Abstract
KsgA is an rRNA methyltransferase important to the process of small subunit biogenesis in bacteria. It is ubiquitously found in all life including archaea and eukarya, where the enzyme is referred to as Dim1. Despite the emergence of considerable data addressing KsgA function over the last several years, details pertaining to RNA recognition are limited, in part because the most accessible substrate for in vitro studies of KsgA is the 900,000 Daltons 30S ribosomal subunit. To overcome challenges imposed by size and complexity we adapted recently reported techniques to construct in vivo assembled mutant 30S subunits suitable for use in in vitro methyltransferase assays. Using this approach, numerous 16S rRNA mutants were constructed and tested. Our observations indicate that the 790 loop of helix 24 plays an important role in overall catalysis by KsgA. Moreover, the length of helix 45 also is important to catalysis. In both cases loss of catalytic function occurred without an increase in the production of N6-methyladenosine, a likely indication that there was no critical reduction in binding strength. Both sets of observations support a ‘proximity’ mechanism of KsgA function. We also report that several of the mutants constructed failed to assemble properly into 30S subunits, while some others did so with reduced efficiency. Therefore, the same technique of generating mutant 30S subunits can be used to study ribosome biogenesis on the whole.
Ribosomes are ubiquitously present in all life and despite some phylogenetic differences, all ribosomes share common architecture, structure, and function that are easily recognizable. On the other hand, ribosome biogenesis, the cellular process of making ribosomes, is highly divergent across domains (compare recent reviews, for example (1) and (2)). In all cases, numerous trans-acting ribosome biogenesis factors (RBFs) are required to carry out diverse pre-rRNA processing, nucleotide modifications, and chaperone functions. The sole notable RBF common to all life is a dimethyltransferase termed KsgA in bacteria and Dim1 in eukaryotic and archaeal organisms. The enzyme was first described in E. coli, where it catalyzes the conversion of two adjacent adenosines A1518 and A1519, also conserved, in the 30S subunit rRNA to N6, N6-dimethyladenosines (m62A) (3, 4). The methyltransferase has since been described in eubacteria (5), archaebacteria (6), eukaryotes (7, 8) and in cellular organelles (9, 10).
Connolly et al. recently reported that KsgA functions as a late stage RBF and that the methylation triggers release of KsgA from the assembling subunit, allowing it to finally mature and enter the translation cycle (11). Therefore, it appears that KsgA acts as a gatekeeper to prevent immature 30S subunits from erroneously entering the translation cycle. Because of its critical role in ribosome biogenesis, it is important to understand the biochemical and structural properties of the KsgA/Dim1 family of enzymes.
A proposed binding location of KsgA was determined by site-directed hydroxyl radical probing (12). Accordingly, KsgA primarily binds to a region of helix 44 and the 790 loop of 16S rRNA with little apparent direct interaction with the stem of helix 45; the location of the dimethylated adenosines resides in the loop of this helix. These observations have prompted the idea that KsgA relies upon a ‘proximity’ model to recognize and dimethylate A1518 and A1519 (13). In this model, KsgA remains anchored to helix 44/ 790 loop until a certain assembly threshold event occurs at which time the target adenosines of helix 45 are brought into close proximity to the active site of KsgA. An alternative binding model was recently proposed, which suggests that KsgA binds directly to helix 45 (14).
We examined the 16S rRNA sites implicated by each model using site-directed mutagenesis and in vivo assembly of mutant 30S subunits (Figure 1). Although these mutations were designed primarily to test the ‘proximity’ model for KsgA action, the results are discussed in the context of both proposed models (see Discussion). Since members of the KsgA/Dim1 family show considerable structural and functional similarities, it is expected that observations made for KsgA’s mechanism of dimethylation can be extended to other KsgA/Dim1 members as well.
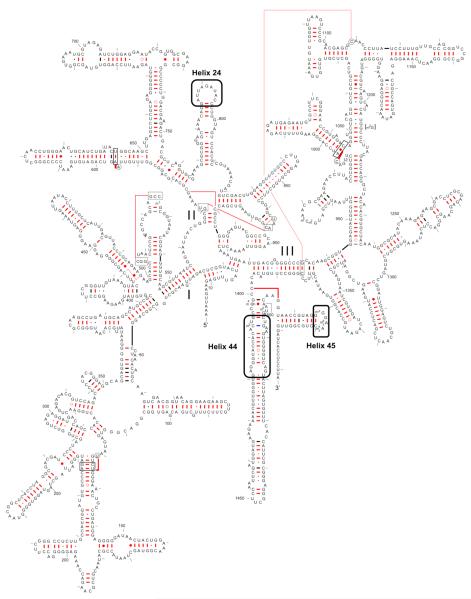
Figure 1.
Annotated 16S rRNA secondary structure from E. coli. Boxed regions are segments predicted to interact with KsgA in the context of a fully assembled 30S subunit. The figure of 16S rRNA is adapted from the work of Gutell, R. R. (35).
EXPERIMENTAL PROCEDURE
Bacterial strains and plasmids
The bacterial strains used in this study were E. coli MRE600, DH5α (Invitrogen), and BL21-DE3 (Invitrogen).
BL21-DE3 cells were transformed with pET15b-KsgA plasmid for the overexpression of His-tagged KsgA protein (15). The plasmid pGST-MS2 (gift from Prof. Rachel Green, Johns Hopkins University) which expresses the GST-MS2 fusion protein in large quantities was also introduced into BL21-DE3 cells. Growth and induction was carried out according to Youngman and Green (16).
Plasmids pSpurMS2 (AmpR) and pcI857 (KanR) were also a generous gift from Prof. Green. pSpurMS2 had been constructed by inserting a tag that allows specific binding to the coat protein of the MS2 bacteriophage in the E. coli rrnB operon of plasmid pLK35 (16). pSpurMS2 carries the rrnB operon under the control of the lambda operator/promoter, thus allowing inducible expression of tagged ribosomes (16, 17). Mutations A1418C, A1483C, 790-loop−, G791A, G791C, G791U, add2bp (adding two G-C base pairs in helix 45), del2bp (deleting two G-C base pairs in helix 45), and Hel45− were introduced in the 16S rRNA of pSpurMS2 by sited directed mutagenesis (QuikChange XL, Stratagene and Phusion, Finnzymes). DH5α cells were first transformed with pcI857, which contains the temperature-sensitive cI857 allele of the λ repressor gene. The λ cI857 repressor completely represses transcription from rrnB of pSpurMS2 at 30 °C, but is inactivated at 42 °C allowing transcription from rrnB (18).
To obtain 30S subunits unmethylated at A1518 and A1519 for KsgA methylation assays, a standard procedure of selecting strains resistant to the antibiotic kasugamycin (>600 μg/ml) was followed (6). One such resistant DH5α strain (DH5αR + pcI857) was then transformed with non-mutant or mutant pSpurMS2 to obtain tagged, unmethylated 30S subunits that, respectively, are mutation-free (30STag,UM) or bear one of the above mentioned mutations (30STag,UM[mutation]).
Purification of His-tagged KsgA
KsgA was affinity purified on a HiTrap Chelating column (Amersham Pharmacia) as previously described (6). Purity was assessed by SDS-PAGE. Purified protein was dialyzed into storage buffer containing 50 mM Tris [pH 7.4], 400 mM NH4Cl, 6 mM BME and 10% glycerol. Concentration was estimated using the Bradford method. KsgA was then aliquoted and stored at 4 °C.
Purification of GST-MS2 fusion protein
GST-MS2 fusion protein was purified as previously described (16). Briefly, clarified lysate from two liters of induced cell culture was mixed with 10 ml Glutathione-Sepharose resin (Amersham Biosciences) pre-equilibrated in PBS buffer (140 mM NaCl, 2.7 mM KCl, 10.2 mM Na2HPO4, and 1.8 mM KH2PO4) and stirred at room temperature for 30 min. The mixture was then loaded onto a gravity flow column and the settled column was washed with 100 ml PBS buffer. GST-MS2 protein was eluted with 40 ml elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM reduced glutathione), dialyzed against three 1 L changes of storage buffer (1x PBS, 20% glycerol), and stored in aliquots at −80 °C.
Purification of submethylated 30S ribosomal subunits
Untagged, A1518-A1519 unmethylated 30S subunits (30SUnTag,UM) were purified from kasugamycin resistant E. coli MRE600 strain via ultracentrifugation across a sucrose gradient using standard techniques (19).
In order to obtain pure, in vivo-derived 30S ribosomal subunits for our assays, we used the MS2 affinity purification system developed by Youngman and Green (16). Briefly, a single colony of kasugamycin resistant DH5α cells transformed with the plasmids pcI857 and either non-mutant or mutant pSpurMS2 was used to inoculate 10 ml LB media with Amp (100 μg/ml), Kan (100 μg/ml) and Ksg (400 μg/ml) and grown to saturation at 30 °C. To obtain natively methylated tagged 30S subunits for the negative control, wild-type (kasugamycin sensitive) DH5α cells carrying pcI857 and non-mutant pSpurMS2 were inoculated in media lacking kasugamycin. These cultures were then diluted 100-fold into LB media containing Amp (100 μg/ml) and expression of tagged ribosomes was induced by incubation at 42 °C with vigorous shaking until an OD600 of 0.6-0.7 was reached. Total crude ribosomes were obtained by pelleting clarified cell lysate through a sucrose cushion. To isolate tagged 30S subunits from the crude ribosomal mixture, a 5 ml GSTrap FF FPLC column (Amersham Biosciences) was used. Per 70 mg crude ribosomes, 3 mg GST-MS2 was loaded onto the column and washed with 10 ml binding buffer (20 mM Tris-HCl [pH 7.5], 100 mM NH4Cl, 0.3 mM MgCl2, 6 mM BME). Crude ribosomes were then applied to the column and washed with 40 ml binding buffer. Due to the low concentration of Mg+2 (0.3 mM) in the binding buffer, 70S ribosomes separate into constituent subunits (19), allowing selective retention of tagged 30S subunits on the column. These tagged 30S subunits were eluted using 20 ml elution buffer (50 mM Tris-HCl [pH 7.5], 100 mM NH4Cl, 0.3 mM MgCl2, 10 mM reduced glutathione, 6 mM BME) and dialyzed against buffer K (40 mM Tris-HCl [pH 7.4], 40 mM NH4Cl, 4 mM MgOAc, 6 mM BME). The subunits were then concentrated to at least 1 nmol/ml (1 A260 = 67 pmol) using Amicon Ultra centrifugal filters (Millipore, MWCO 100,000), and stored at −80 °C in aliquots. Alternatively, to assess their structural integrity, eluted 30S subunits were dialyzed into a low magnesium buffer (50 mM Tris–HCl [pH 7.5], 150 mM NH4Cl, 0.3 mM MgCl2, and 6 mM BME) and loaded on 10–30% sucrose density gradients prepared in the same buffer. For some mutants, the fractions containing 30S subunits or precursor particles were pooled, dialyzed against buffer K and stored at −80 °C to be used later for in vitro methylation assays.
In vitro methylation assay of KsgA
The in vitro assays were carried out according to O’Farrell et al. (6). Reactions contained 200 nM ribosomal subunits (10 pmol/50μl reaction), 200 nM KsgA (10 pmol/50 μl reaction), 0.02 mM 3H-methyl-SAM (780 cpm/pmol; PerkinElmer), and reaction buffer K (40 mM Tris [pH 7.4], 40 mM NH4Cl, 4 mM MgOAc, 6 mM BME). Total reaction volume and components were adjusted based on the number of time points intended. Volume for each time point was typically 50 μl. Buffer and subunits were preheated to 37 °C for 5 min to avoid any lag in reaction onset. Reactants were mixed and the reaction was initiated by addition of SAM. At each designated time point, 50 μl reaction mixture was withdrawn and quenched by adding to a pre-chilled tube containing 10 μl of 100 mM unlabeled SAM (Sigma-Aldrich). After the last time point, the quenched reactions were deposited onto DE81 filter papers (Whatman), washed twice with ice-cold 5% TCA, and rinsed briefly with ethanol. Filters were air-dried for 1 h, placed into scintillation fluid, and counted in a Packard 1500 Tri-Carb Liquid Scintillation Analyzer. Alternatively, at each designated time point, 50 μl reaction mixture was withdrawn and added to pre-chilled phenol/chloroform/isoamyl alcohol mixture to begin extraction of 16S rRNA for HPLC nucleoside analysis (see below).
HPLC analysis
Labeled 16S rRNA was extracted from 30S subunits, digested to individual nucleosides, and separated on an HPLC system as previously described (6). Each sample was spiked with non-labeled N6-methyladenosine and N6, N6-dimethyladenosine nucleoside standards (Sigma-Aldrich) for ease of identification in the UV chromatogram. Prior to subjecting the samples to HPLC analysis, the concentration of total nucleosides in each was determined by UV spectroscopy. For each sample, 80 μl nucleoside mixture was applied onto the column.
RESULTS
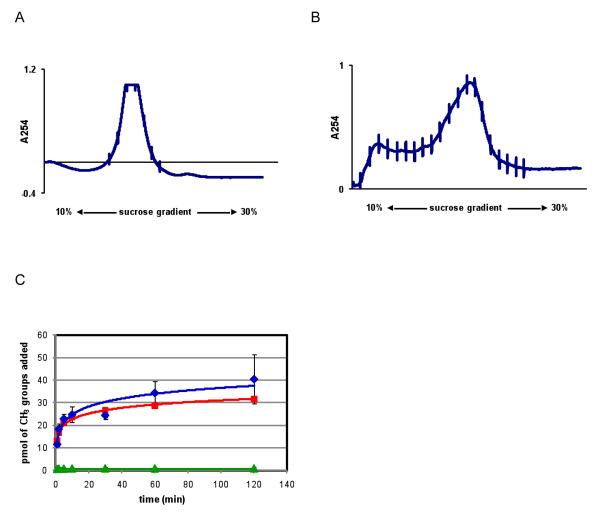
Youngman and Green developed a method of preparing a subset of cellular ribosomal subunits in E. coli that can contain site mutations, deletions, and insertions in the rRNA (16). The design of these engineered particles features a tag RNA sequence inserted into the rRNA, which is located at the ‘spur’ (helix 6) of 16S rRNA. The presence of the tag can be recognized by a fusion protein composed of the MS2 coat protein and GST (glutathione-S-transferase), thus allowing tagged subunits to be purified from a mixture of all cellular ribosomal subunits. Using reagents and protocols from the laboratory of Rachel Green we replicated the production of tagged 30S subunits (16). However, to use 30S particles with KsgA in in vitro methylation assays, we needed to produce similarly tagged 30S subunits from an E. coli strain lacking active KsgA. To remain as close as possible to the published protocol, we did this by selecting the parent strain resistant to the antibiotic kasugamycin. Using this modified strain, we produced tagged 30S subunits lacking methylation at A1518 and A1519 (30STag,UM). Sucrose density gradient analysis reveals that small ribosomal subunits produced in this manner retain overall structural integrity despite migrating as heavier species, as reported by Youngman and Green (16), than their untagged counterparts (30SUnTag,UM) purified from kasugamycin resistant MRE600 strain (Figure 2A, B). Further, 30STag,UM subunits show comparable methylation to the untagged control in in vitro KsgA activity assay (Figure 2C). Therefore, tagged 30S subunits isolated from this kasugamycin resistant strain provide a robust system to test the role of specific 16S rRNA nucleotides in KsgA methylation. The most obvious shortcoming of this method is that on the basis of sucrose gradient analysis it appears that some mutants are incapable of forming stable 30S subunits. Therefore, some desired mutants could not be successfully prepared and tested. On the other hand, we have identified new 16S rRNA residues critical to 30S assembly.
Figure 2.
Sucrose density gradient profiles and in vitro time course KsgA methylation assay of 30S subunit substrates. (A) Sucrose gradient profile of 30S subunits isolated from MRE600 E. coli strain made kasugamycin resistant (30SUnTag,UM) (B) Sucrose gradient analysis to determine the structural integrity of 30S subunits isolated from kasugamycin resistant DH5α strain expressing MS2 tagged 16S rRNA (30STag,UM). (C) The three curves are time courses of KsgA transferring tritiated methyl groups from S-adenosyl-L-methionine to three types of 30S subunits. Blue Diamonds: 30SUnTag,UM. Red Squares: 30STag,UM. Green Triangles: Same as Red Squares, but from a strain sensitive to kasugamycin, therefore natively methylated at A1518 and A1519 (30STag).
Creation and testing of helix 44 mutants
The proposed mechanism of KsgA action according to the proximity model posits that KsgA binds principally to helix 44 and helix 24 of 16S rRNA (12) and remains anchored there until A1518 and A1519 are brought into close proximity to the active site of KsgA during the course of 30S assembly and maturation.
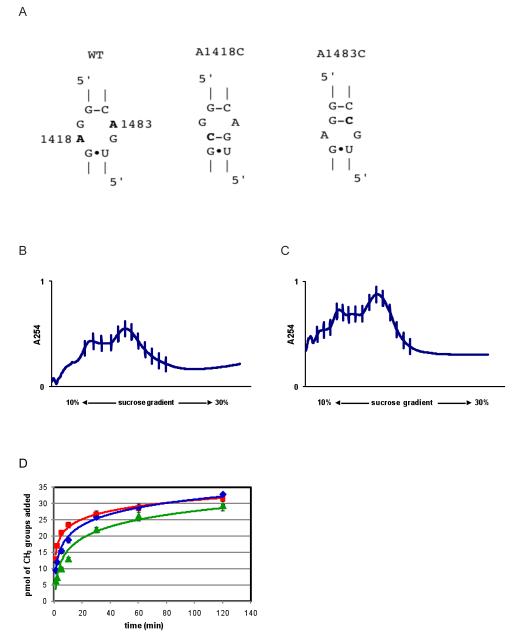
The above model predicts that KsgA interacts extensively with nucleotides 1406 - 1420 and 1482 – 1497 (E. coli numbering) of helix 44 near the decoding region (12). Moreover, most of these RNA/protein interactions are predicted to involve the backbone of the rRNA and not the nucleobases themselves. Therefore, to disrupt a local region of RNA backbone conformation predicted to interact with a positively charged patch of residues (R221, R222, K223, and R248) on the C-terminus domain of KsgA, we mutated the tandem, sheared G/A base pairs formed by G1417:A1483 and A1418:G1482 (Figure 3A). We generated tagged 30S particles containing the single mutations A1418C (30STag,UM[A1418C]) and A1483C (30STag,UM[A1483C]), in which the corresponding sheared G/A pairing was replaced by a G-C base pair. In doing so, the backbone is expected to be significantly altered as the remaining sheared pairing has a high propensity to adopt a Watson-Crick edge from being a single G/A pairing flanked by Watson-Crick base pairs (20). Structural integrity and assembly completeness of the particles were assessed by ultracentrifugation through a sucrose gradient (Figure 3B, C). Results of this analysis indicated that full 30S particles, as well as pre-30S particles, are produced. In vitro time course KsgA activity assays indicated that the mutant 30S subunits served as substrates for KsgA, exhibiting only slightly less activity compared to 30STag,UM subunits (Figure 3D).
Figure 3.
Disruption of sheared base pairings in helix 44 and KsgA activity. (A) The A1418C and A1483C mutations in helix 44 were constructed and tested for substrate activity. (B) and (C), respectively, represent the sucrose gradient profiles of A1418C and A1483C mutants. (D) Time course activity assays for KsgA catalyzed transfer of methyl groups to 30STag,UM subunits (red) and the mutant subunits, 30STag,UM[A1418C] (green) and 30STag,UM[A1483C] (blue).
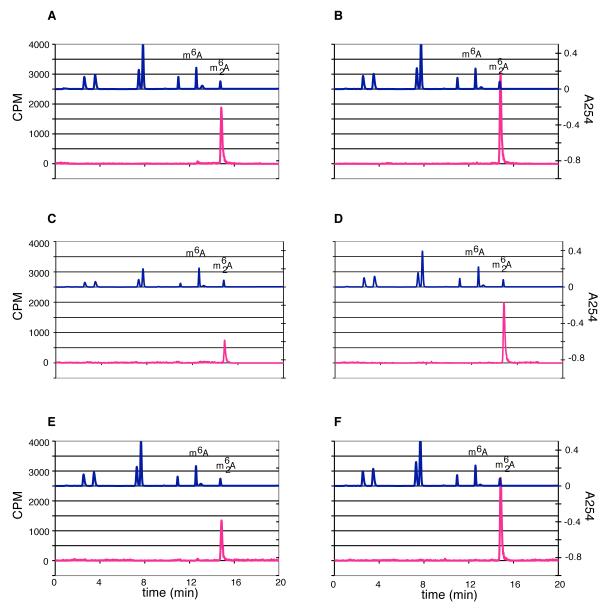
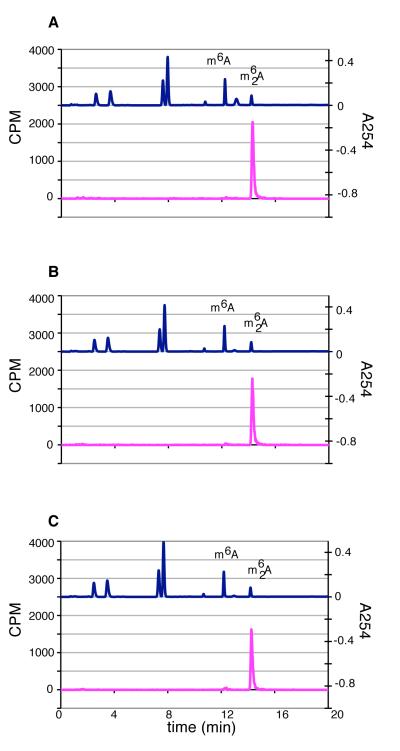
There are several possibilities for the reduced methylation of the A1418C and A1483C mutants by KsgA. First, total affinity purified material was used, which included the small fraction of pre-30S particles that might not serve as a substrate. Thus, the overall amount of suitable substrate present might have been lower. Second, KsgA might have reduced affinity for the mutant 30S subunits, thereby reducing catalytic rate. Third, substrate binding might be unaffected, but catalytic efficiency might be reduced. To gain further insight, we relied on the observation that reduced KsgA affinity for substrate can generate larger quantities of monomethyladenosine (Unpublished data. O’Farrell, H. C., Culver, G. M., and Rife, J. P.). Therefore, we monitored the ratios of monomethyladenosine vs. dimethyladenosine at various time points during the reaction (Figure 4). As can be seen, vanishingly small amounts of monomethyladenosine are produced at early and late time points. While KsgA might have reduced affinity for the 30S mutant subunits, the threshold of disrupting KsgA’s processive nature was not achieved. Therefore, the reduced activity likely comes from reduced amounts of active substrate or overall reduced catalytic efficiency.
Figure 4.
HPLC nucleotide analysis of KsgA methylated substrates mutated at A1418 and A1483. (A), (C), and (E), respectively, represent the traces of 30STag,UM, 30STag,UM[A1418C] and 30STag,UM[A1483C] reactions at the 2 minute time point. (B), (D), and (F) represent products involving the same substrates, but at the end of 2 hr. The top trace in each panel is measured absorbance at 254 nm of a collection of nucleoside standards including m6A and m62A, while the bottom trace in each panel is measured 3H levels.
Helix 24 mutations
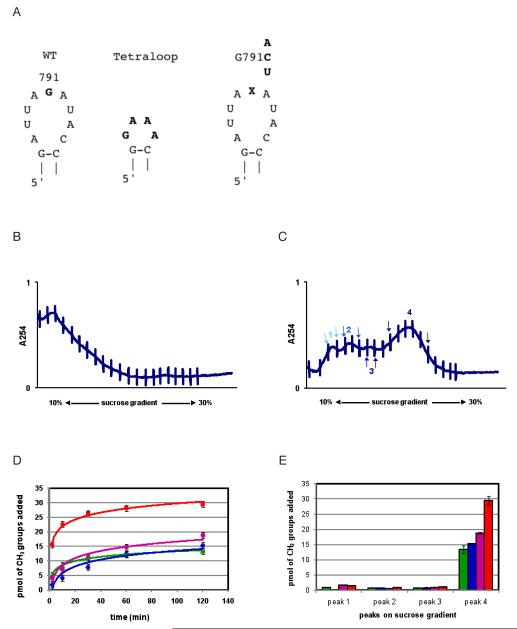
A second prediction made by the Xu et al. binding model is that significant interactions occur between KsgA and loop nucleotides of helix 24, also known as the ‘790-loop’ (12). Detailed understanding of loop 790/KsgA interactions are lacking because in the model by Xu et al. this loop region clashes with KsgA. Incompatibility in this region is likely an artifact arising from the fact that KsgA was docked onto a crystal structure of the 30S subunit in its translationally active conformation. Nevertheless, the prediction was made that the 790 loop makes direct interaction with KsgA (12, 21). Two strategies were adopted to probe the importance of the 790 loop to KsgA catalysis, the first being a large scale change and the second a series of point mutations involving G791 (Figure 5A). G791 is predicted to make direct interactions with the target adenosine pocket of KsgA (12). Further, G791 was previously mutated by others to the three remaining nucleotides and shown to be at least partially active in translation (22).
Figure 5.
Effects of helix 24 loop mutations on 30S assembly efficiency and methylation by KsgA. (A) The loop nucleotides (787-795) were replaced with GAAA, a member of the GNRA tetraloop family. G791 was mutated to each of A, C, and U. (B) and (C), respectively, show the sucrose gradient analysis for the 790-loop− mutant and the G791U mutant, which is representative of all three single G791 mutations. For G791 mutants, material corresponding to the 30S peak (peak 4 in G791U profile) was used for KsgA time course activity assays, while sucrose gradient fractions of earlier peaks (peaks 1, 2, and 3) were used in a 2-hr end point methylation assay. Fractions pooled for each peak are indicated by arrows matching in color to the peak number. (D) Time course activity assays for 30STag,UM subunits (red) and the three mutant subunits, 30STag,UM[G791A] (green), 30STag,UM[G791C] (blue), and 30STag,UM[G791U] (purple). (E) 2-hr end point methylation assay of sucrose gradient fractions of 30STag,UM (red), 30STag,UM[G791A] (green), 30STag,UM[G791C] (blue), and 30STag,UM[G791U] (purple).
The 790-loop construct wherein the entire 790 loop (nucleotides 785-797) was replaced with the GNRA tetraloop GAAA yielded no fully formed 30S subunits (Figure 5B), indicating that this loop is critical to the 30S ribosomal biogenesis and assembly pathways. However, we were able to produce 30S particles containing the G791A (30STag,UM[G791A]), G791C (30STag,UM[G791C]), and G791U (30STag,UM[G791U]) mutations (Figure 5C). In each case, several precursor 30S particles were observed in addition to 30S subunits. To better understand the suitability of the precursor particles and fully assembled 30S subunits as substrates, individual peaks isolated from the sucrose gradient were tested for their ability to support KsgA activity. Precursor peaks 1-3 for all three mutants did not support methylation, while the fourth peak, corresponding to the fully assembled 30S subunit, was clearly a substrate for KsgA (Figure 5D, E). For all three mutants, overall activity was reduced by about 50-60%. Again, to understand the nature of this reduced activity we determined the amount of monomethyladenosine generated at the 2-hr end point and, like with the helix 44 mutations, no significant amount of monomethyladenosine was produced (Figure 6). Therefore, reduction in catalytic activity is likely the result of reduced catalytic efficiency and not a significant reduction in substrate binding. According to the Xu et al. model, the 790 loop is poised to interact with structure motif VIII which forms part of the target adenosine binding site, which may explain why mutation of G791 leads to reduced methylation by KsgA (12, 23).
Figure 6.
HPLC nucleoside analysis of KsgA methylated substrates mutated in the loop of helix 24. (A), (B), and (C), respectively, are chromatograms involving nucleosides derived from 30STag,UM[G791A], 30STag,UM[G791C], and 30STag,UM[G791U] substrates of the 2-hr end point reaction. The top trace in each panel is the measured absorbance at 254 nm of a collection of nucleoside standards including m6A and m62A, while the bottom trace in each panel is measured 3H levels.
Helix 45 mutations
The third group of mutations was located within helix 45, the terminal loop of which contains the target adenosines A1518 and A1519. The stem of helix 45 is composed of nine base pairings, where eight are Watson-Crick base pairs and one is an internal G•U wobble pairing. The terminal loop itself contains the sequence GGAA, where the two adenosines are dimethylated by KsgA and the leading guanosine is methylated by an unknown methyltransferase.
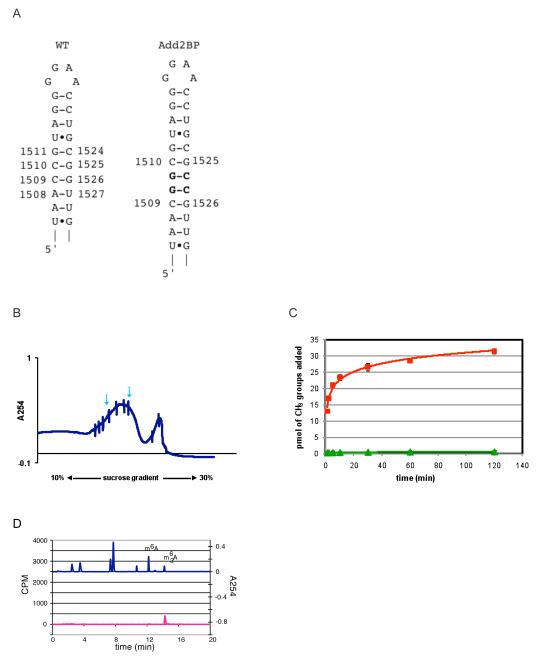
Several mutations were constructed and tested, including the radical deletion of helix 45 to test the ability of KsgA to bind mutant 30S subunits that lacked helix 45 altogether. Unfortunately, removal of helix 45 prevents detectable 30S particle assembly. Two mutants were created to test the importance of helix 45 length on KsgA activity. The ‘add2bp’ construct (30STag,UM[add2bp]) contained two additional G-C base pairs between the base pairs C1510:G1525 and G1511:C1524 (Figure 7A). We also constructed a ‘del2bp’ mutation to test the consequences of shortening helix 45 by removing the C1509:G1526 and C1510:G1525 base pairs of helix 45. Shortening helix 45 by two base pairs resulted in no detectable 30S particles being formed, while lengthening the same helix by two base pairs allowed for the formation of 30S subunits (Figure 7B). Therefore, helix 45 appears to be important to the formation of the full 30S subunit. In case of add2bp, particles corresponding to full size 30S subunits showed very small amounts of KsgA methylation (Figure 7C), revealed primarily to be the dimethylated product by nucleoside analysis (Figure 7D). From this data, it appears that KsgA function is dependent on the length of helix 45, which is consistent with the proximity model.
Figure 7.
Effects of lengthening helix 45 by two base pairs on methylation by KsgA. (A) A mutant composed of the addition of two base pairs in helix 45 was constructed and tested for substrate activity. (B) Sucrose gradient analysis for the add2bp mutant. Material corresponding to the 30S peak, indicated by arrows, was used for the activity assay. (C) KsgA time course activity assay of 30STag,UM (red) and mutant 30STag,UM[add2bp] (green) subunits. (D) Nucleoside analysis of the 2-hr end point reaction product. The top trace is measured absorbance at 254 nm of a collection of nucleoside standards including m6A and m62A, while the bottom trace is measured 3H levels.
DISCUSSION
Many rRNA modification enzymes require very complex substrates, which makes structure/function studies at the level of RNA sequence difficult. For example, in vitro reconstitutions suggest that the smallest substrate identified to date for KsgA is all of 16S rRNA and eight ribosomal proteins (24). In vivo, methylation by KsgA appears to require something like the size and complexity of the 30S subunit and therefore approximately 0.9 million Daltons (25). To study the effect of rRNA mutations on KsgA activity, we chose to use a tagged ribosome system developed by Youngman and Green (16) to circumvent shortcomings associated with other methods of creating and testing rRNA mutations. Other systems suffer from either the lack of cell viability when critical residues are altered (26) or low reconstitution yields when in vitro transcribed rRNA is used (27). We have successfully used a tagged ribosome method to create mutant 30S subunit substrates by using a strain lacking active KsgA. In doing so we confirmed that KsgA is sensitive to the composition of the loop of helix 24.
When considering only 16S rRNA methyltransferases from E. coli, it is anticipated that all but one enzyme will require their substrates to have present at least multiple ribosomal proteins in the pre-30S complex or 30S subunit. It is important to note that this method is only suitable for enzymes that require nearly fully formed or fully formed 30S subunits. Fortunately, at least 8 of 10 16S rRNA methyltransferases in E. coli, can utilize fully formed 30S subunits (28-33). Therefore, generating substrates with rRNA mutations can be readily accomplished for the study of many other rRNA methyltransfersases.
The most significant shortcoming of this strategy is that some mutations, including several documented here, lead to aborted ribosome biogenesis. An unanticipated outcome of this work is the identification of previously unreported residues of 16S rRNA critical to the assembly of 30S subunits. Therefore, this method is clearly well suited to the study of mutational effects on ribosomal assembly.
KsgA binding and the proximity model
Any satisfactory model of KsgA activity needs to explain how KsgA can sense the overall 30S conformation, as well as provide insight into how two adjacent adenosines can gain access into the active site pocket of KsgA in a processive manner. This binding mode, and the understanding that KsgA can sense the degree of 30S assembly, led us to propose a proximity model for KsgA (13). In this model, KsgA binds pre-16S rRNA at some early to intermediate time point in the ribosome biogenesis cascade and awaits a late assembly threshold event. This assembly threshold is governed by the close approach of helix 45 and specifically A1518 and A1519 to the active site of KsgA. Work presented supports the involvement of the 790 loop in substrate recognition and catalytic function, and reinforces the importance of the length of helix 45. On the other hand, mutations to two helix 44 G/A pairings only marginally reduced over all methylation activity, suggesting that this region is of less importance to KsgA function or that the binding model of Xu et al. requires refinement (12).
At about the time of completion of the experiments reported here, there was only one experimentally derived binding model for KsgA, one in which KsgA binds principally to helix 44 and the 790 loop (12). A competing binding model was recently published by Tu et al. and is based on the crystal structure of KsgA bound to an RNA homoduplex composed of an oligonucleotide encompassing helix 45 of 16S rRNA (14). Seriously confounding these results is the fact that KsgA is binding to a structural artifact that results from a dimerization reaction commonly observed in the attempted crystallization of short RNA hairpins. The high concentration of RNA typically found in crystallization solutions sometimes favors the energetically accessible duplex form, which was reported by Tu et al. For proteins that bind to the helix region of a hairpin, a shift to a duplex in the crystal lattice is not a problem because the binding site is preserved (28). However, in the Tu et al. structure, KsgA binds principally to the internal loop that is only present in the duplex form of the RNA (14). The authors note that although their structure is not a catalytically relevant assembly, it shows how KsgA can bind to double stranded RNA. The experiments described herein were not specifically designed to address the question of whether or not KsgA binds principally to helix 45. Thus, future work should address this competing model directly.
ACKNOWLEDGEMENTS
We thank Prof. Rachel Green for kindly providing us with the pSpurMS2, pcI857, and pGST-MS2 constructs, and Dr. Heather O’Farrell for extending to us access to her unpublished data and critically reading the manuscript.
ABBREVIATIONS
- RBF
Ribosome biogenesis factor
- RNP
Ribonucleoprotein
- SAM
S-adenosyl-L-methionine
- Amp
Ampicillin
- Kan
Kanamycin
- Ksg
Kasugamycin
- BME
β-mercaptoethanol
- GST
Glutathione-S-transferase
- m6A
N6-methyladenosine
- m62A
N6, N6-dimethyladenosine
- UM
Unmethylated
Footnotes
This work was supported by the National Institutes of Health (1R01GM066900)
REFERENCES
- 1.Kaczanowska M, Ryden-Aulin M. Ribosome Biogenesis and the Translation Process in Escherichia Coli. Microbiol. Mol. Biol. Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The Post-Transcriptional Steps of Eukaryotic Ribosome Biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helser TL, Davies JE, Dahlberg JE. Change in Methylation of 16S Ribosomal RNA Associated with Mutation to Kasugamycin Resistance in Escherichia Coli. Nat. New Biol. 1971;233:12–14. doi: 10.1038/newbio233012a0. [DOI] [PubMed] [Google Scholar]
- 4.Poldermans B, Roza L, Van Knippenberg PH. Studies on the Function of Two Adjacent N6, N6-Dimethyladenosines Near the 3′ End of 16 S Ribosomal RNA of Escherichia Coli. III. Purification and Properties of the Methylating Enzyme and Methylase-30 S Interactions. J. Biol. Chem. 1979;254:9094–9100. [PubMed] [Google Scholar]
- 5.Van Buul CP, Damm JB, Van Knippenberg PH. Kasugamycin Resistant Mutants of Bacillus Stearothermophilus Lacking the Enzyme for the Methylation of Two Adjacent Adenosines in 16S Ribosomal RNA. Mol. Gen. Genet. 1983;189:475–478. doi: 10.1007/BF00325912. [DOI] [PubMed] [Google Scholar]
- 6.O’Farrell HC, Pulicherla N, Desai PM, Rife JP. Recognition of a Complex Substrate by the KsgA/Dim1 Family of Enzymes has been Conserved Throughout Evolution. RNA. 2006;12:725–733. doi: 10.1261/rna.2310406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lafontaine D, Delcour J, Glasser AL, Desgres J, Vandenhaute J. The DIM1 Gene Responsible for the Conserved m6(2)Am6(2)A Dimethylation in the 3′-Terminal Loop of 18 S rRNA is Essential in Yeast. J. Mol. Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 8.Housen I, Demonte D, Lafontaine D, Vandenhaute J. Cloning and Characterization of the KlDIM1 Gene from Kluyveromyces Lactis Encoding the m2(6)A Dimethylase of the 18S rRNA. Yeast. 1997;13:777–781. doi: 10.1002/(SICI)1097-0061(19970630)13:8<777::AID-YEA140>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 9.Seidel-Rogol BL, McCulloch V, Shadel GS. Human Mitochondrial Transcription Factor B1 Methylates Ribosomal RNA at a Conserved Stem-Loop. Nat. Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 10.Tokuhisa JG, Vijayan P, Feldmann KA, Browse JA. Chloroplast Development at Low Temperatures Requires a Homolog of DIM1, a Yeast Gene Encoding the 18S rRNA Dimethylase. Plant Cell. 1998;10:699–711. doi: 10.1105/tpc.10.5.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly K, Rife JP, Culver G. Mechanistic Insight into the Ribosome Biogenesis Functions of the Ancient Protein KsgA. Mol. Microbiol. 2008;70:1062–1075. doi: 10.1111/j.1365-2958.2008.06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, O’Farrell HC, Rife JP, Culver GM. A Conserved rRNA Methyltransferase Regulates Ribosome Biogenesis. Nat. Struct. Mol. Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- 13.Rife JP. Roles of the Ultra-Conserved Ribosomal RNA Methyltransferase KsgA in Ribosome Biogenesis. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. 1st ed Landes Bioscience; Austin, TX: 2009. pp. 509–523. [Google Scholar]
- 14.Tu C, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural Basis for Binding of RNA and Cofactor by a KsgA Methyltransferase. Structure. 2009;17:374–385. doi: 10.1016/j.str.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Farrell HC, Musayev FN, Scarsdale JN, Wright HT, Rife JP. Crystallization and Preliminary X-Ray Diffraction Analysis of KsgA, a Universally Conserved RNA Adenine Dimethyltransferase in Escherichia Coli. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1490–1492. doi: 10.1107/s0907444903011855. [DOI] [PubMed] [Google Scholar]
- 16.Youngman EM, Green R. Affinity Purification of in vivo-Assembled Ribosomes for in vitro Biochemical Analysis. Methods. 2005;36:305–312. doi: 10.1016/j.ymeth.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Douthwaite S, Powers T, Lee JY, Noller HF. Defining the Structural Requirements for a Helix in 23 S Ribosomal RNA that Confers Erythromycin Resistance. J. Mol. Biol. 1989;209:655–665. doi: 10.1016/0022-2836(89)93000-3. [DOI] [PubMed] [Google Scholar]
- 18.Gourse RL, Takebe Y, Sharrock RA, Nomura M. Feedback Regulation of rRNA and tRNA Synthesis and Accumulation of Free Ribosomes After Conditional Expression of rRNA Genes. Proc. Natl. Acad. Sci. U. S. A. 1985;82:1069–1073. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moazed D, Van Stolk BJ, Douthwaite S, Noller HF. Interconversion of Active and Inactive 30 S Ribosomal Subunits is Accompanied by a Conformational Change in the Decoding Region of 16 S rRNA. J. Mol. Biol. 1986;191:483–493. doi: 10.1016/0022-2836(86)90143-9. [DOI] [PubMed] [Google Scholar]
- 20.Olson WK, Esguerra M, Xin Y, Xiang-Jun L. New Information Content in RNA Base Pairing Deduced from Quantitative Analysis of High-Resolution Structures. Methods. 2009;47:177–186. doi: 10.1016/j.ymeth.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desai PM, Rife JP. The Adenosine Dimethyltransferase KsgA Recognizes a Specific Conformational State of the 30S Ribosomal Subunit. Arch. Biochem. Biophys. 2006;449:57–63. doi: 10.1016/j.abb.2006.02.028. [DOI] [PubMed] [Google Scholar]
- 22.Song WS, Kim HM, Kim JH, Sim SH, Ryou SM, Kim S, Cha CJ, Cunningham PR, Bae J, Lee K. Functional Analysis of the Invariant Residue G791 of Escherichia Coli 16S rRNA. J. Microbiol. 2007;45:418–421. [PubMed] [Google Scholar]
- 23.O’Farrell HC, Scarsdale JN, Rife JP. Crystal Structure of KsgA, a Universally Conserved rRNA Adenine Dimethyltransferase in Escherichia Coli. J. Mol. Biol. 2004;339:337–353. doi: 10.1016/j.jmb.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 24.Thammana P, Held WA. Methylation of 16S RNA during Ribosome Assembly in vitro. Nature. 1974;251:682–686. doi: 10.1038/251682a0. [DOI] [PubMed] [Google Scholar]
- 25.Lowry CV, Dahlberg JE. Structural Differences between the 16S Ribosomal RNA of E. Coli and its Precursor. Nat. New Biol. 1971;232:52–54. doi: 10.1038/newbio232052a0. [DOI] [PubMed] [Google Scholar]
- 26.Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires CL. Construction and Initial Characterization of Escherichia Coli Strains with Few Or no Intact Chromosomal rRNA Operons. J. Bacteriol. 1999;181:3803–3809. doi: 10.1128/jb.181.12.3803-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krzyzosiak W, Denman R, Nurse K, Hellmann W, Boublik M, Gehrke CW, Agris PF, Ofengand J. In vitro Synthesis of 16S Ribosomal RNA Containing Single Base Changes and Assembly into a Functional 30S Ribosome. Biochemistry. 1987;26:2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- 28.Tscherne JS, Nurse K, Popienick P, Ofengand J. Purification, Cloning, and Characterization of the 16 S RNA m2G1207 Methyltransferase from Escherichia coli. The J Biol Chem. 1999;274:924–929. doi: 10.1074/jbc.274.2.924. [DOI] [PubMed] [Google Scholar]
- 29.Lesnyak DV, Osipiuk J, Skarina T, Sergiev PV, Bogdanov AA, Edwards A, Savchenko A, Joachimiak A, Dontsova OA. Methyltransferase that modifies guanine 966 of the 16 S rRNA: functional identification and tertiary structure. J Biol Chem. 2006;282:5880–5887. doi: 10.1074/jbc.M608214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basturea GN, Rudd KE, Deutscher MP. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA. 2006;12:426–434. doi: 10.1261/rna.2283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen NM, Douthwaite S. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J Mol Biol. 2006;359:777–786. doi: 10.1016/j.jmb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, Suzuki Y, Ochi K. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol. 2007;63:1096–1106. doi: 10.1111/j.1365-2958.2006.05585.x. [DOI] [PubMed] [Google Scholar]
- 33.Kimura S, Suzuki T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010;38:1341–1352. doi: 10.1093/nar/gkp1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jovine L, Hainzl T, Oubridge C, Scott WG, Sixma TK, Wonacott A, Skarzynski T, Nagai K. Crystal Structure of the Ffh and EF-G Binding Sites in the Conserved Domain IV of Escherichia coli 4.5S RNA. Structure. 2000;8:527–540. doi: 10.1016/s0969-2126(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 35.Gutell RR. 2008 http://www.rna.ccbb.utexas.edu/