Abstract
Recent studies have demonstrated that polymorphisms near the interleukin-28B (IL-28B) gene could predict the response to Peg-IFN-a/RBV combination therapy in HCV-infected patients. The aim of the study was to correlate the serum level of IL28B in HCV-infected patients with virus genotype/subgenotype and disease progression. IL28B serum level was detected and variations at five single nucleotide polymorphisms (SNPs) in IL28B gene region were genotyped and analyzed. The variation of IL28B genetic polymorphisms was found to be strongly associated with HCV infection when healthy control group was compared to HCV-infected patients with all P values <0.0001. Functional analysis revealed that subjects carrying rs8099917-GG genotype had higher serum level of IL28B than those with GT or TT genotypes (P = 0.04). Also, patients who were presented with cirrhosis (Cirr) only or with cirrhosis plus hepatocellular carcinoma (Cirr+HCC) had higher levels of serum IL28B when compared to chronic HCV-infected patients (P = 0.005 and 0.003, resp.). No significant association was found when serum levels of IL28B were compared to virus genotypes/subgenotypes. This study indicates that variation at SNP rs8099917 could predict the serum levels of IL28B in HCV-infected patients. Furthermore, IL28B serum level may serve as a useful marker for the development of HCV-associated sequelae.
1. Introduction
Approximately, 170 to 180 million people (~3% of the world population) are estimated to be infected with hepatitis C virus (HCV) [1]. Prolonged and persistent HCV infection may lead to cirrhosis and hepatocellular carcinoma (HCC) [2, 3]. Up until recently, the treatment of chronic HCV infection involved a 24- or 48-week course of pegylated interferon-alpha (PEG-IFNα) in combination with ribavirin (RBV) [4, 5]. Unfortunately, the efficacy of this treatment is limited, with only half of the patients are able to achieve a sustained virological response (SVR), particularly in patients with genotype 1 [4, 6]. More recently, newer and more potent antiviral agents have been approved with higher efficacy and tolerability but burdened with a vastly higher cost. Although PEG-IFNα/RBV therapy is not first line therapy nowadays in the majority of clinical scenarios it is still recommended in triple therapy combination regimens in many patients to shorten the duration of therapy. In addition, in many countries worldwide, interferon based therapy is still used since the newer, more potent antiviral agents are not routinely available yet because of the associated high cost (http://www.hcvguidelines.org/) [7].
With the recent advent of genome-wide association studies, three major studies have reported the significance of several single nucleotide polymorphisms (SNPs) in the vicinity of interleukin-28B (IL28B) gene, capable of predicting the response outcome of combination therapy in genotype-1 HCV infected patients. Ge et al. (2009) found that the SNP, rs12979860, which is 3 kb upstream of IL28B gene, was significantly associated with SVR in genotype-1 HCV-infected patients undergoing 48 weeks of PEG-IFNα plus RBV treatment [8]. In a study conducted on Japanese patients infected with HCV genotype-1, Tanaka et al. (2009) found additional SNPs, near IL28B gene, which were strongly associated with null virological response (NVR) and SVR, with rs8099917 being the most significant [9]. A similar study conducted on Australian patients, also concluded a strong association of rs8099917 with SVR in individuals infected with genotype-1 HCV and undergoing combination therapy [10]. Furthermore, a study conducted by Thomas et al. (2009) reported that variations in rs12979860 could play a pivotal role in the spontaneous, natural clearance of HCV [11]. Thus, much effort is being put in order to determine the predictive power of the genetic polymorphisms around the IL28B gene in relation to SVR, before it can be implemented into the current treatment therapy.
IL28B belongs to the cluster of interferon-λ (IFN-λ) genes that belong to the type-III class of IFNs and are associated with natural clearance of HCV, by being directly produced in response to the infection. IFN-λ genes are clustered on chromosome 19 and encode IFN-λ1 (IL29), IFN-λ2 (IL28A), and IFN-λ3 (IL28B) [12, 13]. The lambda-IFNs have a restricted target cell range and they signal via the JAK/STAT pathway, upregulating the expression of a set of interferon-stimulated genes (ISGs), through which they inhibit virus replication [14, 15].
Thus far, not many studies have looked into the correlation of IL28B serum levels with the different IL28B genetic variants in HCV patients [16], and there are few reports analyzing the significance of these variants in the progression of HCV infected patients to advanced stages of the disease. Therefore, the current study aims to determine the correlation of serum IL-28 levels in relation to IL-28B genetic variations, different viral genotypes/subgenotypes, and relation to disease progression. Furthermore, the study aims at understanding the impact of IL28B SNPs in the development of HCV-related cirrhosis and HCC.
2. Materials and Methods
2.1. Patients
This study was conducted at Riyadh area of Saudi Arabia where samples were collected from three major hospitals, namely, King Faisal Specialist Hospital & Research Center, King Khalid University Hospital and Riyadh Military Hospital. The study was approved by the ethics committees from each hospital and conformed to the ethical guidelines of the 2000 declaration of Helsinki. All participating patients signed an informed consent prior to enrollment in the study. The study included 678 patients who were divided into three groups according to their disease progression including chronic (group 1), cirrhosis (group 2), and cirrhosis+HCC (group 3). Group 1: chronically infected patients (detectable HCV RNA >6 months) without cirrhosis or HCC. Patients in this group did not have any clinical or biochemical evidence of cirrhosis and had a fibroscan that shows less than stage 2 fibrosis. Group 2: patients with cirrhosis who are diagnosed based on either liver biopsy or clinical conditions including the presence of the following criteria; platelet count <90,000/L, radiological (ultrasonography [US] or computed tomography [CT]) evidence of cirrhosis, or esophageal varices (demonstrated by endoscopy), and signs of liver dysfunction including; albumin level <30 g/L, INR ≥1.5, or bilirubin level >35 μmol/L. Group 3: patients with HCC on the background of cirrhosis. The diagnosis of HCC was based upon established CT or magnetic resonance imaging (MRI) criteria in published guidelines for the diagnosis and management of HCC [17]. Enhancement of a liver lesion during the arterial phase and contrast washout during the portal phase, in patients with background cirrhosis was considered diagnostic of HCC. Trucut biopsy or fine needle aspiration was obtained only where considerable doubt existed after imaging studies and alpha-fetoprotein (AFP) tests. The study also included 600 uninfected healthy subjects, used as a control group, who were characterized by the absence of any known serological marker for HCV, HCV RNA or any evidence of liver disease.
2.2. Subgenotyping of HCV
HCV RNA was extracted from sera using QIAmp MinElute Virus Spin Kit (QIAGEN, Santa Clarita, CA, USA) following manufacturer's recommendations. Extracted RNA was converted to cDNA as described before by Al-Qahtani et al. [18] and subgenotypes were determined according to the procedures described by Murphy et al. [19]. PCR products were sequenced using BigDye Terminator v3.1 cycle sequencing Kit (Applied Biosystems, Foster City, CA, USA) following manufacturer's recommendations. DNA sequence was then analyzed using BLASTN 2.2.23+ software (http://blast.ncbi.nlm.nih.gov/blast.cgi) against all sequences in the database to determine the subgenotype of each sample. The sequence that showed the lowest e-value and maximum identity was taken as the subgenotype of the sample analyzed.
2.3. Detection of Serum IL-28B by ELISA
IL-28B concentration was determined in patients' sera using a sandwich ELISA according to the manufacturer's recommendations (USCNK, Life Science Inc., Wuhan, China). Briefly, sera or recombinant human IL-28B, as standards, were added to plates coated with a monoclonal antibody against IL-28B and incubated at room temperature for two hours. All analyses and calibrations were carried out in a duplicate. Optical density was determined using a microtiter plate reader (BioTek ELx50, Winooski, VT, USA) at 450 nm with the lower end of sensitivity of the assay was 15.6 pg/mL and the upper limit was 1000 pg/mL. Samples with readings greater than the upper limit were diluted until their absorbance readings fall within the linear range of the assay.
2.4. Statistical Analysis
Statistical analyses were completed using SPSS v. 17.0 (SPSS Inc., Chicago, IL, USA). Chi-square test was used to compare the genotypic distribution of IL28 SNPs among patients and controls. The association results were expressed in terms of odds ratio (OR) and their 95% confidence intervals (CI). A P value of <0.05 was considered to be statistically significant. The SNPs were tested for Hardy-Weinberg equilibrium (HWE) using the DeFinetti program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). All correlation analyses were performed in the R software environment (http://www.r-project.org/).
3. Results
3.1. Association of IL28B Polymorphisms with HCV Infection
Five SNPs in the vicinity of the IL28B gene (rs8105790, rs8099917, rs7248668, rs12979860, and rs12980275) were genotyped in 678 HCV-infected patients and 600 healthy, uninfected control subjects. The demographic and clinical data are shown in Table 1. There were significant differences between the patient groups in all categories except for viral load, ALT, and creatinine levels.
Table 1.
Basic characteristics of all subjects included in this study.
| Variable | Chronic HCV | Cirrhosis+HCC | Control | P value |
|---|---|---|---|---|
| Age (yrs.)¶ | 48.00 (36.00–57.00) | 57.00 (48.00–63.00) | 29.00 (24.00–36.00) | <0.000§ |
| Sex | ||||
| Male count (%) | 274 (50.5%) | 94 (51.4%) | 568 (94.7%) | <0.000ϕ |
| Female count (%) | 269 (49.5%) | 89 (48.6%) | 32 (5.3%) | |
| BMI¶ | 31.88 (26.53–71.43) | 28.72 (24.57–34.13) | 0.014§ | |
| Platelet count (109/L)¶ | 216.00 (169.00–284.50) | 148.00 (102.50–200.50) | <0.000§ | |
| Bil (umol/L)* | 13.21 ± 26.18 | 25.07 ± 73.34 | <0.000ψ | |
| ALT (IU/L)* | 74.53 ± 66.9 | 83.66 ± 62.47 | 0.766ψ | |
| AST (IU/L)* | 45.07 ± 40.299 | 75.16 ± 57.16 | <0.000ψ | |
| AFP (ug/L)* | 6.36 ± 22.03 | 15.97 ± 27.14 | <0.000ψ | |
| ALP (IU/L)* | 106.30 ± 53.07 | 122.65 ± 76.77 | 0.004ψ | |
| Creatinine (umol/L)* | 110 ± 151.31 | 97.72 ± 127.30 | 0.074ψ | |
| HCV load IU/mL (log10)¶ | 5.80 (4.788–6.41) | 6.025 (5.49–6.44) | 0.511§ | |
| HCV genotypes | ||||
| 4 (357) | 249 (70%) | 108 (30%) | <0.0001 | |
| 1 (148) | 115 (77.7%) | 33 (22.3%) | ||
| Others¶¶ (173) | 154 (89%) | 19 (11%) |
BMI: body mass index; Bil: bilirubin; ALT: alanine aminotransferase; AST: aspartate aminotransferase; AFP: α-fetoprotein, ALP: alkaline phosphatase. *Values are expressed as mean ± SD, ¶values are expressed as median-interquartile range (25th–75th), ¶¶includes genotypes 2, 3, and untypable genotypes. §Nonparametric tests, ϕChi-squared tests, and ψindependent t-tests.
When the patient group was compared to the uninfected control subjects, all SNPs were found to have a significant association in relation to HCV infection (Table 2). The risk allele “G” for the SNP rs8099917 was found to be significant when the patient group was compared to the uninfected control group with an OR of 2.55 (95% C.I. 2.062–3.160), χ²-value of 77.16, and P < 0.0001. Under the dominant model, a significant association was observed with an OR of 2.047 (95% C.I. 1.593–2.630), χ²-value of 31.91, and P < 0.0001. While under the recessive model, a significant association was observed with an OR of 0.132 (0.075–0.233), χ²-value of 64.25, and P < 0.0001, indicating that inheriting a homozygous GG genotype would increase the risk of HCV infection by nearly 3 times than those carrying the heterozygous GT genotype. Likewise, a similar significant result was obtained for rs8105790 under the recessive model, with an OR of 0.089 (0.053–0.152), χ²-value of 116.35, and P < 0.0001, indicating that patients who are homozygous to CC genotype would have nearly 5 times increase in their risk of HCV infection than those carrying the heterozygous CT genotype.
Table 2.
Genotypic distribution for IL-28B gene polymorphism when patient group (groups “1+2+3”) was compared to control group.
| SNPs | Genotype/allele distribution | Healthy controls n = 600 |
HCV patients n = 678 |
OR (95% C.I.) | χ 2 | P value |
|---|---|---|---|---|---|---|
| rs8105790 | 2.784 (2.340–3.311) | 137.99 | <0.0001 | |||
| CC | 16 (2.7%) | 159 (23.4%) | ||||
| CT | 233 (38.8%) | 280 (41.3%) | ||||
| TT | 351 (58.5%) | 239 (35.3%) | ||||
| C | 265 (22.1%) | 598 (44.1%) | ||||
| T | 935 (77.9%) | 758 (55.9%) | ||||
| CC + CT versus TT | 2.589 (2.065–3.247) | 69.23 | <0.0001 | |||
| CC versus CT + TT | 0.089 (0.053–0.152) | 116.35 | <0.0001 | |||
|
| ||||||
| rs8099917 | 2.552 (2.062–3.160) | 77.16 | <0.0001 | |||
| GG | 14 (2.3%) | 104 (15.3%) | ||||
| GT | 114 (19.0%) | 138 (20.4%) | ||||
| TT | 472 (78.7%) | 436 (64.3%) | ||||
| G | 142 (11.8%) | 346 (25.5%) | ||||
| T | 1058 (88.2%) | 1010 (74.5%) | ||||
| GG + GT versus TT | 2.047 (1.593–2.630) | 31.91 | <0.0001 | |||
| GG versus GT + TT | 0.132 (0.075–0.233) | 64.25 | <0.0001 | |||
|
| ||||||
| rs7248668 | 1.863 (1.533–2.264) | 39.72 | <0.0001 | |||
| AA | 58 (9.7%) | 94 (13.9%) | ||||
| AG | 79 (13.2%) | 172 (25.4%) | ||||
| GG | 463 (77.2%) | 412 (60.7%) | ||||
| A | 195 (16.3%) | 360 (26.5%) | ||||
| G | 1005 (83.8%) | 996 (73.5%) | ||||
| AA + AG versus GG | 2.182 (1.708–2.788) | 39.65 | <0.0001 | |||
| AA versus AG + GG | 0.665 (0.470–0.941) | 5.35 | 0.0206 | |||
|
| ||||||
| rs12979860 | 1.476 (1.250–1.743) | 21.15 | <0.0001 | |||
| TT | 59 (9.9%) | 73 (10.8%) | ||||
| CT | 230 (38.6%) | 367 (54.1%) | ||||
| CC | 307 (51.5%) | 238 (35.1%) | ||||
| T | 348 (29.2%) | 513 (37.8%) | ||||
| C | 844 (70.8%) | 843 (62.2%) | ||||
| TT + CT versus CC | 1.964 (1.568–2.460) | 34.88 | <0.0001 | |||
| TT versus CT + CC | 0.911 (0.634–1.308) | 0.26 | 0.61213 | |||
|
| ||||||
| rs12980275 | 1.765 (1.488–2.093) | 42.97 | <0.0001 | |||
| GG | 59 (9.8%) | 89 (13.12%) | ||||
| AG | 184 (30.7%) | 327 (48.2%) | ||||
| AA | 357 (59.5%) | 262 (38.6%) | ||||
| G | 302 (25.2%) | 505 (37.2%) | ||||
| A | 898 (74.8%) | 851 (62.8%) | ||||
| GG + AG versus AA | 2.333 (1.863–2.920) | 55.44 | <0.0001 | |||
| GG versus AG + AA | 0.722 (0.509–1.023) | 3.37 | 0.06631 | |||
Risk allele marked in BOLD letter.
Haplotype analysis revealed three blocks that were significantly distributed between patient group and uninfected healthy control subjects. The blocks were for SNPs rs12980275 and rs12979860, respectively, AC (freq. = 0.603, χ² = 19.06, P < 0.0001), GT (freq. = 0.256, χ² = 50.08, P < 0.0001), and AT (freq. = 0.082, χ² = 11.055, P = 0.0009) (Table 3).
Table 3.
Haplotype analysis for SNPs rs12980275 and rs12979860 when patient group (groups “1+2+3”) was compared to control group.
| Haplotype | Freq. | HCV patients, healthy control ratio counts | HCV, healthy control frequencies | Chi-square | P value |
|---|---|---|---|---|---|
| Block 1 | |||||
| AC | 0.603 | 763.3 : 592.7, 777.1 : 422.9 | 0.563, 0.648 | 19.064 | 1.26 × 10−5 |
| GT | 0.256 | 425.3 : 930.7, 229.5 : 970.5 | 0.314, 0.191 | 50.086 | 1.47 × 10−12 |
| AT | 0.082 | 87.7 : 1268.3, 120.9 : 1079.1 | 0.065, 0.101 | 11.055 | 9 × 10−4 |
| GC | 0.06 | 79.7 : 1276.3, 72.5 : 1127.5 | 0.059, 0.060 | 0.032 | 0.857 |
3.2. Association of IL28B Polymorphisms with Disease Progression
In order to determine whether these SNPs play any role in the progression of the HCV infection to liver cirrhosis and HCC, further analysis was done by comparing HCV-infected patients suffering from cirrhosis (group 2) to patients with chronic HCV infection (group 1) and by comparing patients who have progressed to HCC as a result of persistent HCV infection (group 3) to group 1 patients. No significant observations were found for any of the five SNPs in both comparisons (data not shown). Furthermore, these SNPs were also analyzed by comparing groups 2+3 combined against group 1, and none of the SNPs showed any significant associations (Table 4).
Table 4.
Genotypic distribution for IL-28B gene polymorphism when chronic HCV patients (group 1) were compared to individuals with liver cirrhosis and HCC carriers (groups “2+3”).
| SNPs | Genotype/allele distribution | (Chronic HCV) n = 518 |
(Cirr + HCC) n = 160 |
OR (95% C.I.) | χ ² | P value |
|---|---|---|---|---|---|---|
| rs8105790 | 1.049 (0.815–1.350) | 0.14 | 0.710 | |||
| CC | 119 (23%) | 40 (25%) | ||||
| CT | 216 (42%) | 64 (40%) | ||||
| TT | 183 (35%) | 56 (35%) | ||||
| C | 454 (43.8%) | 144 (45%) | ||||
| T | 582 (56.2%) | 176 (55%) | ||||
| CC + CT versus TT | 1.014 (0.700–1.471) | 0.01 | 0.939 | |||
| CC versus CT + TT | 0.895 (0.592–1.351) | 0.28 | 0.596 | |||
|
| ||||||
| rs8099917 | 0.986 (0.739–1.315) | 0.01 | 0.923 | |||
| GG | 80 (15.4%) | 24 (15%) | ||||
| GT | 105 (20.3%) | 33 (20.6%) | ||||
| TT | 333 (64.3%) | 103 (64.4%) | ||||
| G | 265 (25.6%) | 81 (25.3%) | ||||
| T | 771 (74.4%) | 239 (74.7%) | ||||
| GG + GT versus TT | 0.996 (0.688–1.442) | 0 | 0.983 | |||
| GG versus GT + TT | 1.035 (0.631–1.698) | 0.02 | 0.891 | |||
|
| ||||||
| rs7248668 | 1.204 (0.912–1.590) | 1.72 | 0.190 | |||
| AA | 65 (12.5%) | 29 (18.1%) | ||||
| AG | 136 (26.3%) | 36 (22.5%) | ||||
| GG | 317 (61.2%) | 95 (59.4%) | ||||
| A | 266 (25.7%) | 94 (29.4%) | ||||
| G | 770 (74.3%) | 226 (70.6%) | ||||
| AA + AG versus GG | 1.079 (0.752–1.549) | 0.17 | 0.679 | |||
| AA versus AG + GG | 0.648 (0.402–1.046) | 3.18 | 0.074 | |||
|
| ||||||
| rs12979860 | 1.167 (0.903–1.507) | 1.39 | 0.238 | |||
| TT | 50 (9.7%) | 23 (14.4%) | ||||
| CT | 283 (54.6%) | 84 (52.5%) | ||||
| CC | 185 (35.7%) | 53 (33.1%) | ||||
| T | 383 (37%) | 130 (40.6%) | ||||
| C | 653 (63%) | 190 (59.4%) | ||||
| TT + CT versus CC | 1.122 (0.771–1.632) | 0.36 | 0.548 | |||
| TT versus CT + CC | 0.636 (0.375–1.080) | 2.84 | 0.092 | |||
|
| ||||||
| rs12980275 | 0.929 (0.716–1.206) | 0.3 | 0.580 | |||
| GG | 69 (13%) | 20 (12.5%) | ||||
| AG | 252 (49%) | 75 (46.9%) | ||||
| AA | 197 (38%) | 65 (40.6%) | ||||
| G | 390 (38%) | 115 (36%) | ||||
| A | 646 (62%) | 205 (64%) | ||||
| GG + AG versus AA | 0.897 (0.625–1.288) | 0.35 | 0.555 | |||
| GG versus AG + AA | 1.076 (0.631–1.832) | 0.07 | 0.788 | |||
Risk allele marked in BOLD letter.
3.3. Correlation between IL28B Serum Level and HCV Infection
Next, to evaluate if there is a correlation between serum IL28B levels and the different SNP genotypes, a box-plot analysis was performed for the different SNPs against average logarithmic values of IL28B levels. Only rs8099917 showed a significant correlation in relation to IL28B serum levels. The SNP rs8099917 showed an increase of ~0.06 in log10 of serum IL28B levels, with the addition of each G-allele, suggesting that HCV-infected patients with GG genotype have higher serum IL28B levels (Figure 1 and Table 5).
Figure 1.
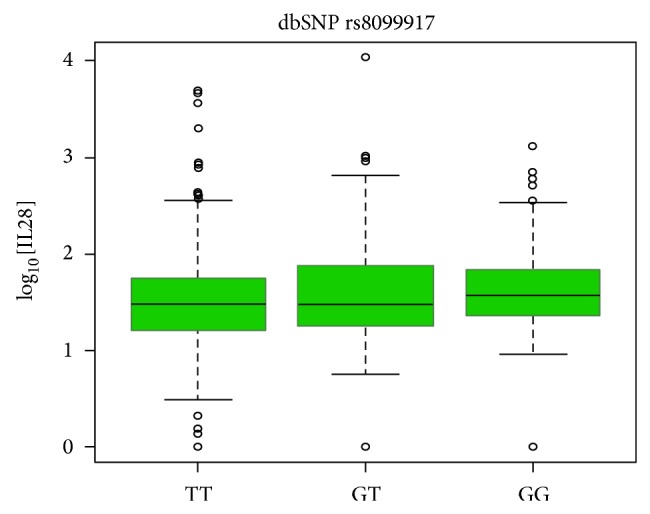
Boxplot analysis of rs8099917 genotypes against average serum IL28B concentration.
Table 5.
Correlation of average IL28B serum levels with the genotypes of rs8099917.
| Value | Std. error | t-value | P value | |
|---|---|---|---|---|
| (Intercept) | 1.586 | 0.025 | 58.82 | 0 |
| rs8099917-GT | 0.057 | 0.049 | 1.16 | 0.25 |
| rs8099917-GG | 0.115 | 0.057 | 2.03 | 0.04 |
Note: results for “intercept” correspond to “GG” genotype; that is, log10 (conc.) = 1.586 for TT genotype, with ~0.06 increase in log10 (conc.) with each additional G-allele.
A similar analysis was done to estimate the correlation of serum IL28B levels with regard to different groups of patients classified according to their severity of the disease. A significant result was obtained when group 2 and groups 2+3 were compared to group 1 with an increase of 0.16 in log10 of IL28B serum level for group 2 and group 2+3 samples, while no additional significant effect was observed for group 3 alone (Figure 2 and Table 6).
Figure 2.
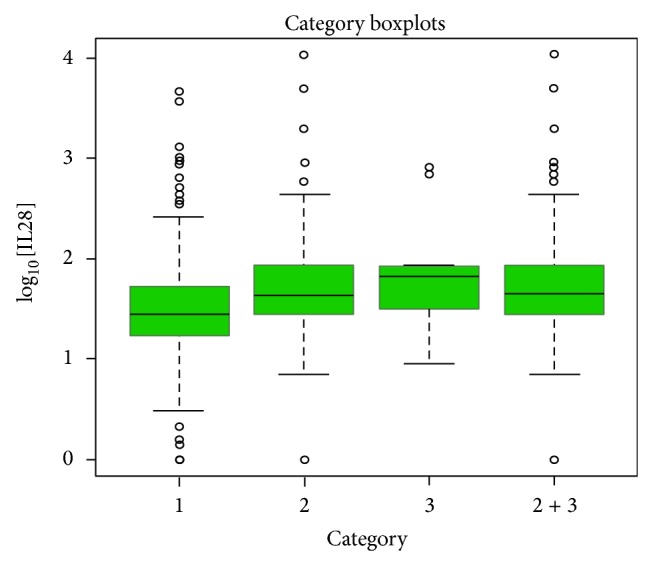
Boxplot analysis of average serum IL28B concentration with different outcomes of HCV infection.
Table 6.
Correlation of average IL28B serum levels with different outcomes of HCV infection.
| Value | Std. error | t-value | P value | |
|---|---|---|---|---|
| (Intercept) | 1.461 | 0.027 | 54.77 | 0 |
| Group 2 | 0.161 | 0.057 | 2.82 | 0.005 |
| Group 3 | 0.126 | 0.161 | 0.79 | 0.43 |
| Groups 2 + 3 | 0.156 | 0.053 | 2.95 | 0.003 |
Note: results for “intercept” correspond to group = 1; that is, log10 (conc.) = 1.46 for group 1 samples, with ~0.16 increase in log10 (conc.) for group 2 samples, and no additional sig. affect in log10 (conc.) for group 3 samples.
Based on a correlation of average log10 of serum IL28 levels, no significant differences were observed between the different HCV genotypes/subgenotypes (classified based on partial NS5B sequence) and serum IL28 levels (Tables 7 and 8).
Table 7.
Linear regression of serum IL28B levels with different factors.
| Factor | Value | Coefficient | P value |
|---|---|---|---|
| rs12979860 | CT | 0.033 | 0.525 |
| rs12979860 | TT | 0.043 | 0.593 |
| rs12980275 | AG | 0.059 | 0.251 |
| rs12980275 | GG | 0.021 | 0.769 |
| rs8105790 | CT | 0.011 | 0.838 |
| rs8105790 | CC | −0.029 | 0.644 |
| rs8099917 | GT | 0.098 | 0.0915 |
| rs8099917 | GG | 0.107 | 0.111 |
| rs7248668 | AG | 0.097 | 0.0889 |
| rs7248668 | AA | 0.034 | 0.602 |
| HCV genotype | 1a | 0.269 | 0.618 |
| HCV genotype | 1b | 0.285 | 0.595 |
| HCV genotype | 1c | −0.176 | 0.815 |
| HCV genotype | 1g | 0.12 | 0.845 |
| HCV genotype | 2 | 0.318 | 0.603 |
| HCV genotype | 2a | 0.217 | 0.738 |
| HCV genotype | 2c | 0.127 | 0.827 |
| HCV genotype | 2k/1b | −0.187 | 0.774 |
| HCV genotype | 3 | −0.097 | 0.881 |
| HCV genotype | 3a | 0.198 | 0.721 |
| HCV genotype | 4 | 0.034 | 0.954 |
| HCV genotype | 4 beta | −0.244 | 0.745 |
| HCV genotype | 4a | 0.214 | 0.688 |
| HCV genotype | 4d | 0.272 | 0.609 |
| HCV genotype | 4l | 0.229 | 0.76 |
| HCV genotype | 4m | −0.219 | 0.736 |
| HCV genotype | 4n | 0.325 | 0.567 |
| HCV genotype | 4o | 0.44 | 0.472 |
| HCV genotype | 4o/4 beta | 0.034 | 0.956 |
| HCV genotype | 4r | 0.366 | 0.502 |
| All versus 1a | 0.028 | 0.787 | |
| All versus 1b | 0.049 | 0.521 | |
| All versus 1a/b | 0.047 | 0.472 | |
| All versus 4a | −0.042 | 0.472 | |
| All versus 4d | 0.042 | 0.482 | |
| All versus 4a/d | −0.001 | 0.985 | |
| Group 2 | 0.245 | 0.0002 | |
| Group 3 | 0.337 | 0.044 | |
| Groups 2 + 3 | 0.255 | 0.00005 |
Table 8.
Robust regression of serum IL28B levels with different factors.
| Factor | Value | Coefficient | P value |
|---|---|---|---|
| rs12979860 | CT | 0.050 | 0.269 |
| rs12979860 | TT | 0.032 | 0.645 |
| rs12980275 | AG | 0.038 | 0.394 |
| rs12980275 | GG | 0.010 | 0.87 |
| rs8105790 | CT | 0.041 | 0.391 |
| rs8105790 | CC | 0.032 | 0.556 |
| rs8099917 | GT | 0.057 | 0.246 |
| rs8099917 | GG | 0.115 | 0.043 |
| rs7248668 | AG | 0.051 | 0.3 |
| rs7248668 | AA | 0.038 | 0.49 |
| HCV genotype | 1a | 0.311 | 0.854 |
| HCV genotype | 1b | 0.219 | 0.897 |
| HCV genotype | 1c | −0.176 | 0.666 |
| HCV genotype | 1g | −0.400 | 0.818 |
| HCV genotype | 2 | 0.318 | 0.853 |
| HCV genotype | 2a | 0.217 | 0.9 |
| HCV genotype | 2c | 0.127 | 0.941 |
| HCV genotype | 2k/1b | −0.187 | 0.914 |
| HCV genotype | 3 | −0.097 | 0.955 |
| HCV genotype | 3a | 0.198 | 0.907 |
| HCV genotype | 4 | 0.489 | 0.776 |
| HCV genotype | 4 beta | −0.244 | 0.551 |
| HCV genotype | 4a | 0.152 | 0.928 |
| HCV genotype | 4d | 0.184 | 0.913 |
| HCV genotype | 4l | 0.229 | 0.575 |
| HCV genotype | 4m | −0.219 | 0.9 |
| HCV genotype | 4n | 0.325 | 0.848 |
| HCV genotype | 4o | 0.006 | 0.997 |
| HCV genotype | 4o/4 beta | −0.189 | 0.995 |
| HCV genotype | 4r | 0.259 | 0.879 |
| All versus 1a | 0.141 | 0.0938 | |
| All versus 1b | 0.042 | 0.507 | |
| All versus 1a/b | 0.089 | 0.096 | |
| All versus 4a | −0.045 | 0.363 | |
| All versus 4d | 0.005 | 0.925 | |
| All versus 4a/d | −0.039 | 0.397 | |
| Group 2 | 0.161 | 0.005 | |
| Group 3 | 0.126 | 0.432 | |
| Groups 2 + 3 | 0.156 | 0.003 |
A linear regression analysis was performed to determine if a significant relationship exists between the different parameters (such as IL28B SNPs, viral subgenotypes, and patient category) and serum IL28 levels (Table 7). No significant relationship was observed either for the IL28B genotypes or the viral subgenotypes, with respect to IL28 levels, while a significant relationship was observed when the different patient groups were analyzed in relation to IL28 levels (for group 2 — r 2 = 0.245, P = 0.0002; for group 3 — r 2 = 0.337; P = 0.045; for group 2+3, r 2 = 0.255; P < 0.0001), suggesting that the advancement of HCV patients to different stages may depend on the serum IL28B levels.
To confirm the results obtained in Table 7, a robust linear regression, by excluding all the outlier values, was performed. This is to determine if the linear regression model failed as a result of these nonconformal outliers or whether no true relationship existed between the various parameters and serum IL28B levels (Table 8). The GG genotype of rs8099917 was found to have a significant relation with serum IL28B levels in the robust model (P value = 0.043, regression coefficient, r 2 = 0.115). A significant relationship was also observed for group 2 and group 2+3 (r 2 = 0.161 and 0.156, P value = 0.005 and 0.003, resp.).
3.4. Receiver Operating Characteristic (ROC) Curve Analysis
ROC analysis was used to establish the specificity and sensitivity of using IL28B serum concentration as a marker for HCV-associated diseases. The best cut-off values were 75.3% sensitivity and 38.5% specificity when chronic patients (group 1) were compared with cirrhosis+HCC patients (groups 2+3). The area under the curve was 0.60 (95% Confidence interval: 0.55–0.65, and P = 0.005). The cut-off value of IL-28B in the serum that could distinguish chronic patients from Cirr+HCC patients was 51.0 pg/mL (Figure 3).
Figure 3.
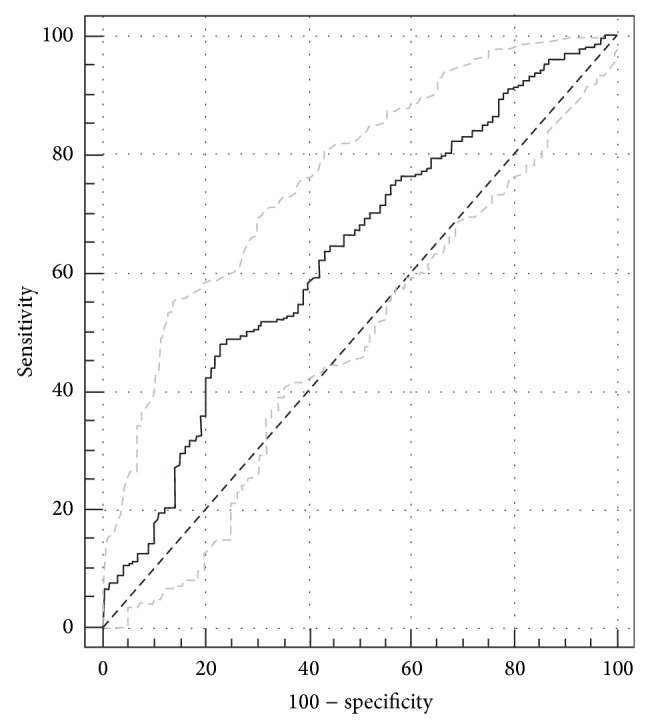
ROC curve of the true positive rate (sensitivity), false positive rate (100-specificity) predictive values of the IL28B serum level concentration among patients infected with HCV and individual with liver cirrhosis+HCC patients.
4. Discussion
IL-28B belongs to type III IFNs, also called IFN-λs, which was discovered in 2002 by two independent groups [12, 13]. Though structurally different from Type-1 IFNs, the IFN-λs exhibit a similar mechanism as that of IFN-α's in terms of their signaling and biological activities. Although both groups of IFNs play a major role in antiviral activities, IFN-λs have recently generated much interest because of their link to the spontaneous resolution and successful treatment of HCV infection [20].
Recent studies have reported that the mRNA expression levels of IL28 in peripheral blood mononuclear cells are lower in patients undergoing treatment and who carry the minor G allele for the SNP rs8099917. This seems theoretically reasonable, as increased levels of IL28B together with the combination therapy would facilitate clearance of the virus [9, 10].
On the contrary, Abe et al. reported that the expression levels of IL28B in liver are lower in PEG-IFN-treated patients having rs8099917 TT genotype [21]. The present study is in agreement with the findings of Abe et al., as the serum IL28B levels were found to decrease with the addition of each T-allele in HCV-infected patients [21]. A plausible explanation of increased expression of IL28B levels in patients undergoing treatment could be for the unique capability of IFN-λs to increase its expression when induced by IFN-α, that is, for patients being treated with PEG-IFN α/β-ribavirin would most likely have increased levels of IL28B levels as a result of its direct stimulation with IFN-α [22, 23]. Further investigations are needed to understand how the serum IL28B levels vary according to the IL28B genetic polymorphisms, since such polymorphisms could affect expression and/or the stability of IL28B mRNA. Also, it is not clear at what level is the expression of IL28B being affected, that is, whether at the transcription and/or the translation levels in HCV-infected patients undergoing treatment with PEG-IFN α/β-ribavirin.
In the current study, rs8099917 was the only SNP that significantly correlated with the IL28B serum levels, while the other four SNPs, including rs12979860, failed to show any correlation with serum IL28B levels. This finding is at variance with the results obtained by Langhans et al., as the study showed correlation of serum level with the C allele of rs12979860 SNP [16]. However, it is unclear if rs8099917 variants were tested in this study as no data were shown. We also observed a significant relationship between IL28B levels and advanced stages of HCV infection as IL28 serum level tends to increase with the severity of the disease; however, further investigations are needed to explain the effect of IL28B levels on disease progression.
IL28B variants have been reported to have a significant effect on fibrosis progression in HCV-infected patients [24, 25]. Recently, a study conducted on Italian population, reported that the HCV-related cirrhotic patients carrying rs12979860-C allele are less likely to undergo liver transplantation or suffer from end-stage liver diseases such as HCC [26]. A similar finding was reported that the C-allele has a protective role in the development of HCV-induced HCC and in HCV recurrence after liver transplantation [27]. On the contrary, other studies have reported that the variants of IL28B may not be involved in the hepatocarcinogenesis of HCV-infected patients [25, 28]. Results reported in the present study were in agreement with that of the latter studies as none of the five IL28B genetic variants investigated in this study had any significant effect on the advancement of HCV infection to cirrhosis and/or HCC; though, we did observe that these SNPs could have a significant role in the establishment of HCV infection as they showed significant differences in distribution between patients and healthy control subjects. However, our results must be interpreted with caution in view of the small number of HCC patients included in the study. Therefore, studies with larger number of cirrhotic and HCC patients are needed to ascertain the role of IL28B polymorphisms in hepatocarcinogenesis among HCV-infected patients.
Although there are several studies reporting the association of HCV virus genotype with treatment response, there are no reports that correlate IL28B serum levels with HCV viral genotypes/subgenotypes. This is the first study that has looked into such a correlation, but, surprisingly, our findings indicate that there is no correlation between IL28B serum level and genotypes/subgenotypes of HCV. This is consistent with other studies that examined serum level of cytokines or chemokines in relation to HCV genotypes [29, 30]. These studies indicated that no correlation was found between HCV genotypes, cytokine/chemokine serum levels, and disease progression or response to treatment in HCV-infected patients.
Furthermore, we have also found that the IL28B serum levels have a significant correlation with the different outcomes of HCV infection, especially among patients suffering from cirrhosis and HCC, where the IL28 serum levels tend to increase with progression of the disease. However, it is unclear why the IL28 serum levels are affected by the different outcomes of HCV infection and by rs8099917-G allele, and therefore additional research is required to establish these correlations.
The analysis of ROC curve revealed the usefulness of the IL28B serum level as a prognostic marker for disease progression in HCV-infected patients. It was possible to differentiate patients with liver complication, such as liver cirrhosis and HCC, from chronic patients. However, these results must be carefully interpreted as the area under the curve was not very high (0.6) and this could be attributed to the low number of patients with cirrhosis and HCC.
5. Conclusions
The T-allele of rs8099917 was found to be correlated with reduced IL28B serum levels. Also, IL28B variants might play a significant role in HCV infection, but they may not be considered as risk factors in the progression of HCV infection to advanced stages such as liver cirrhosis and HCC.
Acknowledgments
This study was supported in part by a grant from King Abdulaziz City for Science and Technology [ARP-30-38] and was approved by the Research Advisory Council (RAC) of King Faisal Specialist Hospital and Research Centre, RAC no. 2090001. The support of the Research Center administration at King Faisal Specialist Hospital & Research Center is highly appreciated. The authors are grateful to Hanan Shaarawi and Maureene Delos-Reyes for secretarial and logistic assistance.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver International. 2009;29(1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Barrera J. M., Bruguera M., Guadalupe Ercilla M., et al. Persistent Hepatitis C viremia after acute self-limiting posttransfusion Hepatitis C. Hepatology. 1995;21(3):639–644. doi: 10.1002/hep.1840210306. [DOI] [PubMed] [Google Scholar]
- 3.Seeff L. B. Natural history of chronic hepatitis C. Hepatology. 2002;36(5, supplement 1):S35–S46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 4.Hadziyannis S. J., Sette H., Jr., Morgan T. R., et al. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Annals of Internal Medicine. 2004;140(5):346–355. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- 5.Manns M. P., McHutchison J. G., Gordon S. C., et al. Peginterferon alfa-2b plus ribavirin compared with interferonalfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. The Lancet. 2001;358(9286):958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 6.McHutchison J. G., Lawitz E. J., Shiffman M. L., et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. The New England Journal of Medicine. 2009;361(6):580–593. doi: 10.1056/nejmoa0808010. [DOI] [PubMed] [Google Scholar]
- 7.Kohli A., Shaffer A., Sherman A., Kottilil S. Treatment of hepatitis C: a systematic review. The Journal of the American Medical Association. 2014;312(6):631–640. doi: 10.1001/jama.2014.7085. [DOI] [PubMed] [Google Scholar]
- 8.Ge D., Fellay J., Thompson A. J., et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka Y., Nishida N., Sugiyama M., et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nature Genetics. 2009;41(10):1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 10.Suppiah V., Moldovan M., Ahlenstiel G., et al. IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nature Genetics. 2009;41(10):1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 11.Thomas D. L., Thio C. L., Martin M. P., et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotenko S. V., Gallagher G., Baurin V. V., et al. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nature Immunology. 2003;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 13.Sheppard P., Kindsvogel W., Xu W., et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nature Immunology. 2003;4(1):63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 14.Marcello T., Grakoui A., Barba-Spaeth G., et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131(6):1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 15.Robek M. D., Boyd B. S., Chisari F. V. Lambda interferon inhibits hepatitis B and C virus replication. Journal of Virology. 2005;79(6):3851–3854. doi: 10.1128/jvi.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langhans B., Kupfer B., Braunschweiger I., et al. Interferon-lambda serum levels in hepatitis C. Journal of Hepatology. 2011;54(5):859–865. doi: 10.1016/j.jhep.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 17.Bruix J., Sherman M., Llovet J. M., et al. Clinical management of hepatocellular carcinoma. Conclusions of the Barcelona-2000 EASL conference. European Association for the Study of the Liver. Journal of Hepatology. 2001;35(3):421–430. doi: 10.1016/s0168-8278(01)00130-1. [DOI] [PubMed] [Google Scholar]
- 18.Al-Qahtani A. A., Rubino S., Al-Ahdal M. N. Sequence variation of the HVR1 region of hepatitis C virus in response to interferon-alpha and ribavirin treatment. Journal of Infection in Developing Countries. 2011;5(5):370–376. doi: 10.3855/jidc.1806. [DOI] [PubMed] [Google Scholar]
- 19.Murphy D. G., Willems B., Deschênes M., Hilzenrat N., Mousseau R., Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5′ untranslated region sequences. Journal of Clinical Microbiology. 2007;45(4):1102–1112. doi: 10.1128/jcm.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly C., Klenerman P., Barnes E. Interferon lambdas: the next cytokine storm. Gut. 2011;60(9):1284–1293. doi: 10.1136/gut.2010.222976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe H., Hayes C. N., Ochi H., et al. IL28 variation affects expression of interferon stimulated genes and peg-interferon and ribavirin therapy. Journal of Hepatology. 2011;54(6):1094–1101. doi: 10.1016/j.jhep.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 22.Ank N., West H., Bartholdy C., Eriksson K., Thomsen A. R., Paludan S. R. Lambda interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. Journal of Virology. 2006;80(9):4501–4509. doi: 10.1128/jvi.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sirén J., Pirhonen J., Julkunen I., Matikainen S. IFN-α regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29. Journal of Immunology. 2005;174(4):1932–1937. doi: 10.4049/jimmunol.174.4.1932. [DOI] [PubMed] [Google Scholar]
- 24.Abe H., Ochi H., Maekawa T., et al. Common variation of IL28 affects gamma-GTP levels and inflammation of the liver in chronically infected hepatitis C virus patients. Journal of Hepatology. 2010;53(3):439–443. doi: 10.1016/j.jhep.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 25.Bochud P.-Y., Bibert S., Kutalik Z., et al. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology. 2012;55(2):384–394. doi: 10.1002/hep.24678. [DOI] [PubMed] [Google Scholar]
- 26.Fabris C., Falleti E., Cussigh A., et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. Journal of Hepatology. 2011;54(4):716–722. doi: 10.1016/j.jhep.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 27.Eurich D., Boas-Knoop S., Bahra M., et al. Role of IL28B polymorphism in the development of hepatitis C virus-induced hepatocellular carcinoma, graft fibrosis, and posttransplant antiviral therapy. Transplantation. 2012;93(6):644–649. doi: 10.1097/TP.0b013e318244f774. [DOI] [PubMed] [Google Scholar]
- 28.Joshita S., Umemura T., Katsuyama Y., et al. Association of IL28B gene polymorphism with development of hepatocellular carcinoma in Japanese patients with chronic hepatitis C virus infection. Human Immunology. 2012;73(3):298–300. doi: 10.1016/j.humimm.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 29.Akbar H., Idrees M., Butt S., et al. High baseline interleukine-8 level is an Independent risk factor for the achievement of sustained virological response in chronic HCV patients. Infection, Genetics and Evolution. 2011;11(6):1301–1305. doi: 10.1016/j.meegid.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L., Miao L., Han F., Dou X.-G. Cytokine levels in serum of patients with chronic hepatitis C and its significance. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2011;27(3):301–303. [PubMed] [Google Scholar]


