30 | 24.65/26.45 | 6.56/7.04 | 13.85/14.86 |
| Fech | 1.44/1.39 | 1.93/1.87 | 1.53/1.48 | 2.63/2.54 | 3.01/2.91 | 2.52/2.43 | 2.80/2.71 |
| Txnl4a | 2.14/1.46 | 1.78/1.21 | 2.04/1.39 | 2.85/1.94 | 2.95/2.01 | 2.74/1.87 | 2.84/1.94 |
| Usp14 | 2.14/1.40 | 1.97/1.29 | 2.61/1.72 | 2.01/1.32 | 2.37/1.55 | 3.88/2.55 | 3.33/2.19 |
| Yipf5 | 1.17/1.04 | 2.22/1.96 | 1.78/1.57 | 1.98/1.75 | 3.09/2.73 | 2.12/1.87 | 2.77/2.44 |
| (B) Genes down-regulated between two- and fivefold in TCs as compared with others | |||||||
| Dpysl3 | 226.34/161.65 | 4.86/3.47 | 35.54/25.38 | 220.54/157.51 | 38.85/27.75 | 2.99/2.13 | 17.08/12.20 |
| Lims2 | 2.16/2.42 | 28.92/32.46 | 6.04/6.77 | 119.74/134.38 | 174.63/195.98 | 37.64/42.24 | 393.49/441.60 |
| Tubb6 | 5.38/3.70 | 3.96/2.73 | 25.62/17.62 | 457.27/314.51 | 267.70/184.12 | 16.55/11.38 | 9.71/6.68 |
Table 8.
The down-regulated fold of specific genes in TCs as compared with others in chromosome 18
| Folds(TC5 vs others/TC10 vs others) | |||||||
|---|---|---|---|---|---|---|---|
| (A) Genes up-regulated between one- and twofold in TCs as compared with others | |||||||
| Gene symbol | Fibroblast | Stem | ATII | CD8_T_BL | CD8_T_LL | Basal_cell | Duct_cell |
| 1700034H14Rik | 2.01/2.59 | 1.13/1.46 | 2.11/2.72 | 11.70/15.08 | 10.71/13.80 | 6.98/9.00 | 5.12/6.59 |
| 2210409D07Rik | 1.00/1.54 | 1.01/1.55 | 4.12/6.31 | 3.73/5.72 | 6.93/10.63 | 16.66/25.54 | 8.70/13.34 |
| 5430411K18Rik | 1.94/2.09 | 1.26/1.36 | 2.71/2.92 | 4.24/4.56 | 4.50/4.85 | 2.92/3.15 | 2.54/2.74 |
| Arhgap12 | 1.08/1.58 | 3.11/4.55 | 1.29/1.89 | 2.58/3.78 | 3.17/4.64 | 10.67/15.62 | 6.19/9.06 |
| Arhgap26 | 1.13/1.17 | 1.49/1.54 | 8.78/9.06 | 18.96/19.57 | 13.15/13.57 | 70.65/72.91 | 35.92/37.07 |
| Arsi | 1.07/1.18 | 1.14/1.26 | 25.60/28.26 | 21.69/23.95 | 56.02/61.85 | 43.99/48.58 | 17.26/19.06 |
| Hspa9 | 2.83/3.57 | 2.15/2.70 | 1.08/1.36 | 1.38/1.74 | 1.43/1.80 | 1.82/2.30 | 2.93/3.69 |
| Impa2 | 3.99/4.10 | 1.36/1.39 | 1.30/1.34 | 1.65/1.69 | 1.66/1.71 | 2.94/3.02 | 3.17/3.26 |
| Iws1 | 2.27/2.98 | 1.52/2.00 | 3.88/5.11 | 12.77/16.79 | 13.43/17.66 | 10.93/14.36 | 7.67/10.08 |
| Map3k2 | 1.48/1.47 | 2.36/2.34 | 3.77/3.74 | 9.79/9.72 | 11.64/11.55 | 9.24/9.17 | 6.01/5.96 |
| Mapk4 | 1.08/1.16 | 1.06/1.13 | 4.58/4.89 | 4.69/5.01 | 6.72/7.17 | 3.73/3.98 | 8.17/8.71 |
| Matr3 | 2.04/3.30 | 1.17/1.89 | 8.32/13.47 | 19.84/32.15 | 23.19/37.58 | 7.76/12.57 | 9.33/15.11 |
| Nfatc1 | 2.44/1.73 | 1.56/1.11 | 16.01/11.35 | 35.31/25.03 | 34.71/24.61 | 4.90/3.47 | 4.92/3.49 |
| Pcdhb10 | 1.92/2.19 | 1.55/1.77 | 5.62/6.41 | 1.55/1.77 | 15.45/17.62 | 1.47/1.67 | 1.95/2.23 |
| Pcdhb15 | 2.23/1.77 | 3.33/2.65 | 5.09/4.05 | 3.80/3.02 | 6.70/5.34 | 1.60/1.28 | 4.69/3.74 |
| Pcdhb8 | 1.96/2.06 | 1.27/1.33 | 2.23/2.34 | 12.19/12.82 | 5.11/5.38 | 6.13/6.45 | 6.10/6.42 |
| Seh1l | 1.63/2.35 | 1.53/2.21 | 1.74/2.51 | 1.19/1.71 | 1.50/2.16 | 5.80/8.39 | 5.60/8.10 |
| Snx24 | 2.08/1.96 | 1.10/1.04 | 131.64/124.43 | 1.36/1.29 | 1.71/1.62 | 84.17/79.56 | 26.83/25.36 |
| Stard6 | 2.97/1.83 | 1.71/1.05 | 10.62/6.54 | 11.65/7.18 | 14.98/9.23 | 7.01/4.32 | 18.24/11.24 |
| Tmed7 | 1.92/1.86 | 1.34/1.30 | 31.81/30.96 | 20.57/20.03 | 22.11/21.53 | 31.42/30.59 | 25.99/25.30 |
| (B) Genes down-regulated between two- and fivefold in TCs as compared with others | |||||||
| Scgb3a2 | 2.14/3.14 | 2.16/3.17 | 4083.95/5996.46 | 2.25/3.30 | 6.29/9.24 | 6608.77/9703.65 | 3851.22/5654.77 |
| Zfp397 | 2.34/3.46 | 2.52/3.71 | 2.24/3.31 | 2.78/4.11 | 2.89/4.26 | 8.71/12.84 | 3.70/5.46 |
In chromosome 17 the number of up- and down-regulated genes more than one-fold in TC5 was 332 and 250, 330 and 252, 184 and 398, 232 and 350, 223 and 359,153 and 429, or 192 and 390, as compared with MSCs, Fbs, ATII, ABCs, PACs, T-BL or T-LL, respectively. The number of up- and down-regulated genes more than one-fold in TC10 was 226 and 356, 245 and 337, 139 and 443, 194 and 388,177 and 405, 120 and 462, or 146 and 436, as compared with MSCs, Fbs, ATII, ABCs, PACs, T-BL or T-LL, respectively. The number of up- and down-regulated genes in TC5 and TC10 was 220 and 244, 229 and 236, 134 and 393, 185 and 341,170 and 352, 112 and 421, or 142 and 386, as compared with MSCs, Fbs, ATII, ABCs, PACs, T-BL or T-LL, respectively (Table 9). In chromosome 18 the number of up- and down-regulated genes in TC5 was 190 and 77, 173 and 94, 73 and 194, 92 and 175, 85 and 182, 54 and 213, or 58 and 209, as compared with MSCs, Fbs, ATII, ABCs, PACs, T-BL or T-LL, respectively. The number of up- and down-regulated genes in TC10 was 152 and 115, 140 and 127, 58 and 209, 69 and 198, 74 and 193, 44 and 223, or 49 and 218, as compared with MSCs, Fbs, ATII, ABCs, PACs, T-BL or T-LL, respectively. The number of up- and down-regulated genes in TC5 and TC10 was 143 and 68, 130 and 84, 52 and 188, 66 and 172, 69 and 177, 41 and 210, or 48 and 208, as compared with MSCs, Fbs, ATII, ABCs, PACs, T-BL or T-LL, respectively (Table 10). The detailed genes were shown in Additional files 3 and 4.
Table 9.
The number of genes specifically up- or down-regulated in pulmonary telocytes, as compared with other cells respectively in chromosome 17
| Compaired pairs | Up > 1 | Up > 2 | Up > 5 | Down > 1 | Down > 2 | Down > 5 | Down > 10 |
|---|---|---|---|---|---|---|---|
| TC5 vs. stem | 332 | 106 | 34 | 250 | 75 | 10 | 3 |
| TC10 vs. stem | 226 | 90 | 25 | 356 | 132 | 25 | 6 |
| TCs vs. stem | 220 | 68 | 19 | 244 | 68 | 8 | 3 |
| TC5 vs. fibroblast | 330 | 136 | 50 | 252 | 87 | 14 | 3 |
| TC10 vs. fibroblast | 245 | 95 | 36 | 337 | 124 | 22 | 4 |
| TCs vs. fibroblast | 229 | 84 | 31 | 236 | 79 | 9 | 2 |
| TC5 vs. ATII | 184 | 97 | 28 | 398 | 283 | 160 | 92 |
| TC10 vs. ATII | 139 | 71 | 19 | 443 | 333 | 193 | 114 |
| TCs vs. ATII | 134 | 61 | 15 | 393 | 227 | 151 | 85 |
| TC5 vs. CD8BL | 232 | 139 | 60 | 350 | 269 | 160 | 105 |
| TC10 vs. CD8BL | 194 | 113 | 40 | 388 | 300 | 175 | 115 |
| TCs vs. CD8BL | 185 | 100 | 37 | 341 | 261 | 149 | 97 |
| TC5 vs. CD8LL | 223 | 135 | 55 | 359 | 262 | 158 | 104 |
| TC10 vs. CD8LL | 177 | 105 | 43 | 405 | 301 | 170 | 122 |
| TCs vs. CD8LL | 170 | 99 | 39 | 352 | 254 | 150 | 95 |
| TC5 vs. basal cell | 153 | 77 | 28 | 429 | 332 | 201 | 120 |
| TC10 vs. basal cell | 120 | 59 | 23 | 462 | 377 | 238 | 150 |
| TCs vs. basal cell | 112 | 55 | 16 | 421 | 327 | 194 | 111 |
| TC5 vs. duct cell | 192 | 100 | 36 | 390 | 288 | 169 | 98 |
| TC10 vs. duct cell | 146 | 75 | 25 | 436 | 332 | 205 | 119 |
| TCs vs. duct cell | 142 | 67 | 20 | 386 | 282 | 165 | 88 |
Table 10.
The number of genes specifically up- or down-regulated in pulmonary telocytes, as compared with other cells respectively in chromosome 18
| Compaired pairs | Up > 1 | Up > 2 | Up > 5 | Down > 1 | Down > 2 | Down > 5 | Down > 10 |
|---|---|---|---|---|---|---|---|
| TC5 vs. stem | 190 | 83 | 18 | 77 | 21 | 0 | 0 |
| TC10 vs. stem | 152 | 49 | 15 | 115 | 31 | 4 | 1 |
| TCs vs. stem | 143 | 42 | 9 | 68 | 17 | 0 | 0 |
| TC5 vs. fibroblast | 173 | 81 | 17 | 94 | 31 | 1 | 1 |
| TC10 vs. fibroblast | 140 | 46 | 11 | 127 | 45 | 5 | 1 |
| TCs vs. fibroblast | 130 | 44 | 10 | 84 | 24 | 0 | 0 |
| TC5 vs. ATII | 73 | 35 | 14 | 194 | 148 | 77 | 43 |
| TC10 vs. ATII | 58 | 25 | 12 | 209 | 165 | 99 | 56 |
| TCs vs. ATII | 52 | 22 | 11 | 188 | 146 | 73 | 41 |
| TC5 vs. CD8BL | 92 | 50 | 27 | 175 | 133 | 84 | 49 |
| TC10 vs. CD8BL | 69 | 42 | 20 | 198 | 153 | 94 | 59 |
| TCs vs. CD8BL | 66 | 36 | 18 | 172 | 131 | 80 | 47 |
| TC5 vs. CD8LL | 85 | 59 | 23 | 182 | 142 | 94 | 46 |
| TC10 vs. CD8LL | 74 | 39 | 18 | 193 | 160 | 104 | 63 |
| TCs vs. CD8LL | 69 | 37 | 17 | 177 | 138 | 90 | 45 |
| TC5 vs. basal cell | 54 | 31 | 10 | 213 | 178 | 116 | 66 |
| TC10 vs. basal cell | 44 | 23 | 9 | 223 | 194 | 140 | 84 |
| TCs vs. basal cell | 41 | 19 | 8 | 210 | 175 | 112 | 63 |
| TC5 vs. duct cell | 58 | 37 | 11 | 209 | 167 | 106 | 54 |
| TC10 vs. duct cell | 49 | 27 | 11 | 218 | 186 | 126 | 70 |
| TCs vs. duct cell | 48 | 22 | 9 | 208 | 164 | 102 | 52 |
Hierarchical clustering of genes in chromosomes 17 and 18 was performed by TIGR Multi-experiment Viewer (MeV v4.9), respectively (Figure 1A and B). The physical and functional interaction of specific genes was further evaluated by String Network analysis (www.string-db.org) in chromosomes 17 and 18 (Figure 2). Twenty six genes presented close associations with each other. TCs-specific genes which were up- or down-regulated in TC5 and TC10 were selected as gene clusters. Figures 3 and 4 demonstrated differential changes of TCs-specific genes in TCs and other cells. Top 20% up- or down-regulated genes in TC5 or TC10 were extracted and compared with other cells via the normalize gene expression data in either chromosomes 17 (Figure 5) or 18 (Figure 6). The result showed that high or low expressed genes in TCs had few similarities with Fbs, MSCs, ATII, ABCs, PACs, T-BL, or T-LL, respectively.
Figure 1.
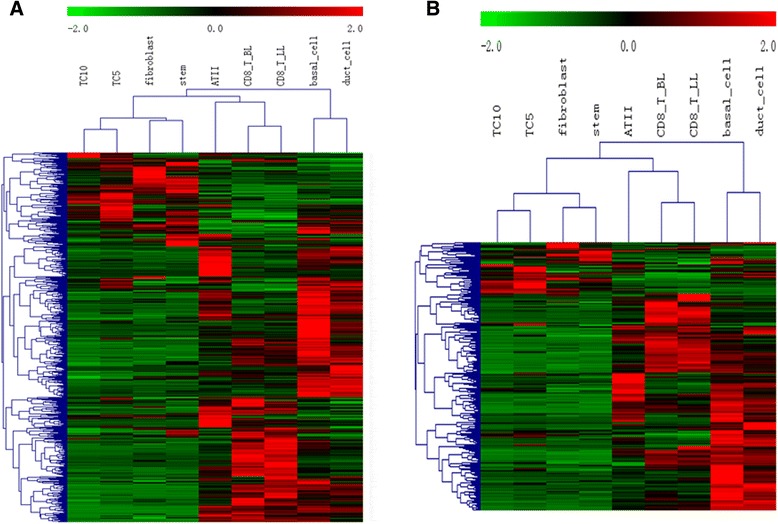
Hierarchical clustering of 582 genes in chromosomes 17 (A) and 267 genes 18 (B) among murine pulmonary telocytes (TCs), mesenchymal stem cells (MSCs), fibroblasts (Fbs), alveolar type II cells (ATII), airwaybasal cells (ABCs), proximal airway cells (PACs), CD8 + T cells from bronchial lymph nodes (T-BL) and CD8 + T cells from lungs (T-LL).
Figure 2.
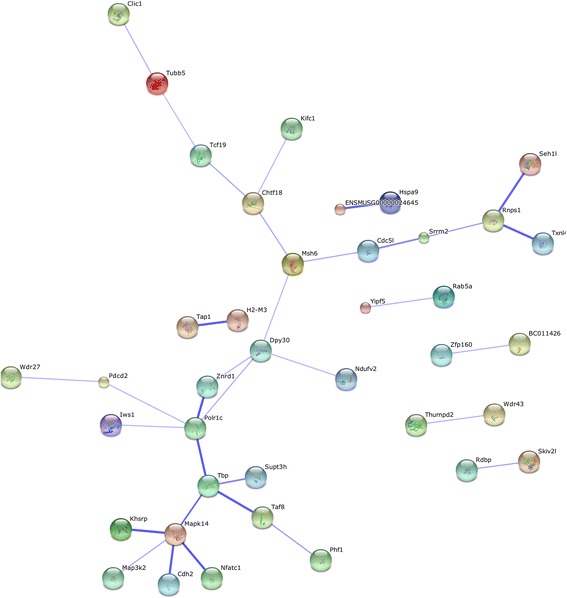
The physical and functional interaction of specific genes by String Network analysis in chromosome 17 and 18.
Figure 3.
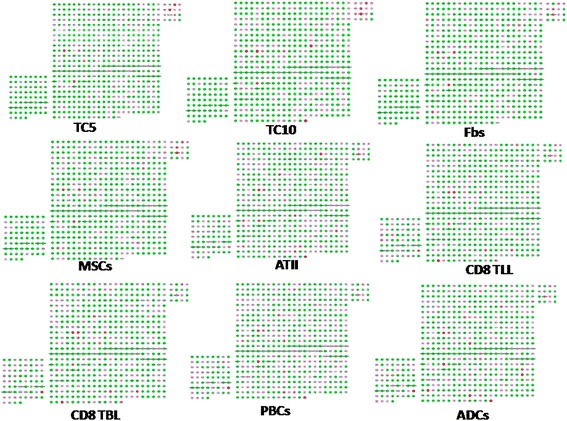
TCs-specific genes in chromosome 17 were selected as a specific group. Each node stands for each given gene. The upper right rectangular group represents up-regulated TCs-specific genes while the bottom left one represents down-regulated TCs-specific genes. Red nodes show genes up-regulated while green nodes show genes down-regulated.
Figure 4.

TCs-specific genes in chromosome 18 were selected as a specific group. Each node stands for each given gene. The upper right rectangular group represents up-regulated TCs-specific genes while the bottom left one represents down-regulated TCs-specific genes. Red nodes show genes up-regulated while green nodes show genes down-regulated.
Figure 5.
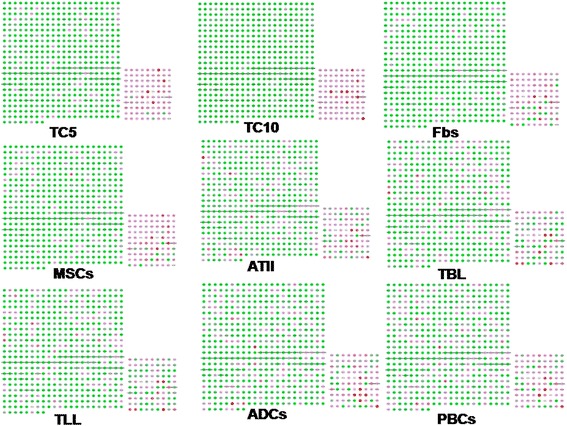
Top 20% up-regulated genes in TC10 was extracted and compared with the other cells via the normalize gene expression data in chromosome 17. Each node stands for each given gene. The bottom right rectangular group represents top 20% up-regulated TC10 genes.Red nodes show genes up-regulated while green nodes show genes down-regulated.
Figure 6.
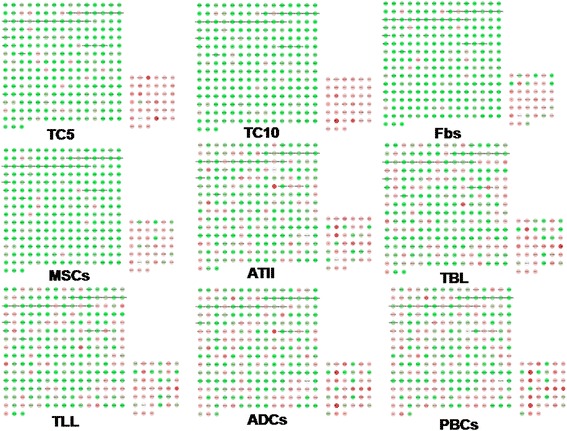
Top 20% up-regulated genes in TC10 was extracted and compared with the other cells via the normalize gene expression data in chromosome 18. Each node stands for each given gene. The bottom right rectangular group represents top 20% up-regulated TC10 genes.Red nodes show genes up-regulated, while green nodes show genes down-regulated.
Discussion
Chromosome 17 represents between 2.5 and 3 percent of human genome while chromosome 18 approximately 2.7% [30,31]. It has been reported that chromosomes 17 and 18 contained many disease-associated genes [32-37]. The present study initially demonstrated TCs-specific genes, of which 16 and 10 up-regulated and 68 and 22 down-regulated, in chromosomes 17 and 18, respectively. Of them, data from the present study indicated Mapk14, Trem2, MCFD2, or Dpysl3 as the representative of up-regulated genes and E4F1 and PDCD2 as the representative of down-regulated genes, though the further study is needed.
Mapk14 is the encoding gene of p38 to regulate different cellular functions, promote the production of pro-inflammatory cytokines, and be involved in the response to stress and metabolic pathways. p38 plays important roles in the maintenance of homoeostasis and related pathologies [38-40]. Trem2 is the member of Trem family which is highly conserved in evolution in different specials [41,42]. Trem2 negatively regulates cytokine synthesis and plays an inhibitory regulator in inflammatory response [43,44]. Telocytes were found to connect with immune cells and regulate the immune response [19]. Over-expressed Mapk14 gene in TCs may promote the production and secretion of cytokines or other signal materials to induce inflammation, while up-regulated Trem2 gene tends to inhibit inflammatory cytokine synthesis to counteract the activation of p38. It seems that TCs play a complicated and dual role in immune surveillance and immune regulation. During the physical process, TCs can be activated to maintain homeostasis to induce proliferation, differentiation and tissue regeneration. On the other hand, TCs initiate the tissue inflammation to induce pathogenesis under some challenges.
MCFD2 encodes proteins involved in the transport of FV and FVIII from the endoplasmic reticulum to the Golgi apparatus [45]. It is suggested that TCs play an important role as an autocrine/paracrine factor in maintaining stem cell potential and self-renewal [46,47]. TCs were recently discovered to join the stem cells in the regeneration and repair from myocardial infarction [48]. We speculate that MCFD2 up-regulated in TCs may promote positive interaction and intercommunication between TCs and stem cells, and contribute to tissue injury and repair by maintaining tissue homoeostasis. CRMP4 protein encoded by Dpysl3 is strongly expressed in the developing nervous system, and play a critical role in neuronal outgrowth and polarity, axon guidance and axonal protection, and regeneration [49,50]. The features of neural axon have great similarity with Tps of TCs on morphology and biological function. It is possible that up-regulated Dpysl3 is indispensable for TCs self-proliferation and cell-cell network forming.
E4F1 controls mammalian embryonic and somatic cell proliferation and survival, and is a key posttranslational regulator of p53, which modulates its effector functions involved in cell growth arrest or apoptosis [51,52]. The low expression of E4F1 in TCs has a positive effect on cell proliferation and differentiation and maintains the viability and activity of TCs in tissues. Programmed cell death 2(PDCD2) is a highly conversed protein and expressed in embryonic and adult tissues widely. The transfection of a construct expressing PDCD2 could induce apoptosis in human cell lines [53]. Equally, TCs down-regulate the PDCD2 to balance apoptosis and proliferation to maintain the homeostasis.
Conclusion
The study globally analyzed the variation of genes in chromosome 17 and 18 among pulmonary TCs, MSCs, Fbs, ATII, ABCs, PACs, T-BL, and T-LL for the first time. Approximately 15% and 12% genes in chromosomes 17 and 18 were identified as TCs-specific genes. The specific up-regulated genes, i.e. Mapk14, Trem2, MCFD2 and Dpysl3, and specific down-regulated genes, i.e. E4F1 and PDCD2, in chromosome 17 and 18 made us have a deeper insight into biological features and functions of TCs. It has been found that TCs present morphological contact with immune cells to form a cellular interaction network to participate in immune regulation [19]. Our study revealing variation of immune-associated genes in chromosome 17 and 18 gave an additional support to the essential role of TCs in immune surveillance and immune homeostasis which may protect from immune disorder in acute and chronic pulmonary diseases. TCs also played a vital role in tissue proliferation, differentiation and regeneration to maintain the tissue homeostasis.
Reviewers’ comments
Reviewer 1: Dragos Cretoiu
This topic is original and the study provides new cues within the field of the telocytes (TCs), a distinct type of stromal cells. There are some minor issues which, if addressed, could improve the manuscript.
1) Introduction – There are more papers already published regarding the presence of telocytes in different organs, which might be worth mentioning. Some characteristics of the immunophenotype and ultrastructure should also be described.
Authors’ response: We have now described the presence of TCs in different organs and the characteristics of the immunophenotype and ultrastructure in introduction paragraph.
The aim of the study is not clearly stated and therefore a strong justification for the purpose of this study must be included. Also, some discussion about the chromosome 2 and 3 gene expression profile in TCs should be mentioned.
Authors’ response: We revised and highlighted the aim of our study to investigate features and patterns of TCs-specific gene profiles and explore potential function of TCs by focusing in chromosomes 17 and 18. It was not only the first time but also a new method to mine the TCs-specific gene profiles in chromosome 17 and 18. We now also have made a discussion about the study for the chromosome 2 and 3 gene expression profiles in TCs, which had not been published before we submitted this article.
2) Results – It would be useful for the readers to describe name of gene (or the encoded protein) at least for the most significant ones.
Authors’ response: We completely agree with this suggestion and have listed these significant genes in Tables 1, 2, 3, 4, 5, 6, 7 and 8.
3) Conclusion – I would suggest that this paragraph should be re-written pointing out the specific novelty of this study and the major findings leading to some functional hypotheses. Anyway, phrases like “Data from the present study demonstrated TC-specific genes in chromosomes 17 and 18, although the mechanism of TCs-specific genes in biological process of TCs needs to be furthermore explored” must be removed since is hampering the clarity and the significance of the study. TCs diversity according to organ location is quite intriguing, even considering the proposed roles of this cell subset in immune surveillance/tissue homeostasis. The authors should comment more on this.
Authors’ response: We thank the reviewer’s advice and have re-written the conclusion paragraph to highlight the specific novelty of our study and TCs-specific genes identification with its potential function in chromosomes 17 and 18. Also, negative phrases like “Data from the present study demonstrated TC-specific genes in chromosomes 17 and 18, although the mechanism of TCs-specific genes in biological process of TCs needs to be furthermore explored” have been removed.
Besides, TCs was isolated from murine lung tissue in this study. We have described the proposed roles of TCs in immune surveillance/tissue homeostasis which may protect from immune disorder in acute and chronic pulmonary diseases. We think it’s really meaning to differentiate the proposed roles of TCs subsets from different organ location in further study, as reviewer suggested.
4) An English language revision is mandatory. Many sentences should be rephrased. For example “The novel feature of telocytes under TEM has extremely long TPs with podomers and podoms, to contact with neighboring or distant effectors like axinal of neuron”.
Quality of written English: Needs some language corrections before being published.
Authors’ response: We have revised the article by an English native specialist. I hope you are satisfied with the revised version of the manuscript and hope it suitable for the publication.
Reviewer 2: Qing Kay Li, Department of Pathology, Johns Hopkins Medical Institutions
Although telocytes (TCs) have been found in a variety of organs, TCs-specific biomarkers still need to be evaluated. This manuscript studied the gene expression profiles of murine pulmonary TCs and compared their profiles with a variety of cell types, including mesenchymal stem cells, fibroblasts, alveolar type II cells, airway basal cells, proximal airway cells, CD8+ T cells from bronchial lymph nodes, and CD8+ T cells from the lung. In the study, genes of chromosome 17 and 18 were extracted and analyzed using bioinformatics tools, in order to identify TCs-specific genes and their functional networks. They found that 16 and 10 of TCs-specific genes were up-regulated, and 68 and 22 were down-regulated in the chromosome 17 and 18, respectively. Particularly, Mapk14, Trem2, MCFD2 and Dpysl3 were up-regulated, whereas the E4F1 and PDCD2 were down-regulated in TCs. Their data demonstrated that the differential expression of subset of genes in chromosomes 17 and 18 were unique features of pulmonary TCs, and they might be used as biomarkers to distinguish TCs from the other type of cells. Their findings were also suggestive of that TCs might play critical roles in immune surveillance and other intracellular functions.
Quality of written English: Acceptable.
Authors’ response: We appreciated Dr. Qing Kay Li’s encouraging comment on this article.
Acknowledgements
The work was supported by Zhongshan Distinguished Professor Grant (XDW), The National Nature Science Foundation of China (91230204, 81270099, 81320108001, 81270131, 81300010), The Shanghai Committee of Science and Technology (12JC1402200, 12431900207, 11410708600, 14431905100), Operation funding of Shanghai Institute of Clinical Bioinformatics, and Ministry of Education, Academic Special Science and Research Foundation for PhD Education (20130071110043).
Abbreviation
- ABCs
Airway basal cells
- ATII
Alveolar type II cells
- DPYSL3
Dihydropyrimidinase-like 3
- E4F1
E4F transcription factor 1
- Fbs
Fibroblasts
- GEO
Gene Expression Omnibus
- MAPK14
Mitogen-activated protein kinase 14
- MCFD2
Multiple coagulation factor deficiency 2
- MSCs
Mesenchymal stem cells
- NCBI
National Center for Biotechnology Information
- PACs
Proximal airway cells
- PDCD2
Programmed cell death 2
- T-BL
CD8+ T cells from bronchial lymph nodes
- TCs
Telocytes
- TC5
TCs on days 5
- TC10
TCs on days 10
- TEM
Transmission electron microscopy
- T-LL
CD8+ T cells from lungs
- Tps
Telopodes
- TREM2
Triggering receptor expressed on myeloid cells 2
Additional files
Data profiles for all genes in chromosome 17.
Data profiles for all genes in chromosome 18.
Details of up-regulated and down-regulated gene expression variations in chromosome 17.
Details of up-regulated and down-regulated gene expression variations in chromosome 18.
Footnotes
Jian Wang and Ling Ye contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JW and LY analyzed data and wrote the manuscript, MLJ contributed to data collection and normalization, and XDW made study plans, overviewed dataand revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jian Wang, Email: jianwang@hotmail.com.
Ling Ye, Email: yeling22@163.com.
Meiling Jin, Email: jinml232@gmail.com.
Xiangdong Wang, Email: Xiangdong.wang@clintransmed.org.
References
- 1.Zheng Y, Li H, Manole CG, Sun A, Ge J, Wang X. Telocytes in trachea and lungs. J Cell Mol Med. 2011;15(10):2262–8. doi: 10.1111/j.1582-4934.2011.01404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rusu MC, Jianu AM, Mirancea N, Didilescu AC, Manoiu VS, Paduraru D. Tracheal telocytes. J Cell Mol Med. 2012;16(2):401–5. doi: 10.1111/j.1582-4934.2011.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rusu MC, Nicolescu MI, Jianu AM, Lighezan R, Manoiu VS, Paduraru D. Esophageal telocytes and hybrid morphologies. Cell Biol Int. 2012;36(12):1079–88. doi: 10.1042/CBI20120007. [DOI] [PubMed] [Google Scholar]
- 4.Vannucchi MG, Traini C, Manetti M, Ibba-Manneschi L, Faussone-Pellegrini MS. Telocytes express PDGFRalpha in the human gastrointestinal tract. J Cell Mol Med. 2013;17(9):1099–108. doi: 10.1111/jcmm.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao J, Wang F, Liu Z, Yang C. Telocytes in liver: electron microscopic and immunofluorescent evidence. J Cell Mol Med. 2013;17(12):1537–42. doi: 10.1111/jcmm.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi G, Lin M, Xu M, Manole CG, Wang X, Zhu T. Telocytes in the human kidney cortex. J Cell Mol Med. 2012;16(12):3116–22. doi: 10.1111/j.1582-4934.2012.01582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cretoiu D, Hummel E, Zimmermann H, Gherghiceanu M, Popescu LM. Human cardiac telocytes: 3D imaging by FIB-SEM tomography. J Cell Mol Med. 2014;18(11):2157–64. doi: 10.1111/jcmm.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gherghiceanu M, Popescu LM. Cardiac telocytes - their junctions and functional implications. Cell Tissue Res. 2012;348(2):265–79. doi: 10.1007/s00441-012-1333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rusu MC, Mirancea N, Manoiu VS, Valcu M, Nicolescu MI, Paduraru D. Skin telocytes. Ann Anat. 2012;194(4):359–67. doi: 10.1016/j.aanat.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Luesma MJ, Gherghiceanu M, Popescu LM. Telocytes and stem cells in limbus and uvea of mouse eye. J Cell Mol Med. 2013;17(8):1016–24. doi: 10.1111/jcmm.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gevaert T, De Vos R, Van Der Aa F, Joniau S, van den Oord J, Roskams T, et al. Identification of telocytes in the upper lamina propria of the human urinary tract. J Cell Mol Med. 2012;16(9):2085–93. doi: 10.1111/j.1582-4934.2011.01504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vannucchi MG, Traini C, Guasti D, Del PG, Faussone-Pellegrini MS. Telocytes subtypes in human urinary bladder. J Cell Mol Med. 2014;18(10):2000–8. doi: 10.1111/jcmm.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cretoiu SM, Cretoiu D, Popescu LM. Human myometrium - the ultrastructural 3D network of telocytes. J Cell Mol Med. 2012;16(11):2844–9. doi: 10.1111/j.1582-4934.2012.01651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corradi LS, Jesus MM, Fochi RA, Vilamaior PS, Justulin LJ, Goes RM, et al. Structural and ultrastructural evidence for telocytes in prostate stroma. J Cell Mol Med. 2013;17(3):398–406. doi: 10.1111/jcmm.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz-Flores L, Gutierrez R, Saez FJ, Diaz-Flores LJ, Madrid JF. Telocytes in neuromuscular spindles. J Cell Mol Med. 2013;17(4):457–65. doi: 10.1111/jcmm.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popescu LM, Faussone-Pellegrini MS. TELOCYTES - a case of serendipity: the winding way from Interstitial Cells of Cajal (ICC), via Interstitial Cajal-Like Cells (ICLC) to TELOCYTES. J Cell Mol Med. 2010;14(4):729–40. doi: 10.1111/j.1582-4934.2010.01059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popescu LM, Gherghiceanu M, Cretoiu D, Radu E. The connective connection: interstitial cells of Cajal (ICC) and ICC-like cells establish synapses with immunoreactive cells. Electron microscope study in situ. J Cell Mol Med. 2005;9(3):714–30. doi: 10.1111/j.1582-4934.2005.tb00502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popescu LM, Gherghiceanu M, Suciu LC, Manole CG, Hinescu ME. Telocytes and putative stem cells in the lungs: electron microscopy, electron tomography and laser scanning microscopy. Cell Tissue Res. 2011;345(3):391–403. doi: 10.1007/s00441-011-1229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, Bai C, Wang X. Telocyte morphologies and potential roles in diseases. J Cell Physiol. 2012;227(6):2311–7. doi: 10.1002/jcp.23022. [DOI] [PubMed] [Google Scholar]
- 20.Cretoiu SM, Cretoiu D, Marin A, Radu BM, Popescu LM. Telocytes: ultrastructural, immunohistochemical and electrophysiological characteristics in human myometrium. Reproduction. 2013;145(4):357–70. doi: 10.1530/REP-12-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suciu L, Popescu LM, Gherghiceanu M, Regalia T, Nicolescu MI, Hinescu ME, et al. Telocytes in human term placenta: morphology and phenotype. Cells Tissues Organs. 2010;192(5):325–39. doi: 10.1159/000319467. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Y, Bai C, Wang X. Potential significance of telocytes in the pathogenesis of lung diseases. Expert Rev Respir Med. 2012;6(1):45–9. doi: 10.1586/ers.11.91. [DOI] [PubMed] [Google Scholar]
- 23.Zheng YH, Li H, Ge JB, Gao HJ, Wang XD. Location of telocytes in mouse bronchial and pulmonary tissues. Zhonghua Bing Li Xue Za Zhi. 2012;41(3):172–5. doi: 10.3760/cma.j.issn.0529-5807.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Zhang M, Qian M, Wang L, Cismasiu VB, Bai C, et al. Genetic comparison of mouse lung telocytes with mesenchymal stem cells and fibroblasts. J Cell Mol Med. 2013;17(4):567–77. doi: 10.1111/jcmm.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Y, Cretoiu D, Yan G, Cretoiu SM, Popescu LM, Wang X. Comparative proteomic analysis of human lung telocytes with fibroblasts. J Cell Mol Med. 2014;18(4):568–89. doi: 10.1111/jcmm.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cismasiu VB, Radu E, Popescu LM. miR-193 expression differentiates telocytes from other stromal cells. J Cell Mol Med. 2011;15(5):1071–4. doi: 10.1111/j.1582-4934.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun X, Zheng M, Zhang M, Qian M, Zheng Y, Li M, et al. Differences in the expression of chromosome 1 genes between lung telocytes and other cells: mesenchymal stem cells, fibroblasts, alveolar type II cells, airway epithelial cells and lymphocytes. J Cell Mol Med. 2014;18(5):801–10. doi: 10.1111/jcmm.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng M, Sun X, Zhang M, Qian M, Zheng Y, Li M, et al. Variations of chromosomes 2 and 3 gene expression profiles among pulmonary telocytes, pneumocytes, airway cells, mesenchymal stem cells and lymphocytes. J Cell Mol Med. 2014;18(10):2044–60. doi: 10.1111/jcmm.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gene Expression Omnibus (GEO). http://www.ncbi.nlm.nih.gov/geo/.
- 30.Genetics Home Reference. http://ghr.nlm.nih.gov.
- 31.Nusbaum C, Zody MC, Borowsky ML, Kamal M, Kodira CD, Taylor TD, et al. DNA sequence and analysis of human chromosome 18. Nature. 2005;437(7058):551–5. doi: 10.1038/nature03983. [DOI] [PubMed] [Google Scholar]
- 32.Jiang H, Bai X, Meng F, Zhang C, Zhang X. Evaluation of chromosome 17 polysomy in breast cancer by FISH analysis of whole nuclei, and its clinicopathological significance. Oncol Lett. 2014;7(6):1954–8. doi: 10.3892/ol.2014.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinholz MM, Bruzek AK, Visscher DW, Lingle WL, Schroeder MJ, Perez EA, et al. Breast cancer and aneusomy 17: implications for carcinogenesis and therapeutic response. Lancet Oncol. 2009;10(3):267–77. doi: 10.1016/S1470-2045(09)70063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L, Lin J, Du H, Hu C, Huang Z, Lv Z, et al. Gender-specific DNA methylome analysis of a Han Chinese longevity population. Biomed Res Int. 2014;2014:396727. doi: 10.1155/2014/396727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert F. Disease genes and chromosomes: disease maps of the human genome. Chromosome 17. Genet Test. 1998;2(4):357–81. doi: 10.1089/gte.1998.2.301. [DOI] [PubMed] [Google Scholar]
- 36.Gilbert F. Disease genes and chromosomes: disease maps of the human genome. Chromosome 18. Genet Test. 1997;1(1):69–71. doi: 10.1089/gte.1997.1.69. [DOI] [PubMed] [Google Scholar]
- 37.Cody JD, Carter EM, Sebold C, Heard PL, Hale DE. A gene dosage map of Chromosome 18: a map with clinical utility. Genet Med. 2009;11(11):778–82. doi: 10.1097/GIM.0b013e3181b6573d. [DOI] [PubMed] [Google Scholar]
- 38.Oeztuerk-Winder F, Ventura JJ. The many faces of p38 mitogen-activated protein kinase in progenitor/stem cell differentiation. Biochem J. 2012;445(1):1–10. doi: 10.1042/BJ20120401. [DOI] [PubMed] [Google Scholar]
- 39.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429(3):403–17. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 40.Schieven GL. The p38alpha kinase plays a central role in inflammation. Curr Top Med Chem. 2009;9(11):1038–48. doi: 10.2174/156802609789630974. [DOI] [PubMed] [Google Scholar]
- 41.Sharif O, Knapp S. From expression to signaling: roles of TREM-1 and TREM-2 in innate immunity and bacterial infection. Immunobiology. 2008;213(9–10):701–13. doi: 10.1016/j.imbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 42.Viertlboeck BC, Schmitt R, Gobel TW. The chicken immunoregulatory receptor families SIRP, TREM, and CMRF35/CD300L. Immunogenetics. 2006;58(2–3):180–90. doi: 10.1007/s00251-006-0091-8. [DOI] [PubMed] [Google Scholar]
- 43.Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, et al. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol. 2006;177(6):3520–4. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 44.Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, Lanier LL. Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J Immunol. 2006;177(4):2051–5. doi: 10.4049/jimmunol.177.4.2051. [DOI] [PubMed] [Google Scholar]
- 45.Elmahmoudi H, Wigren E, Laatiri A, Jlizi A, Elgaaied A, Gouider E, et al. Analysis of newly detected mutations in the MCFD2 gene giving rise to combined deficiency of coagulation factors V and VIII. Haemophilia. 2011;17(5):e923–7. doi: 10.1111/j.1365-2516.2011.02529.x. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Zhao B, Chen Y, You D, Liu R, Rong M, et al. Multiple coagulation factor deficiency protein 2 contains the ability to support stem cell self-renewal. Faseb J. 2013;27(8):3298–305. doi: 10.1096/fj.13-228825. [DOI] [PubMed] [Google Scholar]
- 47.Toda H, Tsuji M, Nakano I, Kobuke K, Hayashi T, Kasahara H, et al. Stem cell-derived neural stem/progenitor cell supporting factor is an autocrine/paracrine survival factor for adult neural stem/progenitor cells. J Biol Chem. 2003;278(37):35491–500. doi: 10.1074/jbc.M305342200. [DOI] [PubMed] [Google Scholar]
- 48.Popescu LM, Gherghiceanu M, Manole CG, Faussone-Pellegrini MS. Cardiac renewing: interstitial Cajal-like cells nurse cardiomyocyte progenitors in epicardial stem cell niches. J Cell Mol Med. 2009;13(5):866–86. doi: 10.1111/j.1582-4934.2009.00758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marques JM, Rodrigues RJ, Valbuena S, Rozas JL, Selak S, Marin P, et al. CRMP2 tethers kainate receptor activity to cytoskeleton dynamics during neuronal maturation. J Neurosci. 2013;33(46):18298–310. doi: 10.1523/JNEUROSCI.3136-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagai J, Goshima Y, Ohshima T. CRMP4 mediates MAG-induced inhibition of axonal outgrowth and protection against Vincristine-induced axonal degeneration. Neurosci Lett. 2012;519(1):56–61. doi: 10.1016/j.neulet.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 51.Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127(4):775–88. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 52.Paul C, Lacroix M, Iankova I, Julien E, Schafer BW, Labalette C, et al. The LIM-only protein FHL2 is a negative regulator of E4F1. Oncogene. 2006;25(40):5475–84. doi: 10.1038/sj.onc.1209567. [DOI] [PubMed] [Google Scholar]
- 53.Baron BW, Hyjek E, Gladstone B, Thirman MJ, Baron JM. PDCD2, a protein whose expression is repressed by BCL6, induces apoptosis in human cells by activation of the caspase cascade. Blood Cells Mol Dis. 2010;45(2):169–75. doi: 10.1016/j.bcmd.2010.04.007. [DOI] [PubMed] [Google Scholar]


