Abstract
USP7 is a protein deubiquitinase with an essential role in development. Here, we provide evidence that USP7 regulates the activity of Polycomb repressive complex 1 (PRC1) in coordination with SCML2. There are six versions of PRC1 defined by the association of one of the PCGF homologues (PCGF1 to PCGF6) with the common catalytic subunit RING1B. First, we show that SCML2, a Polycomb group protein that associates with PRC1.2 (containing PCGF2/MEL18) and PRC1.4 (containing PCGF4/BMI1), modulates the localization of USP7 and bridges USP7 with PRC1.4, allowing for the stabilization of BMI1. Chromatin immunoprecipitation (ChIP) experiments demonstrate that USP7 is found at SCML2 and BMI1 target genes. Second, inhibition of USP7 leads to a reduction in the level of ubiquitinated histone H2A (H2Aub), the catalytic product of PRC1 and key for its repressive activity. USP7 regulates the posttranslational status of RING1B and BMI1, a specific component of PRC1.4. Thus, not only does USP7 stabilize PRC1 components, its catalytic activity is also necessary to maintain a functional PRC1, thereby ensuring appropriate levels of repressive H2Aub.
INTRODUCTION
Polycomb group (PcG) proteins are transcriptional repressors with key roles in development (1), which usually form distinctive multisubunit complexes: Polycomb repressive complex 1 (PRC1), PRC2, Pho repressive complex (PhoRC) (2), Polycomb repressive deubiquitinase (PR-DUB) (3), and dRING-associated factors (dRAFs) (4). However, not all PcG proteins are stable components of these complexes, as in the case of Drosophila Sex Comb on Midleg (SCM), which cooperates with PRC1 (5). SCM has been shown to repress transcription and to be recruited to PcG target genes independently of PRC1 (6), but its mechanism of action remains unclear. Similar to SCM, its two mammalian homologues, SCMH1 and SCMH2, comprise an MBT domain at the N-terminal region, a domain of unknown function (DUF3588), and a C-terminal SPM domain that mediates interactions with PRC1 (7). Mice harboring a strong hypomorphic mutation of SCMH1 show homeotic transformations and defective spermatogenesis (8), and SCMH1 cooperates with PRC1 in the ubiquitination of geminin (9). The MBT repeats of SCML2 have been shown to bind to monomethylated lysines in histones (10, 11), and we have recently shown that two different isoforms of SCML2 exhibit distinct functions in the regulation of the activation of CDK2/CYCE complexes and in gene repression by PRC1 (12, 13).
There are different versions of PRC1 in mammals, and they mediate the monoubiquitination of histone H2A at lysine 119 (H2Aub), a modification associated with transcriptionally repressed chromatin (14). The different species of PRC1 share an E3 ubiquitin protein ligase, RING1B. PRC1.4 and PRC1.2 are equivalent to Drosophila PRC1 and are composed of RING1B bound to either BMI1 or MEL18 (PCGF4 and PCGF2, respectively), along with different CBX and PHC proteins (15, 16). Both SCML2 and SCMH1 are found associated with these complexes, similar to the case of SCM in Drosophila. Additional versions of PRC1 (PRC1.1, PRC1.3, PRC1.5, and PRC1.6) comprise RING1B bound to RYBP or YAF1 and either PCGF1, PCGF3, PCGF5, or PCGF6 and do not contain CBX, PHC proteins, or SCML2/SCMH1 (15, 17).
Ubiquitin-specific protease 7 (USP7) is a deubiquitinase with an essential role in development from fruit flies to mammals (18, 19). In Drosophila, USP7 is mainly found as a stable complex with GMPS and targets ubiquitinated histone H2B, a modification associated with transcriptional activation. The action of the complex leads to histone H2B deubiquitination, thereby contributing to gene silencing in both flies and mammals (19, 20). However, a number of other USP7 partners have been identified in mammals, and USP7 displays additional roles such as the regulation of the p53-MDM2 axis (21, 22). In the latter case, both p53 and MDM2 are stabilized by USP7, although deletion of USP7 results in the net stabilization of p53 due to the loss of its deubiquitinase MDM2. It was proposed previously that a USP7 knockout results in embryonic lethality in mice due to indirect p53 stabilization. However, a double knockout of USP7 and p53 did not rescue lethality, indicating that USP7 has key roles in development independent of p53 (18, 23).
Interestingly, although USP7 does not encode a PcG protein, its mutation in Drosophila enhances the Polycomb phenotype (19). USP7 has been reported to be associated with PRC1, although there are conflicting results regarding the specific PRC1 components that interact with USP7. On the one hand, USP7 was recovered in a purification of RING1B-associated proteins (24), and we have shown that USP7 is associated with PRC1.1 and PRC1.3 (15, 24). In contrast, Maertens et al. (25) also recovered small amounts of USP7 by purification of MEL18. Also, direct binding of USP7 to the RING finger domains of RING1B, BMI1, and MEL18 has been detected in vitro (25, 26). However, the integrity of PRC1.4 and PRC1.2 involves the heterodimerization of the RING domains of RING1B and BMI1 or MEL18, which are then bound by the E2 ubiquitin-conjugating enzyme UbcH5c (27, 28). Thus, it is not clear how USP7 can access the RING finger domains of these proteins within PRC1 in cells.
In this report, we investigated the actions of USP7 on PRC1 components. We found an interaction between USP7 and the PcG protein SCML2 that facilitates USP7 binding to PRC1.4 and that USP7 occupies genes targeted by SCML2 and BMI1. Inhibition of USP7 alters the posttranslational modifications of several PRC1.4 components and results in a reduction of H2Aub levels, showing that USP7 regulates both the stability of PRC1 components and the activity of the complex.
MATERIALS AND METHODS
Cell lines, extract preparation, transfections, and treatments.
HCT116 wild-type, HCT116 USP7 knockout (USP7-KO) (kindly provided by Bert Vogelstein), HeLa, and 293T-REx cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS), penicillin (100 IU/ml), streptomycin (100 μg/ml), and glutamine (300 μg/ml). P22077 (Tocris) and HBX19818 (Chem Scene) were dissolved in dimethyl sulfoxide (DMSO), and cells were incubated for the indicated times in the presence of the inhibitor or an equivalent amount of DMSO. Whole-cell extracts were prepared by lysing cells in a solution containing 50 mM Tris (pH 7.5), 8 M urea, and 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). Cytosolic and nuclear extracts were prepared as described previously (29), with the following modifications: nuclei were resuspended in 600 mM KCl, and a short sonication was applied to shear the chromatin. Acid-extracted proteins were obtained by resuspending the cells in phosphate-buffered saline (PBS) containing 0.5% Triton X-100, 2 mM phenylmethylsulfonyl fluoride (PMSF), and 0.02% NaN3. After 10 min of lysis on ice and centrifugation for 10 min at 2,000 rpm, the pellet was washed and then resuspended in 0.2 M HCl overnight. After extraction and centrifugation for 10 min at 2,000 rpm, the supernatant containing the acid-extracted proteins was collected.
Transient transfection of HCT116 cells with small interfering RNAs (siRNAs) for human SCML2 (siRNAs 1 [5′-CCAAACGATCTCCTCAGCAAA], 2 [5′-CAGTATGTATTGCTACGGTTA], 3 [5′-GTTATATAGCTGTGTACCTGA], and 4 [5′-CAGGAGATATTTATACTACGA]) was performed by using Lipofectamine RNAimax (Invitrogen) according to the manufacturer's instructions. Transient transfection of HCT116 cells with plasmids encoding green fluorescent protein (GFP), GFP-SCML2A, GFP-SCML2B, GFP-SCML2AΔRBR, and GFP-SCML2BΔRBR was performed by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Stable transfection of 293T-REx cells was carried out with polyethylenimine (PEI). Cells were grown in 10-cm-diameter dishes and transfected with 5 μg of plasmid diluted in 250 μl of 150 mM NaCl, and 30 μl of PEI was diluted in 250 μl of 150 mM NaCl. Diluted plasmids and PEI were mixed, incubated for 15 min at room temperature, and then added to the cells and incubated overnight. The medium was then replaced, and on the following day, the cells were trypsinized and seeded at a 1:10 dilution. The next day, 5 μg/ml blasticidin, 2 μg/ml puromycin (InvivoGen), 100 μg zeocin (Invitrogen), or 1 mg/ml G418 (Sigma) was added to select resistant clones. The expression of the recombinant proteins or the short hairpin RNA (shRNA) was induced by adding 1 μg/ml doxycycline for 24, 48, or 72 h.
Antibodies.
Rabbit antibody against SCML2 was generated by using a glutathione S-transferase (GST) fusion protein of a central region of SCML2 and affinity purified. Antibodies against USP7 (catalogue number A300-033A; Bethyl), BMI1 (catalogue number A301-694A; Bethyl), CBX2 (catalogue number A302-524A; Bethyl), CBX8 (catalogue number A300-882A; Bethyl), RING1B (catalogue number A302-869A; Bethyl), CDK2 (catalogue number sc-163; Santa Cruz), H2A (catalogue number 05-1352; Millipore), H2B (catalogue number L88A6; Cell Signaling Technology), EZH2 (Reinberg laboratory), p53 (catalogue number sc-126; Santa Cruz), MDM2 (catalogue number sc-813; Santa Cruz), p84 (catalogue number GTX70220; GeneTex), and FLAG (M2; Sigma) were used for Western blot analysis, immunoprecipitation, chromatin immunoprecipitation (ChIP), and immunofluorescence.
Plasmids and sequences.
The construction of the backbones for the pINTO plasmids was described previously (15). SCML2A and SCML2B were cloned from human cDNA and inserted into pINTO plasmids by using EcoRI and AgeI restriction sites; truncations and deletions were obtained by PCR.
Protein purification.
His-tagged proteins were expressed in BL21(DE3) cells and purified by Ni-nitrilotriacetic acid (NTA) chromatography. After washing with 30 mM imidazole, the protein was eluted with 250 mM imidazole. His-SCML2B was further purified by using a MonoS column and eluted with a salt gradient. His-USP7-CDC was further purified by size exclusion chromatography using an S200 column. Purified proteins were dialyzed against a solution containing 50 mM Tris (pH 7.5), 100 mM NaCl, and 10% glycerol. In the case of His-USP7-CDC, 1 mM dithiothreitol (DTT) was also added.
GST fusion proteins or fragments were expressed in BL21(DE3) cells for 16 h at 16°C and purified by using a glutathione-Sepharose column. The column was washed with a solution containing 50 mM Tris (pH 7.5) and 50 mM NaCl, and the proteins were eluted in the presence of 10 mM glutathione. Purified proteins were dialyzed against a solution containing 50 mM Tris (pH 7.5), 50 mM NaCl, and 10% glycerol.
Immunoprecipitation.
Five hundred micrograms of nuclear extract at 1 mg/ml in a solution containing 50 mM Tris (pH 7.9) and 200 mM NaCl was centrifuged for 10 min at 20,000 × g at 4°C, and the supernatant was incubated with specific antibodies at 4°C overnight. The samples were then centrifuged for 10 min at 20,000 × g at 4°C, and protein G-Dynabeads (Invitrogen), preblocked in the presence of 1 mg/ml bovine serum albumin (BSA), were added to the supernatant. After 1 h of incubation at 4°C, the beads were washed 5 times with a solution containing 50 mM Tris (pH 7.9), 200 mM NaCl, and 0.05% Igepal CA630 (Sigma). Proteins were eluted in SDS loading buffer. For benzonase treatment, nuclear extracts were incubated in the presence of 50 U/mg benzonase for 16 h at 30°C prior to incubation with the antibodies.
In vitro pulldown.
Purified proteins were incubated in a solution containing 50 mM Tris (pH 7.5) and 200 mM NaCl for 20 min at room temperature. The proteins were then pulled down by using glutathione-Sepharose resin. The beads were washed 5 times with a solution containing 50 mM Tris (pH 7.5), 200 mM NaCl, and 0.05% Igepal CA630, and bound proteins were extracted in SDS loading buffer. Pulldowns were analyzed by Western blotting.
Fluorescence microscopy.
Forty-eight hours after transfection, cells were washed 3 times and fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After washing twice with PBS and twice with water, the cells were mounted by using SlowFade with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) and imaged by using an Axiovert 200M inverted microscope (Zeiss, Germany).
SCML2 pulldown from 293T-REx cells.
After induction of SCML2 with 1 μg/ml doxycycline for 24 h, cells were collected, and nuclear extracts were obtained as described above. The nuclear extract was diluted to 1 mg/ml in a solution containing 50 mM Tris (pH 7.9) and 200 mM NaCl and incubated with Strep-Tactin (IBA) beads for 1 h at 4°C. The beads were washed with a solution containing 50 mM Tris (pH 7.9), 200 mM NaCl, and 0.05% Igepal CA630 (Sigma-Aldrich) and then washed with a solution containing 50 mM Tris (pH 7.9) and 200 mM NaCl. SCML2 and associated proteins were eluted with 2 mM biotin in a solution containing 50 mM Tris (pH 7.9) and 200 mM NaCl.
Deubiquitinase assay.
Deubiquitinase activity was assayed by using ubiquitin with 7-amino-4-methylcoumarin (Ub-AMC) (Boston Biochem) as a substrate, as described previously (30), with small modifications. Briefly, 10 nM His-USP7-CDC was incubated with different concentrations of Ub-AMC in a solution containing 50 mM Tris (pH 7.5), 100 mM NaCl, 1 mM EDTA, 5 mM DTT, and 0.05% Tween 20 at 25°C. The assays were performed with a final volume of 75 μl in black 96-well plates (Corning). Fluorescence emission was measured with a SpectrMax M5 plate reader (Molecular Devices) at excitation and emission wavelengths of 355 nm and 460 nm, respectively.
Chromatin immunoprecipitation.
Cells were fixed in DMEM containing 10 mM HEPES (pH 7.6), 1% formaldehyde, 15 mM NaCl, 0.15 mM EDTA, and 0.075 mM EGTA for 10 min at room temperature. The reaction was quenched with 0.125 M glycine for 5 min at room temperature, and cells were washed with PBS and lysed in a solution containing 50 mM HEPES, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Igepal CA630, and 0.25% Triton X-100. After centrifugation, isolated nuclei were washed once in a solution containing 10 mM Tris (pH 8.0), 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA and then resuspended in a solution containing 10 mM Tris (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, and 0.5% N-lauryl sarcosine and sonicated by using a Diagenode Bioruptor instrument to an average chromatin size of 200 bp. Chromatin was diluted to 150 to 200 μg/300 μl in a solution containing 10 mM Tris (pH 8.0), 1 mM EDTA, 600 mM NaCl, and 3% Triton X-100, and antibodies were added (4 to 8 μg) and incubated at 4°C overnight. Protein G-Dynabeads (Invitrogen) were blocked with BSA, and 20 to 40 μl of beads was added to each immunoprecipitation mixture. After 1 h of incubation at 4°C, the beads were washed 5 times with a solution containing 750 μl of 25 mM HEPES (pH 7.6), 1 mM EDTA, 0.1% N-lauryl sarcosine, 1% NP-40, and 0.5 M LiCl. After a final wash with 750 μl of a solution containing 10 mM Tris (pH 8.0), 1 mM EDTA, and 50 mM NaCl, the beads were resuspended in a solution containing 100 mM sodium bicarbonate, 200 mM NaCl, and 1% SDS. One microgram of proteinase K was added to each sample, chromatin was incubated for 15 min at room temperature, and cross-linking was reversed at 65°C for 16 h. Ten percent of the input was treated in parallel. The DNA was extracted by using a PCR purification kit (Qiagen).
Library construction.
Libraries for ChIP sequencing (ChIP-seq) were prepared according to the manufacturer's instructions (Illumina) and as described previously (31). Briefly, immunoprecipitated DNA (∼5 ng) was first end repaired by using an End-It repair kit (Epicenter), tailed with deoxyadenine by using Klenow Exo− (New England BioLabs [NEB]), and ligated to custom adapters with LigaFast (Promega). Fragments of 300 ± 50 bp were size selected and subjected to ligation-mediated PCR (LM-PCR) amplification using Phusion DNA polymerase (catalogue number M0530; NEB). Libraries were quantified by quantitative PCR (qPCR) using primers annealing to the adapter sequence and sequenced at a concentration of 7 pM on an Illumina Genome Analyzer IIx instrument or at a concentration of 10 pM on an Illumina HiSeq instrument. In some cases, barcoding was utilized to sequence more than one sample per lane.
ChIP-seq analysis.
ChIP-seq analysis was performed as described previously, with modifications (15). Sequenced reads from ChIP-seq experiments were mapped with Bowtie, using the parameters −v2 −m1 (32). Normalized genome-wide read densities were computed with HOMER (33) and visualized on the UCSC genome browser (http://genome.ucsc.edu/). Bound regions (BRs) were identified by using MACS 1.40rc2 (34) with default parameters and then filtered for enriched regions (ERs) with at least 10 tags and an unadjusted P value of <10−10. SCML2 ERs were associated to gene targets by using HOMER, with the restriction that the distance between the ER and the associated gene must be <5 kb.
RESULTS
USP7 interacts with PRC1.4 and SCML2.
A proteomic analysis of the interactome of USP7 did not recover any of the core subunits of PRC1 and instead suggested an interaction of USP7 with SCML2, one of the homologues of the Drosophila PcG protein SCM (35). In support of this interaction, our purification of the different isoforms of SCML2 recovered USP7 as its strongest interactor (12). Reciprocal immunoprecipitation of endogenous SCML2 and USP7 performed with nuclear extracts of HCT116 cells confirmed their association (Fig. 1A). As expected, SCML2 was found in the pulldown of BMI1 or RING1B (Fig. 1A) (12). Additionally, we confirmed the interaction of USP7 with BMI1 and RING1B (Fig. 1A and B), in agreement with data from a previous report (26). None of these interactions were disrupted upon benzonase treatment of the nuclear extracts, indicating that neither DNA nor RNA is bridging these interactions (Fig. 1C). Our results show that USP7 interacts with components of PRC1.4 and SCML2, a PRC1.4-associated protein.
FIG 1.
USP7 interacts with PRC1 and SCML2. (A) Immunoprecipitation of SCML2, BMI1, RING1B, and USP7 was performed with nuclear extracts from HCT116 cells. Pulldown material was analyzed by Western blotting with specific antibodies against USP7 and SCML2. Nonspecific IgG served as a control, and 2% of the input (In) is shown. (B) USP7 immunoprecipitation in nuclear extracts from HCT116 cells. Pulldown material was analyzed by Western blotting with specific antibodies against RING1B, BMI1, KDM2B, EZH2, and histones H2A and H2B. Nonspecific IgG served as a control, and 2% of the input is shown. (C) USP7 immunoprecipitation in either control nuclear extracts (−Benz) or nuclear extracts treated with benzonase (50 U/mg) (+Benz) for 16 h prior to immunoprecipitation. Nonspecific IgG served as a control, and 2% of the input is shown. (D) Schematic representation of the domains in USP7 and the fragments used in the in vitro pulldown experiments. (E) Western blot analysis of the in vitro pulldown of His-SCML2B with GST alone or GST-fused USP7 fragments. (F) Schematic representation of the two human isoforms of SCML2 showing the different domains of the protein and the fragments employed for the in vitro pulldown experiments. (G) Western blot analysis of the in vitro pulldown of His-USP7-N with GST alone or GST fusions of MBT-DUF, MBT-DUF lacking the RBR, or the RBR alone.
To test for a direct interaction of SCML2 and USP7, we performed in vitro pulldown assays using different functional domains of USP7 (Fig. 1D): the N-terminal TRAF domain, the central catalytic domain (CD), and the C-terminal regulatory domain composed of 5 ubiquitin-like repeats (HUBL). Our results indicate that both the TRAF domain (USP7-N) and the HUBL repetitions (USP7-C) interact with His-SCML2B (Fig. 1E). The TRAF domain is essential for the nuclear localization of USP7 and has also been proposed to mediate substrate recognition (36–39). In a reciprocal analysis, we used different fragments of SCML2 (Fig. 1F) and showed that the N-terminal region of SCML2 containing its MBT and DUF domains interacted with the TRAF domain in USP7 (USP7-N) (Fig. 1G). We recently reported that there is an RNA binding region (RBR) between the MBT and DUF domains in SCML2 and that deletion of this RBR impaired SCML2 localization to chromatin and abolished the speckled localization of SCML2A within the nucleus (12). Although the RBR alone was not sufficient for binding to the TRAF domain of USP7, its deletion from the MBT-DUF fragment of SCML2 abolished such interactions (Fig. 1G). SCML2 also bound weakly to the USP7 regulatory HUBL domain (Fig. 1E). Within the N-terminal region, both the MBT and DUF domains contributed to the interaction, while in the C terminus, the SPM domain was sufficient to bind the HUBL domain of USP7 (data not shown).
As the interaction of GMPS with the HUBL domain stimulates USP7 deubiquitinase activity (30), we next tested the SCML2 fragments for a similar regulation of USP7 activity. We could not evaluate the effect on full-length USP7, as SCML2 might be a target of USP7 recognized through the TRAF domain and therefore might compete out the substrate. However, recombinant His-CD-HUBL activity has been reported to be similar to that of full-length USP7 (30). Thus, we used this fragment to assess the direct effects of different truncated SCML2 proteins on the regulation of USP7 deubiquitinase activity by the HUBL domains, using ubiquitin-AMC as a substrate. Analysis of the activity of the CD-HUBL fragment at different substrate concentrations revealed kinetics similar to those previously reported (Km, 5.7 ± 0.9 μM; Vmax, 443 ± 30 arbitrary units [AU]) (30). The addition of the MBT-DUF fragment was ineffectual, while the addition of the SPM domain of SCML2A slightly increased the maximum velocity of the reaction (Km, 8.1 ± 0.6 μM; Vmax, 551 ± 21 AU). Our results indicate that SCML2 interacts with this regulatory region of USP7 but does not induce significant changes to its activity in vitro, although we do not rule out that such an effect might occur in cells with physiologically relevant substrates or through an interaction with the N-terminal TRAF domain.
USP7 associates with SCML2A and PRC1 on target genes.
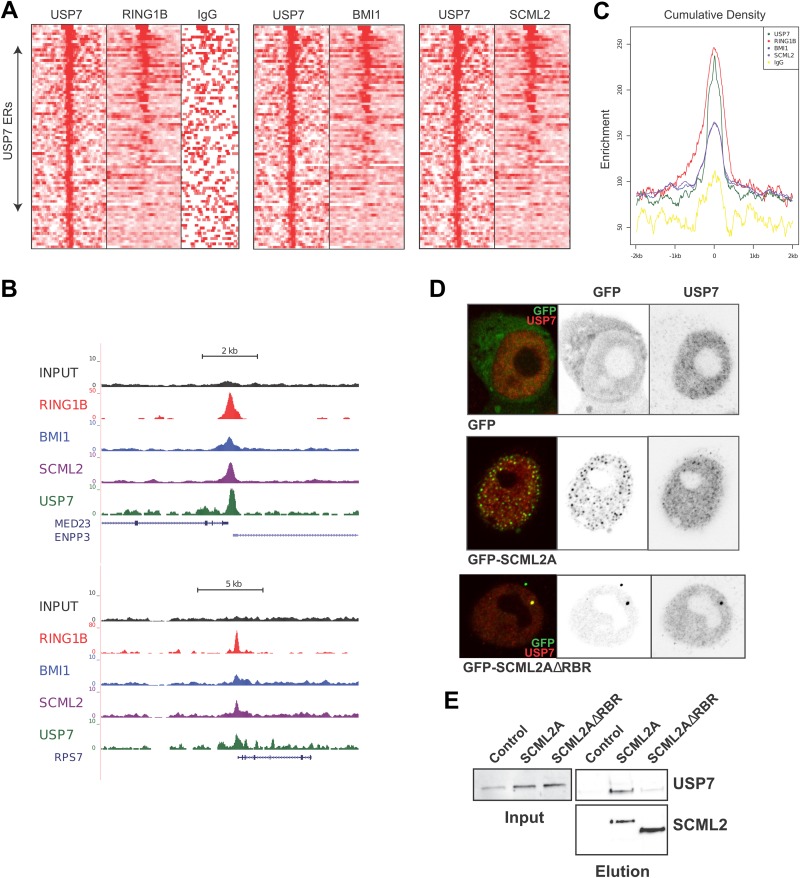
Based on the interaction of USP7 with SCML2, we explored whether USP7 can associate with SCML2/PRC1 target genes. We performed ChIP-seq experiments for USP7 in 293T-REx cells and defined a set of enriched regions for this protein near transcription start sites. Analysis of previously reported data for RING1B (15), BMI1, and SCML2 (12) revealed that ∼50% of the identified USP7-enriched regions also showed binding of these PRC1 components (Fig. 2A). Two examples of USP7-bound promoters that also exhibit peaks for RING1B, BMI1, and SCML2 are shown in Fig. 2B. We confirmed the colocalization of RING1B, BMI1, and SCML2 on USP7-enriched regions by analysis of the cumulative read density for these proteins across these regions (Fig. 2C). RING1B, BMI1, and SCML2 were all significantly enriched in USP7-bound regions. Previous results suggested that the action of USP7 on PRC1 occurs at the chromatin level (25), and our results demonstrate that USP7 cooccupies a subset of PRC1 target genes.
FIG 2.
USP7 colocalizes with SCML2 and PRC1 components on chromatin. (A) Heat map of normalized ChIP-seq density within a 4-kb window of the top 84 high-confidence USP7 enriched regions (ERs), as identified by model-based analysis of ChIP-Seq. RING1B ChIP-seq data were obtained from GEO series accession number GSE34774. (B) Representative read density tracks at 2 genomic locations demonstrating USP7 occupancy. (C) Cumulative read density across all USP7 ERs depicted in panel A. (D) Immunofluorescence analysis of wild-type HCT116 cells transfected with GFP alone, GFP-SCML2A, or GFP-SCML2AΔRBR and stained with an antibody specific for USP7. The individual channels are shown on the right, and the merged image is shown on the left. (E) Immunoprecipitation of SCML2A or SCML2AΔRBR from nuclear extracts of 293T-REx cells by using the Twin-Strep tag. Pulldown material was analyzed by Western blotting.
We further validated the interaction of USP7 with SCML2 by immunofluorescence. GFP-SCML2A localized in discrete foci in HCT116 cells, some of which also contained USP7 (Fig. 2D, middle). Our previous work showed that these speckles represent chromatin-bound SCML2 (12), again supporting that USP7 and SCML2A are found together on chromatin. In contrast to cells expressing GFP-SCML2A, the expression of GFP-SCML2AΔRBR, a mutant that is absent from chromatin, did not support the speckled localization of USP7 in HCT116 cells (Fig. 2D, bottom) (12). These results suggested either that SCML2A is necessary for the localization of USP7 or that mutant SCML2A could still interact with USP7 and act as a dominant negative mutant, driving it away from the speckles. However, pulldown of an RBR deletion mutant of SCML2A (SCML2AΔRBR) from 293T-REx cells recovered markedly reduced amounts of USP7 (Fig. 2E), indicating that the mutant is not acting as a dominant negative mutant and that SCML2A is likely driving the localization of USP7 to chromatin.
SCML2 is required for the action of USP7 on PRC1.4.
USP7 has been reported to directly interact with the ring finger domains of BMI1, MEL18, and RING1B (25, 26), but it is unclear how this interaction can occur in the context of PRC1.4 or PRC1.2 wherein RING1B forms heterodimers with BMI1 or MEL18 through these same domains (27, 28). We hypothesized that SCML2 could act as a bridge between USP7 and PRC1.4. To test this possibility, we first analyzed the levels of different PRC1 components after SCML2 knockdown using two different siRNAs (siRNAs 1 and 2) that target both SCML2 isoforms, in comparison with two different siRNAs (siRNAs 3 and 4) that specifically reduce the levels of SCML2A (Fig. 3A). Knockdown of SCML2 did not alter USP7 levels but reduced the levels of BMI1. In contrast, RING1B was not affected, similar to other USP7 targets such as p53 (Fig. 3A).
FIG 3.
SCML2 is required for the interaction of BMI1 with USP7. (A) Western blot analysis of the expression of SCML2, BMI1, USP7, p53, and RING1B after knockdown with a control siRNA (C), two different siRNAs targeting both SCML2A and SCML2B (siRNAs 1 and 2), and two different siRNAs targeting SCML2A (siRNAs 3 and 4) in HCT116 cells. Ponceau staining is shown as a loading control. (B) Western analysis of pulldown after immunoprecipitation of BMI1 (left) and RING1B (right) performed with nuclear extracts from HCT116 wild-type cells (WT) or USP7-KO cells (KO). (C) Same as for panel B, using nuclear extracts from HCT116 cells transfected with a control siRNA or siRNA 1 targeting both SCML2A and SCML2B (1). (D) 293T cells were transfected with a plasmid encoding HA-ubiquitin together with siRNA 1 for SCML2. After 2 days, the ubiquitinated material was pulled down from nuclear extracts with an anti-HA antibody. The pulled-down material was analyzed by Western blotting with antibodies against BMI1, HA, SCML2, or CDK2 as a loading control. Five percent of the input is shown, the arrows indicate ubiquitinated bands of BMI1, and the bar denotes polyubiquitinated BMI1. (E) Schematic representation of canonical PRC1 showing the interaction domains and the bridging model of SCML2 and USP7. (F) Model for the interaction of USP7 with RING1B in a subcomplex with RYBP.
Next, we addressed whether this effect is due to a bridging activity of SCML2. First, pulldowns of BMI1 and RING1B from nuclear extracts of HCT116 cells, either wild-type or USP7-KO cells, exhibited similar amounts of SCML2, indicating that the SCML2/PRC1 interaction is independent of USP7 (Fig. 3B). In contrast, the absence of SCML2 led to an impaired interaction between the PRC1.4 component BMI1 and USP7 (Fig. 3C), without affecting the amount of USP7 pulled down by RING1B, a subunit common to all versions of PRC1. We next transfected 293T cells with a plasmid encoding hemagglutinin (HA)-ubiquitin and analyzed the ubiquitination of BMI1 in control-treated and SCML2-depleted cells. As expected, the reduction in the level of SCML2 increased the ubiquitination of BMI1 (Fig. 3D), supporting a role of SCML2 in bringing together USP7 and its substrate BMI1. Based on these results, we propose that SCML2 facilitates the association of USP7 with PRC1.4 (Fig. 3E). In the case of other versions of PRC1 that are devoid of BMI1/MEL18, we postulate that USP7 can interact with the RING1B component and thereby bind directly to the complex (Fig. 3F).
USP7 regulates PRC1 activity.
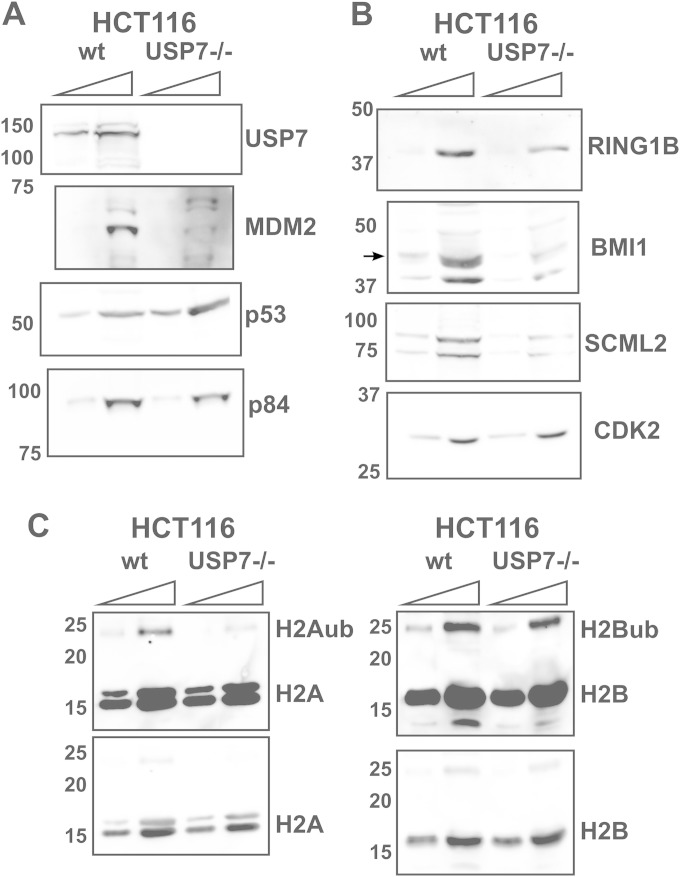
Given that USP7 has been reported to deubiquitinate RING1B, BMI1, and MEL18 (25, 26), we probed for its possible impact on the components of PRC1.4. First, we compared the levels of PRC1.4 components in wild-type HCT116 cells to those in HCT116 cells with a disrupted USP7 gene (USP7-KO), engineered as described previously by Cummins et al. (40). As previously reported, USP7-KO cells have lower levels of MDM2, which result in increased p53 expression levels (Fig. 4A) (40). In line with the reported role of USP7 in stabilizing RING1B (26), its levels were also reduced (Fig. 4B). USP7-KO cells also showed decreased levels of BMI1 and SCML2 (Fig. 4B). In agreement with our data showing a direct interaction between SCML2 and USP7, the levels of both SCML2 isoforms were reduced in the absence of USP7, independent of their association with PRC1. The levels of H2Aub, the main product of PRC1, were reduced in USP7-KO cells, while total levels of histone H2A remained unchanged (Fig. 4C). In contrast, H2B ubiquitination was not affected by the absence of USP7 (Fig. 4C). Interestingly, overexpression of USP7 has been shown to have no effect on the levels of H2Aub or H2Bub (25), supporting the idea that USP7 does not deubiquitinate histone H2A or H2B in vivo and that, in the case of USP7-KO cells, the effect on H2Aub levels is due to decreased levels of PRC1.
FIG 4.
Expression of PRC1 components and histones in HCT116 USP7-KO cells. (A and B) Western blot analysis of the expression of USP7, MDM2, p53, SCML2, and p84 (A) and PRC1 components (B) in whole-cell extracts from wild-type HCT116 (wt) and USP7-KO (USP7−/−) cells. (C) Acid-extracted proteins from HCT116 wild-type and USP7-KO cells were analyzed by Western blotting. Increasing amounts of the extract are loaded for comparison.
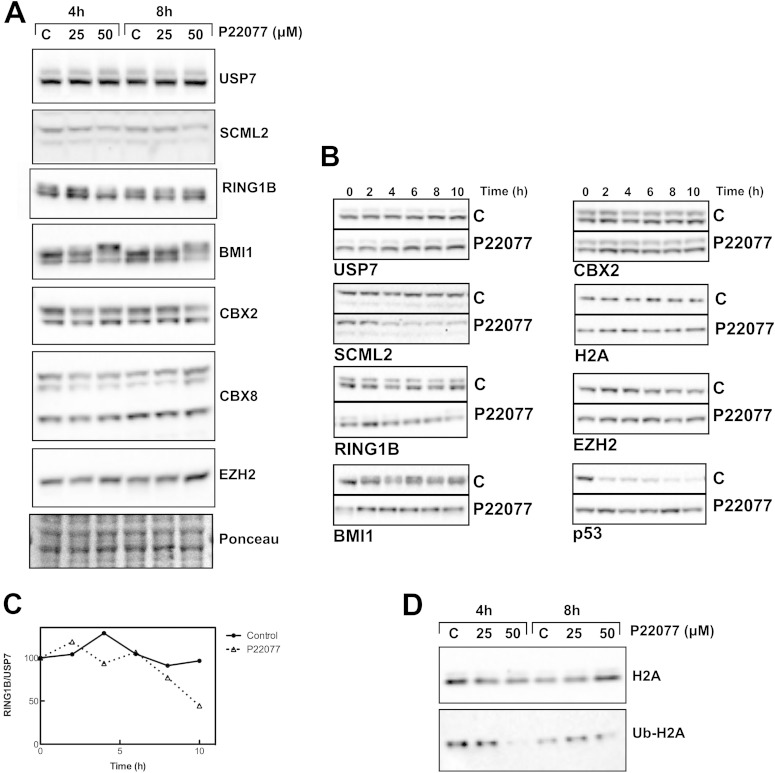
Since the expression of PRC1 is linked with high-level proliferation, while USP7-KO cells show very slow growth, the observed reductions in the levels of PRC1 components could be an indirect phenotype of USP7 depletion. To directly gauge the role of USP7 in the stabilization of PRC1, we took advantage of a recently developed inhibitor of USP7, P22077, which targets mainly USP7 and also has some activity against USP47 (41). Treatment of HCT116 cells with P22077 for 4 or 8 h did not change the levels of USP7 itself or those of EZH2, the catalytic subunit of PRC2 (Fig. 5A). Similarly, no changes were observed for CBX2 or CBX8 (Fig. 5A). In contrast, the levels of SCML2 were slightly reduced at 4 h, and the effect was stronger at 8 h of treatment (Fig. 5A). Interestingly, the inhibition of USP7 resulted in a change in the electrophoretic mobility of the catalytic subunits of PRC1. While the mobility of RING1B was increased, that of BMI1 was reduced (Fig. 5A). Thus, USP7 appears to modulate the posttranslational modification state of two key components of PRC1: RING1B, the catalytic subunit of all the complexes, and BMI1, which enhances RING1B activity (27, 28).
FIG 5.
Effect of USP7 inhibition on PRC1 levels and stability. (A) Western blot analysis of the expression of USP7, SCML2, RING1B, BMI1, CBX2, CBX8, and the PRC2 component EZH2 after treatment with the USP7 inhibitor P22077 for 4 or 8 h at the indicated concentrations. (B) HCT116 cells were treated with DMSO or 50 μM P22077 for 6 h and then treated with cycloheximide and either DMSO or P22077. Cells were collected every 2 h, and whole-cell extracts were analyzed by Western blotting for the expression of USP7, SCML2, RING1B, BMI1, CBX2, and the PRC2 component EZH2. p53 and histone H2A are shown as controls. (C) Changes in the levels of RING1B were quantified and normalized by the amount of USP7. The means of data from 2 independent experiments are shown. (D) Analysis by Western blotting of histone H2A and ubiquitinated histone H2A under the same conditions as those described above for panel A.
The depletion of USP7 has been shown to affect the levels of BMI1 and MEL18, suggesting that USP7 stabilizes both proteins (25). Thus, we analyzed whether the changes elicited by USP7 inhibition lead to reduced stability of PRC1.4 components. We measured the half-life of PRC1 components in HCT116 cells after P22077 treatment in the presence of cycloheximide. USP7 is very stable in both control and P22077-treated cells, similar to the case of CBX2 or histone H2A (Fig. 5B). While p53 was stabilized upon USP7 inhibition, as expected, the stability of SCML2 was reduced (Fig. 5B). In addition to the changes in RING1B mobility noted above, treatment with P22077 also led to a slight reduction in its stability (Fig. 5B and C). A similar effect was seen for BMI1, although the effect on protein stability was much weaker (Fig. 5B). Interestingly, the mobility of BMI1 gradually decreased in control cells starting at 2 h of cycloheximide treatment, similar to the effect seen upon the inhibition of USP7, although the protein remained stable during the incubation time (Fig. 5B). Importantly, USP7 inhibition led to decreased H2Aub levels, indicative of reduced PRC1 activity that might reflect disruptions to the putative posttranslational modifications of its components (Fig. 5D).
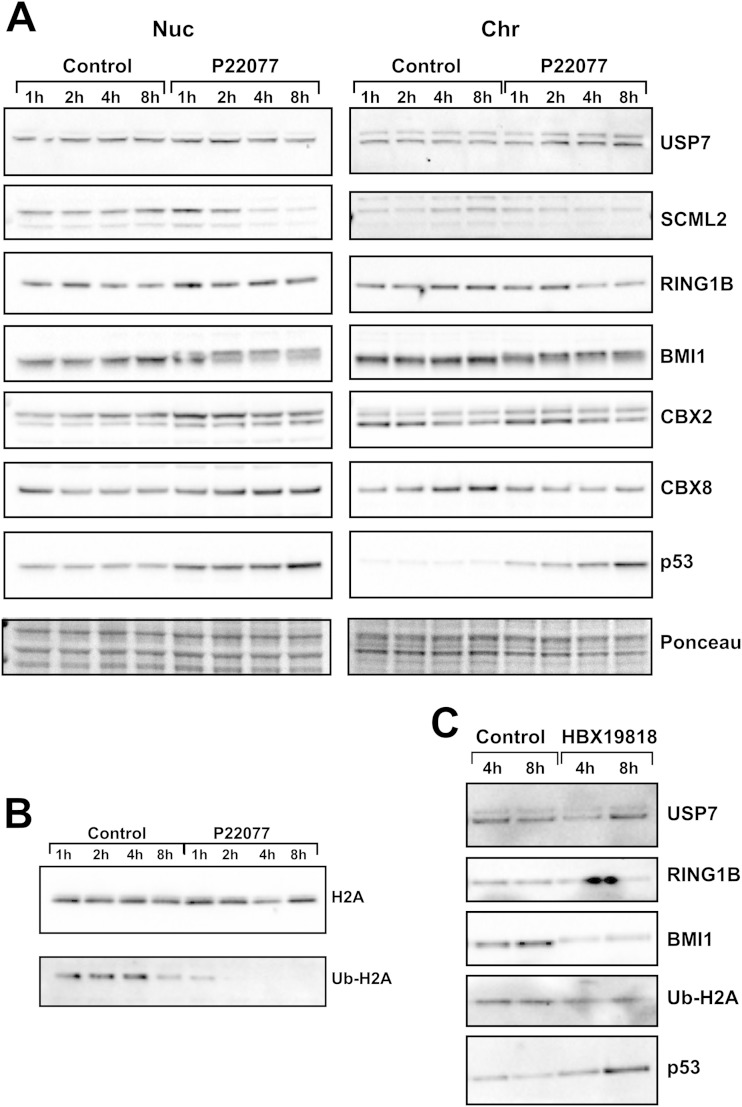
Although the global levels of PRC1 components were not changed after USP7 inhibition, it is possible that the effect of USP7 is specific to chromatin, as previously described (25). We performed fractionation of the soluble nuclear and chromatin fractions from HCT116 cells treated with P22077. USP7 inhibition elicited similar effects on both fractions (Fig. 6A). No changes were observed for USP7 itself, CBX2, or CBX8, while the levels of SCML2 were decreased after 4 h of treatment (Fig. 6A). The change in BMI1 mobility was observed in both fractions, being slightly faster in the case of fractions containing chromatin (Fig. 6A). Regarding RING1B, no change in the soluble nuclear fraction was detected, but a slight reduction in the level of chromatin-bound RING1B was observed after 4 h of USP7 inhibition (Fig. 6A). In contrast, the levels of H2Aub were already reduced after 2 h of P22077 treatment (Fig. 6B). A similar effect was observed by employing a structurally unrelated inhibitor, HBX19818 (Fig. 6C). These results further support that USP7 activity is necessary to maintain the functionality of PRC1 components, independently of its role in the stabilization of PRC1 components.
FIG 6.
USP7 inhibition affects PRC1 on chromatin. (A) HCT116 cells were treated with 50 μM P22077, and the expression levels of USP7, SCML2, RING1B, BMI1, CBX2, CBX8, and p53 were analyzed by Western blotting of the nuclear soluble (Nuc) and chromatin (Chr) fractions. (B) Analysis of the expression of histone H2A and the levels of Ub-H2A by Western blotting of the chromatin fraction of samples treated as described above for panel A. (C) Western blot analysis of the chromatin fraction of HCT116 cells after treatment with the USP7 inhibitor HBX19818 for 4 or 8 h at 50 μM. Expression levels of USP7, RING1B, BMI1, and Ub-H2A were analyzed.
DISCUSSION
Although initially thought to exclusively regulate protein stability, lysine ubiquitination has been shown to play a wide variety of roles in the regulation of protein localization, activity, and interactions. Protein ubiquitination is the result of equilibrium between the actions of E3 ubiquitin ligases and protein deubiquitinases. USP7 is a member of the ubiquitin-specific protease family and has an essential role during development (18). Interestingly, USP7 has been reported to stabilize several RING finger-containing E3 ubiquitin ligases (21, 25, 26, 40, 42–44), suggesting a broad role for this protein in the regulation of protein ubiquitination pathways and not just specific substrates.
Among the E3 ligases targeted by USP7, the ubiquitination of several components of PRC1 is regulated by USP7 (22). The complexity of PRC1 greatly increases from flies to mammals. All the different versions of PRC1 share the catalytic subunit (RING1B or RING1A) and mediate the ubiquitination of histone H2A but also exhibit distinctions in specific associated proteins. USP7 has been consistently recovered in association with RING1B complexes (15, 24). Our previous work has shown that USP7 is part of PRC1.1 and PRC1.3 and is not found in PRC1.2 or PRC1.4 (15). In contrast, Maertens et al. (25) recovered small amounts of USP7 in the purification of PRC1.2 but not in the purification of PRC1.4. Different reports have confirmed the direct binding of USP7 to the RING domain in RING1B, MEL18, and BMI1 in vitro (25, 26), which led to the hypothesis that USP7 binds to PRC1.2 and PRC1.4 directly. However, the structure of these complexes makes it highly unlikely for these interactions to take place in vivo. The minimally active PRC1.2 and PRC1.4 are formed by the heterodimerization of RING1B with MEL18 and BMI1, respectively, through their RING domains, which also constitutes the docking site for UbcH5c (27, 28). Therefore, such interactions would preclude USP7 binding to BMI1, MEL18, or RING1B within PRC1.2 or PRC1.4. We postulated that an additional protein may mediate USP7 binding in the case of these complexes.
Our data reported here along with previously reported proteomic information regarding USP7-associated proteins (12, 35) indicated that SCML2 could be the link between USP7 and PRC1.2 or PRC1.4. In fact, our results demonstrate that SCML2 directly interacts with USP7 and regulates its localization, independent of PRC1. SCML2A, the longest isoform of SCML2 (12), can interact with the TRAF domain in USP7 through its MBT-DUF region and with PRC1.4 through its C-terminal SPM domain, which mediates binding to PHC, a component of PRC1.4 (Fig. 3E). This model predicts that USP7 binding to BMI1 would be abrogated upon the depletion of SCML2, leading to increased ubiquitination of BMI1, as we have shown here (Fig. 3C and D). As a result, the levels of BMI1 are reduced, in agreement with a role of USP7 in BMI1 stabilization (25). The work by Maertens et al. (25) does not report the presence of SCML2 or any other SCM homologue in their purification of MEL18-associated proteins. As SCMH1 and SCML2 have been consistently found together with PRC1.2 and PRC1.4, we surmise that SCML2 also might have been recovered in their purification and not identified as an interactor, thereby explaining the presence of USP7 in the pulldown. The functional relevance of the interaction between SCML2 and USP7 is reflected by the cooccurrence of USP7 with SCML2 and BMI1 on target genes, supporting a role for the USP7-SCML2 module in the regulation of PRC1.4 functions.
In contrast to PRC1.2 and PRC1.4, the RING domain of RING1B is expected to be accessible in other versions of PRC1, suggesting that its direct interaction with USP7 is tenable in these cases, which is consistent with the abundant presence of USP7 upon RING1B purification (15). It is possible that other components of noncanonical PRC1, such as RYBP or YAF2, foster USP7 binding and stabilization of RING1B in a manner similar to that of SCML2. Given the central role of RING1B in the formation and stability of PRC1, it is also likely that its stabilization is not established within the whole complex but rather in the pool of free RING1B or in subcomplexes with other proteins like RYBP (Fig. 3F).
Our results also show that USP7 regulates the stability of SCML2 and RING1B and, perhaps, the state of posttranslational modification of BMI1 and RING1B. Previous reports have shown that USP7 stabilizes BMI1, MEL18, and RING1B (25, 26); in contrast, we detected only a mild destabilization of RING1B upon USP7 inhibition. The lack of a stronger effect upon the inhibition of USP7 is likely due to the long half-life of these proteins (>8 h). Treatment with P22077 induces cell death after 8 h, precluding incubation for the times required to clearly determine if USP7 regulates BMI1 or RING1B stability. Nonetheless, the destabilization of RING1B after USP7 inhibition takes place on chromatin, as previously observed for BMI1 and MEL18 after USP7 depletion (25).
Interestingly, even if the level or the localization of PRC1 is not grossly affected, USP7 inhibition reduces the amount of global H2Aub, the main product of PRC1, underscoring the essential role of USP7 in the regulation of PRC1 function. Our data suggest that the activity of USP7 is necessary to sustain the functionality of PRC1, most likely through the regulation of RING1B. Whether the modification of RING1B affects its intrinsic catalytic activity or the formation of the complex remains to be explored. In contrast, Maertens et al. (25) did not detect a reduction in the level of H2Aub upon USP7 depletion. As the levels of RING1B remaining after the depletion of USP7 were not addressed in that study, it is difficult to predict the extent to which the levels of H2Aub should be decreased. As RYBP-containing PRC1 components are responsible for the bulk of H2Aub, reductions in BMI1 and MEL18 levels would not be expected to alter global H2Aub levels. Based on all of these results, we propose that USP7 ensures PRC1 function through the direct regulation of the activity of the complex and the stability of the catalytic subunits RING1B, BMI1, and MEL18. Furthermore, a recent report showed that USP7 may also cooperate with HSCARG to negatively regulate the activity of PRC1 (45), indicating that the actions of USP7 on PRC1 may be context dependent.
In Drosophila, USP7 has been reported to target H2Bub in association with GMPS, thereby facilitating PcG target gene repression (19). Other studies did not find changes in H2Bub levels when USP7 expression was reduced (25), and we have not detected any changes in H2Bub levels in cells without USP7, suggesting that USP7 is not essential for maintaining global levels of H2B ubiquitination. However, a specific effect on target genes cannot be ruled out. Also, previously reported results revealed an interaction of USP7 with EZH2 in vitro (26). Our data show that the effect of USP7 is restricted to PRC1, as the levels of EZH2 were not changed upon USP7 inhibition.
PRC1 is essential for embryonic development, as reflected by embryonic lethality upon the deletion of the RING1B gene (46). While the elimination of individual components of PRC1.2 and PRC1.4 is not lethal, double knockouts, such as BMI1 and MEL18, BMI1 and CBX2, or PHC1 and PHC2 knockouts, lead to embryonic lethality (47–49). The deletion of RYBP, a common subunit of PRC1.1, PRC1.3, PRC1.5, and PRC1.6, is also embryonic lethal in early stages postimplantation (50). The deletion of USP7 results in a disorganized embryo with reduced proliferation and no signs of apoptosis at embryonic day 6.5 (E6.5) (18), a phenotype very similar to that observed in the absence of RYBP (50). Our results show that USP7 is essential to preserving PRC1 activity and appropriate levels of H2Aub. A recent report showed that, among the proposed mechanisms for PRC1-mediated gene repression, ubiquitination of H2A is essential for the maintenance of the undifferentiated state of embryonic stem cells and that the loss of the catalytic activity of RING1B and RING1A derepresses a subset of PRC1 target genes (14). It has been proposed that the stabilization of p53 in USP7-KO animals is responsible for its phenotype. However, the concomitant deletion of p53 and USP7 does not rescue lethality (18). Additional USP7 targets must then be essential for its functions. Functionally, PRC1 and USP7 converge on several processes, and our results support a model in which USP7 regulation of development is mediated, in part, by PRC1 stabilization.
In this work, we have addressed various aspects of the interplay between USP7 and PRC1. We have shown that SCML2 directly interacts with USP7 and drives its localization. SCML2 connects USP7 to PRC1.4, allowing for the stabilization of BMI1, and USP7 is found on SCML2 and BMI1 target genes. Additionally, USP7 modulates the stability of SCML2 and appears to regulate posttranslational modifications of BMI1 and RING1B. The activity of USP7 is required to ensure full PRC1 action and maintain H2Aub, and the absence of such an activity might underlie the embryonic lethality found in the case of the USP7 knockout. As a whole, our results confirm that USP7 is a key regulator of PRC1 that affects not only its stability but also its activity.
ACKNOWLEDGMENTS
We thank Bert Vogelstein for providing the different HCT116 cell lines. We are grateful to Lynne Vales for comments on the manuscript.
This work was supported by an international outgoing fellowship from the Marie-Curie Actions FP7 from the European Commission to E.L., NIH grants GM064844 and GM37120 to D.R., and HHMI (to D.R.).
REFERENCES
- 1.Simon JA, Kingston RE. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz YB, Pirrotta V. 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 3.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. 2010. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP. 2008. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Jahren N, Miller EL, Ketel CS, Mallin DR, Simon JA. 2010. Comparative analysis of chromatin binding by Sex Comb on Midleg (SCM) and other polycomb group repressors at a Drosophila Hox gene. Mol Cell Biol 30:2584–2593. doi: 10.1128/MCB.01451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornemann D, Miller E, Simon J. 1996. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development 122:1621–1630. [DOI] [PubMed] [Google Scholar]
- 8.Tomotsune D, Takihara Y, Berger J, Duhl D, Joo S, Kyba M, Shirai M, Ohta H, Matsuda Y, Honda BM, Simon J, Shimada K, Brock HW, Randazzo F. 1999. A novel member of murine Polycomb-group proteins, Sex comb on midleg homolog protein, is highly conserved, and interacts with RAE28/mph1 in vitro. Differentiation 65:229–239. doi: 10.1046/j.1432-0436.1999.6540229.x. [DOI] [PubMed] [Google Scholar]
- 9.Luo L, Yang X, Takihara Y, Knoetgen H, Kessel M. 2004. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature 427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- 10.Bonasio R, Lecona E, Reinberg D. 2010. MBT domain proteins in development and disease. Semin Cell Dev Biol 21:221–230. doi: 10.1016/j.semcdb.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sathyamurthy A, Allen MD, Murzin AG, Bycroft M. 2003. Crystal structure of the malignant brain tumor (MBT) repeats in Sex Comb on Midleg-like 2 (SCML2). J Biol Chem 278:46968–46973. doi: 10.1074/jbc.M306469200. [DOI] [PubMed] [Google Scholar]
- 12.Bonasio R, Lecona E, Narendra V, Voigt P, Parisi F, Kluger Y, Reinberg D. 2014. Interactions with RNA direct the Polycomb group protein SCML2 to chromatin where it represses target genes. eLife 3:e02637. doi: 10.7554/eLife.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lecona E, Rojas LA, Bonasio R, Johnston A, Fernandez-Capetillo O, Reinberg D. 2013. Polycomb protein SCML2 regulates the cell cycle by binding and modulating CDK/CYCLIN/p21 complexes. PLoS Biol 11:e1001737. doi: 10.1371/journal.pbio.1001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, Vidal M, Bernstein BE, Koseki H. 2012. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet 8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. 2002. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol 22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W. 2010. Inactivation of HAUSP in vivo modulates p53 function. Oncogene 29:1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. 2005. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell 17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Sarkari F, Sanchez-Alcaraz T, Wang S, Holowaty MN, Sheng Y, Frappier L. 2009. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication. PLoS Pathog 5:e1000624. doi: 10.1371/journal.ppat.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Brooks CL, Kon N, Gu W. 2004. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 13:879–886. doi: 10.1016/S1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson B, Suresh Kumar KG. 2011. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochem Biophys 60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- 23.Kon N, Zhong J, Kobayashi Y, Li M, Szabolcs M, Ludwig T, Canoll PD, Gu W. 2011. Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell Death Differ 18:1366–1375. doi: 10.1038/cdd.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. 2007. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics 6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. 2010. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J 29:2553–2565. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Bie P, Zaaroor-Regev D, Ciechanover A. 2010. Regulation of the Polycomb protein RING1B ubiquitination by USP7. Biochem Biophys Res Commun 400:389–395. doi: 10.1016/j.bbrc.2010.08.082. [DOI] [PubMed] [Google Scholar]
- 27.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. 2006. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J 25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM. 2006. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem 281:20643–20649. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- 29.Lecona E, Barrasa JI, Olmo N, Llorente B, Turnay J, Lizarbe MA. 2008. Upregulation of annexin A1 expression by butyrate in human colon adenocarcinoma cells: role of p53, NF-Y, and p38 mitogen-activated protein kinase. Mol Cell Biol 28:4665–4674. doi: 10.1128/MCB.00650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. 2011. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell 44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 31.Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, Bowman C, Kluger Y, Dynlacht BD. 2011. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci U S A 108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. 2006. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol 4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarkari F, La Delfa A, Arrowsmith CH, Frappier L, Sheng Y, Saridakis V. 2010. Further insight into substrate recognition by USP7: structural and biochemical analysis of the HdmX and Hdm2 interactions with USP7. J Mol Biol 402:825–837. doi: 10.1016/j.jmb.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 38.Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L. 2006. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol 13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- 39.Zapata JM, Pawlowski K, Haas E, Ware CF, Godzik A, Reed JC. 2001. A diverse family of proteins containing tumor necrosis factor receptor-associated factor domains. J Biol Chem 276:24242–24252. doi: 10.1074/jbc.M100354200. [DOI] [PubMed] [Google Scholar]
- 40.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. 2004. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature 428:486. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 41.Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ, Kumar KG, Konietzny R, Fischer R, Kogan E, Mackeen MM, McGouran J, Khoronenkova SV, Parsons JL, Dianov GL, Nicholson B, Kessler BM. 2011. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol 18:1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 42.Canning M, Boutell C, Parkinson J, Everett RD. 2004. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J Biol Chem 279:38160–38168. doi: 10.1074/jbc.M402885200. [DOI] [PubMed] [Google Scholar]
- 43.Nathan JA, Sengupta S, Wood SA, Admon A, Markson G, Sanderson C, Lehner PJ. 2008. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic 9:1130–1145. doi: 10.1111/j.1600-0854.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaman MM, Nomura T, Takagi T, Okamura T, Jin W, Shinagawa T, Tanaka Y, Ishii S. 2013. Ubiquitination-deubiquitination by the TRIM27-USP7 complex regulates tumor necrosis factor alpha-induced apoptosis. Mol Cell Biol 33:4971–4984. doi: 10.1128/MCB.00465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu B, Li S, Zhang X, Zheng X. 2014. HSCARG, a novel regulator of H2A ubiquitination by downregulating PRC1 ubiquitin E3 ligase activity, is essential for cell proliferation. Nucleic Acids Res 42:5582–5593. doi: 10.1093/nar/gku230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. 2003. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci U S A 100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akasaka T, van Lohuizen M, van der Lugt N, Mizutani-Koseki Y, Kanno M, Taniguchi M, Vidal M, Alkema M, Berns A, Koseki H. 2001. Mice doubly deficient for the Polycomb group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 128:1587–1597. [DOI] [PubMed] [Google Scholar]
- 48.Bel S, Core N, Djabali M, Kieboom K, Van der Lugt N, Alkema MJ, Van Lohuizen M. 1998. Genetic interactions and dosage effects of Polycomb group genes in mice. Development 125:3543–3551. [DOI] [PubMed] [Google Scholar]
- 49.Isono K, Fujimura Y, Shinga J, Yamaki M, O-Wang J, Takihara Y, Murahashi Y, Takada Y, Mizutani-Koseki Y, Koseki H. 2005. Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes. Mol Cell Biol 25:6694–6706. doi: 10.1128/MCB.25.15.6694-6706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pirity MK, Locker J, Schreiber-Agus N. 2005. Rybp/DEDAF is required for early postimplantation and for central nervous system development. Mol Cell Biol 25:7193–7202. doi: 10.1128/MCB.25.16.7193-7202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]