Abstract
Epidemiological studies suggest that allergy risk is preferentially transmitted through mothers. This can be due to genomic imprinting, where the phenotype effect of an allele depends on its parental origin, or due to maternal effects reflecting the maternal genome's influence on the child during prenatal development. Loss-of-function mutations in the filaggrin gene (FLG) cause skin barrier deficiency and strongly predispose to atopic dermatitis (AD). We investigated the 4 most prevalent European FLG mutations (c.2282del4, p.R501X, p.R2447X, and p.S3247X) in two samples including 759 and 450 AD families. We used the multinomial and maximum-likelihood approach implemented in the PREMIM/EMIM tool to model parent-of-origin effects. Beyond the known role of FLG inheritance in AD (R1meta-analysis = 2.4, P = 1.0 x 10−36), we observed a strong maternal FLG genotype effect that was consistent in both independent family sets and for all 4 mutations analysed. Overall, children of FLG-carrier mothers had a 1.5-fold increased AD risk (S1 = 1.50, Pmeta-analysis = 8.4 x 10−8). Our data point to two independent and additive effects of FLG mutations: i) carrying a mutation and ii) having a mutation carrier mother. The maternal genotype effect was independent of mutation inheritance and can be seen as a non-genetic transmission of a genetic effect. The FLG maternal effect was observed only when mothers had allergic sensitization (elevated allergen-specific IgE antibody plasma levels), suggesting that FLG mutation-induced systemic immune responses in the mother may influence AD risk in the child. Notably, the maternal effect reported here was stronger than most common genetic risk factors for AD recently identified through genome-wide association studies (GWAS). Our study highlights the power of family-based studies in the identification of new etiological mechanisms and reveals, for the first time, a direct influence of the maternal genotype on the offspring’s susceptibility to a common human disease.
Author Summary
Most human diseases are caused by a combination of multiple environmental and genetic influences. The widely used case/control approach aims to identify disease risk genes by comparing the genetic constitution of affected and healthy individuals. Although successful, this approach ignores additional mechanisms influencing disease risk. Here, we studied mutations in the filaggrin gene (FLG), which are strong risk factors for atopic dermatitis (AD) and allergies, in a large number of families with AD. We found that FLG mutations in the mother, not the father, increased the AD risk of the children, even if the child did not inherit the mutation. Thus, our study revealed, for the first time, a direct influence of a maternal mutation on the child’s risk for a common disease. The maternal FLG effect was only found when the mothers were allergic, and was absent in families of non-allergic mothers. This finding suggests that FLG-induced changes in the maternal immune response shape the child’s immune system during pregnancy and increase the child’s risk for AD. Our study indicates that maternal FLG mutations act as strong environmental risk factors for the child and highlights the potential of family-based studies in uncovering novel disease mechanisms in medical genetics.
Introduction
Atopic dermatitis (AD, eczema) is a chronic inflammatory skin disease with 10–20% prevalence in industrialized countries. The etiology of AD is complex, with multiple genetic and environmental factors influencing disease risk. Genome-wide association studies (GWAS) have successfully identified common genetic variants predisposing to AD, but the effect of these risk loci is small and altogether only account for a fraction of the disease heritability.
The filaggrin gene (FLG) encodes a structural protein playing a critical role in the terminal differentiation of the epidermis and in skin barrier function [1]. Loss-of-function mutations in FLG were identified as the cause of ichthyosis vulgaris, a common Mendelian trait characterized by dry, scaly skin and frequent AD [2]. Subsequent studies revealed that FLG mutations also strongly predispose to AD [3,4]. This observation has been widely replicated, rendering FLG the strongest and best characterized AD risk locus to date [1]. Overall, evidence from human and animal studies demonstrated that filaggrin deficiency results in altered skin structure, impaired barrier function and enhanced antigen penetration through the skin, leading to the production of allergen-specific IgE antibodies (specific sensitization) and AD [5–7].
Epidemiological studies on allergic diseases have shown that maternal allergy is a stronger risk factor for the child than paternal allergy [8,9], although some conflicting results have been reported for AD [10,11]. The molecular basis of this preferential maternal transmission of allergy risk is currently unknown but it can potentially occur through two different biological mechanisms, genomic imprinting or direct maternal genotype effects. In genomic imprinting, either the maternally or the paternally inherited allele is expressed while the alternate allele is silenced. Thus, the effect of an allele depends on its parental origin resulting in phenotypic differences between reciprocal heterozygotes (parent-of-origin effects) [12,13]. Recent studies have demonstrated that parent-of-origin effects in complex diseases can be due to genetic variation in imprinted genes [12,13]. Alternatively, maternal genotype effects occur when the maternal genotype directly influences the child’s phenotype. This effect is independent of the child’s own genotype and occurs through the maternally provided environment during prenatal development. Maternal genotype effects can lead to phenotypic differences between reciprocal heterozygotes and are thus considered parent-of-origin effects [13,14].
We hypothesized that loss-of-function mutations in FLG may show parent-of-origin effects. Analysis of 2 large family-based cohorts strongly supports that maternal FLG mutations directly increase AD risk in the children.
Results
Allelic heterogeneity and population-specific mutations in FLG
To systematically identify loss-of-function variants at the FLG locus, we used data of the Exome Aggregation Consortium (ExAC [15]), which includes whole exome sequencing results of 61,486 individuals. Filtering by frameshift or non-sense mutations revealed 254 loss-of-function mutations in the gene (S1 Table). The majority of FLG mutations were very rare, 227 of 254 FLG mutations (89,4%) had an allele frequency (AF) < 0.0001. Further analysis revealed the presence of population-specific mutations. For example, the p.L4022X mutation was common in East Asia (AF = 0.02) but absent from all other populations studied. This data confirms and extends previous reports of allelic heterogeneity and population-specific mutations in FLG [1].
In the European (non-Finnish) population ExAC reported 146 loss-of-function mutations with a combined AF of 0.052. Of these, the 4 most prevalent mutations, accounting for 86% of all mutant alleles in this population, were selected for genotyping in the present study: p.761fsX35 (c.2282_2285delCAGT; rs558269137, referred to as c.2282del4), p.R501X (c.1501C>T; rs61816761), p.R2447X (c.7339C>T; rs138726443), and p.S3247X (c.9740C>A; rs150597413).
FLG mutations are strong risk factors for AD
The 4 selected mutations were genotyped in 759 complete nuclear families from Central Europe recruited through one or more children with AD (methods and Table 1). The allele frequencies in the founders were in good agreement with previous studies (0.059, 0.034, 0.01 and 0.002 for c.2282del4, p.R501X, p.R2447X and p.S3247X, respectively) [16,17].
Table 1. Study populations.
| Central European | Northern European | ||
|---|---|---|---|
| Families | Total number of families | 759 | 450 |
| Number of complete nuclear families | 759 | 325 | |
| - families with 1 affected child (Trio) | 323 | 35 | |
| - families with 2 affected siblings (ASP) | 399 | 235 | |
| - families with > 2 affected siblings | 37 | 55 | |
| Number of incomplete families | 0 | 125 | |
| Total number of affected children | 1209 | 680 | |
| Unrelated individuals | Number of cases | 1147 | 0 |
| Number of population-based controls a | 3339 | 1854 |
a All population-based controls were included in the study irrespective of AD status. ASP refers to affected sib pairs.
As previously reported, FLG mutations showed a strong over-transmission from heterozygote parents to AD-affected children in a Transmission Disequilibrium Test (TDT; Table 2 [16,17]). We observed no linkage disequilibrium among FLG mutations, since each mutation was on a different haplotype and 2 different mutations never occurred together in the same haplotype (S2 Table). Since previous studies reported that these 4 loss-of-function FLG mutations have the same effect on AD risk, we decided to merge all variants into a combined genotype [4]. This enabled us to work with one common instead of 4 low-frequency variants. Unless stated otherwise, the results presented below refer to the combined genotype.
Table 2. Results of the transmission disequilibrium test.
| Central European | Northern European | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | A1 a | A2 b | Freq c | T / U d | OR e | P f | Freq c | T / U d | OR e | P f |
| c.2282del4 | del | Wild type | 0.059 | 189 / 91 | 2.08 | < 10−6 | 0.047 | 83 / 40 | 2.07 | 7.26 x 10−5 |
| p.R501X | T | C | 0.034 | 97 / 47 | 2.06 | 5.70 x 10−5 | 0.012 | 27 / 13 | 2.08 | 0.058 |
| p.R2447X | T | C | 0.010 | 29 / 17 | 1.71 | 0.27 | 0.014 | 8 / 22 | 0.36 | 0.01 |
| p.S3247X | A | C | 0.002 | 6 / 1 | 6 | 0.31 | 0.004 | 8 / 6 | 1.33 | 1 |
| Combined | Mutation | Wild type | 0.105 | 316 / 150 | 2.11 | < 10−6 | 0.085 | 125 / 78 | 1.60 | 0.001 |
a A1 is the mutant allele.
b A2 is the wild type allele.
c Allele frequency of A1 in the family founders.
d Number of transmitted (T) and un-transmitted (U) A1 alleles.
e Odds ratio calculated as the ratio of transmitted versus un-transmitted alleles.
f Empirical p value calculated by flipping transmitted/untransmitted status in order to account for multiple affected siblings in each family.
Direct maternal genotype effect of FLG mutations
Testing for parent-of-origin effects was performed with the PREMIM/EMIM software. This tool uses a multinomial model-based maximum-likelihood approach for flexible modelling of parent-of-origin effects (see methods) [18,19]. In order to increase power, we included FLG genotypes of 1147 unrelated AD cases and 3339 population-based controls. This set of unrelated individuals does not provide information on parent-of-origin effects, but increases the power to detect them by improving the estimation of genotype frequencies in the general population (see methods) [18,19].
We performed a step-by-step analysis starting with a basic genetic model and successively including additional risk parameters modelling parent-of-origin effects. The basic scenario ignored the available parental genotypes and tested the effect of the child´s genotype on his own phenotype. As expected, we observed large effects with relative risks of 3.1 for heterozygous (R1 parameter) and 10.5 for homozygous FLG mutation carriers (R2 parameter) (Child Genotype or CG model; R1 = 3.1, R2 = 10.5; P CG = 5.9 x 10−74; Table 3).
Table 3. Parent-of-origin analysis of the combined FLG mutations.
| Children Genotype model (CG) | ||||||
| Study | R1 (CI) | R2 (CI) | S1 (CI) | Im (CI) | P null a | - |
| Central Europe | 3.10 (2.68–3.59) | 10.5 (7.20–15.36) | - | - | 5.9 x 10−74 | - |
| Northern Europe | 2.44 (1.95–3.06) | 7.37 (3.37–15.81) | - | - | 9.23 x 10−17 | - |
| Meta-analysis P meta c | 2.89 (2.56–3.27) 2.8 x 10−65 | 9.80 (6.97–13.77)2.1 x 10−39 | - | - | - | - |
| P het d | 0.08 | 0.41 | ||||
| Maternal Child Genotype model (MCG) | ||||||
| Study | R1 (CI) | R2 (CI) | S1 (CI) | Im (CI) | P null a | P MCG vs CG b |
| Central Europe | 2.57 (2.18–3.04) | 7.97 (5.36–11.87) | 1.55 (1.29–1.87) | - | 2.7 x 10−77 | 5.0 x 10−6 |
| Northern Europe | 2.13 (1.67–2.72) | 5.89 (2.67–12.97) | 1.42 (1.11–1.82) | - | 1.42 x 10−17 | 0.005 |
| Meta-analysis P meta c | 2.43 (2.11–2.78) 1.0 x 10−36 | 7.50 (5.26–10.70) 1.2 x 10−28 | 1.50 (1.29–1.74) 8.4 x 10−8 | - | - | - |
| P het d | 0.21 | 0.50 | 0.58 | |||
| Imprinting model (Im) | ||||||
| Study | R1 (CI) | R2 (CI) | S1 (CI) | Im (CI) | P null a | P Im vs CG b |
| Central Europe | 2.70 (2.19–3.31) | 8.06 (5.09–12.78) | - | 1.30 (1.00–1.69) | 1.2 x 10−73 | 0.047 |
| Northern Europe | 2.19 (1.63–2.95) | 5.97 (2.56–13.92) | - | 1.23 (0.86–1.75) | 3.22 x 10−16 | 0.25 |
| Meta-analysis P meta c | 2.52 (2.13–2.99) 1.4 x 10−26 | 7.53 (5.02–11.28) 1.4 x 10−22 | - | 1.28 (1.03–1.57) 0.02 | - | - |
| P het d | 0.26 | 0.54 | 0.80 | |||
a P value for the comparison of each model versus the null model with no effects.
b P value for the comparison of each model versus the Child Genotype model.
c P value for the meta-analysis of each estimated parameter (see methods). CI indicates 95% confidence interval.
d P value for a test of heterogeneity. All results correspond to the combined FLG mutations.
Next maternal genotype effects were modelled by including an additional parameter, S1, to estimate the relative AD risk of children whose mother carried a FLG mutation. Children of FLG mutation-carrier mothers had a striking 1.55 fold increase in AD risk independently of their own genotype (Mother-Child Genotype model or MCG; R1 = 2.57, R2 = 7.97, S1 = 1.55; P MCG = 2.7 x 10−77; Table 3). A comparison of the Child Genotype and the Mother-Child Genotype models by a likelihood-ratio test, strongly supported the existence of a maternal genotype effect (P MCG vs CG = 5.0 x 10−6). In addition, we observed no evidence of interaction between the child and maternal genotypes, indicating that carrying a mutation and having a mutation carrier mother are independent risk factors with additive effect on disease risk (S3 Table and methods). Thus, children with both risk factors, i.e. carrying a FLG mutation and having a mutation carrier mother, have a nearly 4-fold increased disease risk (R1 x S1 = 3.6).
Importantly, both genomic imprinting and maternal genotype effects can lead to similar patterns of parent-of-origin effects and specific tests need to performed to distinguish them [13,14]. Finally, we tested an imprinting model by including the imprinting parameter, Im, which represents the relative risk of the child when inheriting a mutant allele from the mother as opposed to the father. A comparison with the Child Genotype model provided marginal support for the presence of imprinting (P Im vs CG = 0.047; Table 3). In order to test which parent-of-origin scenario better fits our data we performed comparisons versus a full model containing all risk parameter (R1, R2, S1 and Im). Interestingly, adding the maternal genotype parameter S1 to a model already containing Im resulted in a significantly better model (Table 4; p = 3.2 x 10−6). On the contrary, adding Im to a model already containing S1 provided only a marginal improvement (p = 0.03). These results favour the existence of a direct maternal genotype effect of FLG.
Table 4. Comparison of the MCG and Imprinting models with the full model.
| Parameters included | Central European study | Northern European study | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | R1 | R2 | Im | S1 | lnlik | P null a | P CG b | P full c | lnlik | P null a | P CG b | P full c |
| Null | - | - | - | - | −3378.5 | −1376.3 | ||||||
| CG | + | + | - | - | −3209.89 | 5.9 x 10–74 | −1343.5 | x 10−17 | ||||
| MG | + | + | - | + | −3199.46 | 2.7 x 10−77 | 5.0 x 10−06 | 2.9 x 10−02 | −1339.8 | 1.4 x 10−17 | 0.005 | 0.162 |
| Im | + | + | + | - | −3207.92 | 1.2 x 10−73 | 4.7 x 10−02 | 3.2 x 10−06 | −1342.3 | 3.3 x 10−16 | 0.25 | 0.004 |
| Full | + | + | + | + | −3197.07 | 2.9 x 10−77 | 2.7 x 10−06 | −1339.5 | 3.1 x 10−17 | 0.008 | ||
The “+” and “-” symbols indicate whether a given risk parameter was included or excluded in the corresponding model. lnlik is the maximized ln likelihood for each model.
a P value for the comparison of each model versus the null model with no effects.
b P value for the comparison of each model versus the child genotype model including R1 and R2.
c P value for the comparison with the full model including all risk parameters (R1, R2, S1, S2, and Im).
Replication in an independent sample and meta-analysis
We aimed to replicate our findings by examining the same 4 FLG mutations in an independent Northern European population including 450 AD families and 1854 population-based control individuals (methods and Table 1) [17,20–22]. Step-by-step analysis with PREMIM/EMIM again supported a maternal genotype effect. The genotypes of both children and mothers had an independent effect on AD risk, and children of FLG-carrier mothers showed a 1.4 fold increased risk (R1 = 2.13, R2 = 5.89, S1 = 1.42; P MCG = 1.4 x 10−17; P MCG vs CG = 0.005; Table 3 and S3 Table). Importantly, the results obtained were consistent in both populations studied providing strong support to the existence of maternal genotype effects on FLG.
A meta-analysis was performed using the inverse variance method as implemented in METAL [23], which uses the effects estimates and standard errors from each risk parameter. This revealed a highly significant 1.5 fold increased AD risk in children of FLG-carrier mothers (S1meta-analysis = 1.50; P = 8.4 x 10−8; Table 3).
FLG mutations are more frequent in mothers than in fathers
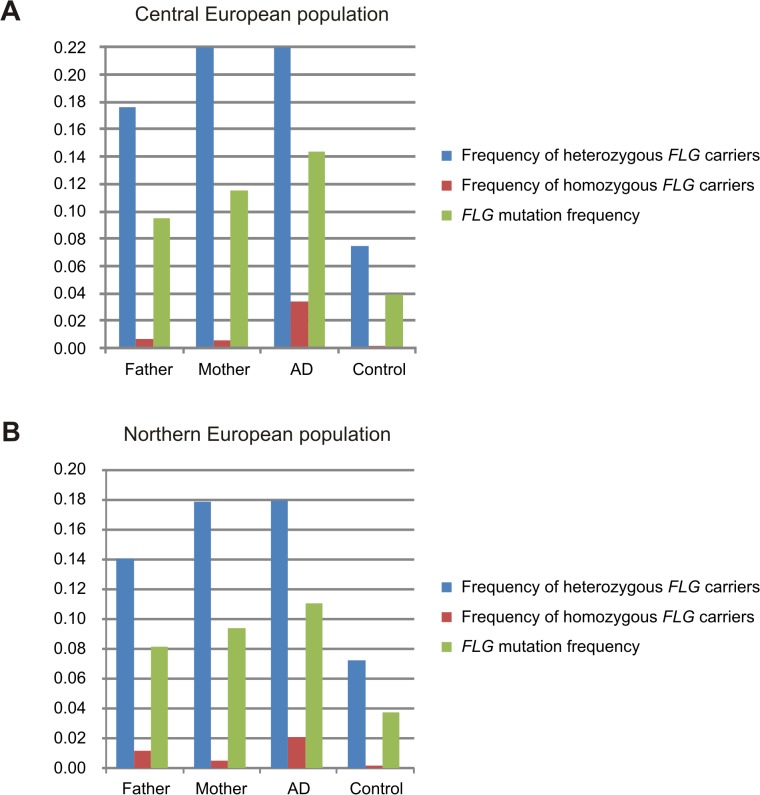
Analysis of parental genotypes revealed a higher prevalence of FLG mutations in mothers than in fathers in both study populations (Fig. 1). This is consistent with the maternal genotype effect observed. Additionally the frequency of FLG mutations in the parental population (mothers and fathers together) was remarkably higher than in population-based controls of unknown phenotype. This is likely due to the recruitment of families with multiple affected children leading to a parental population enriched in strong genetic risk factors.
Fig 1. Frequency of FLG mutations in fathers, mothers, individuals with atopic dermatitis and controls.
Allele and genotype frequencies of the combined FLG-mutations in fathers and mothers were calculated using all available parents. AD refers to the frequency in the AD-affected children including the families and the unrelated AD-cases (available only in the Central European study). Frequency in controls corresponds to population-based individuals with unknown disease status. Results of the Central and Northern European populations are shown in panels A and B, respectively.
Robustness of the FLG maternal effect
At this stage we considered the potential weaknesses of our study in order to discard false positives due to methodological issues and to gain further support to the existence of a FLG maternal genotype effect.
We analysed the FLG c.2282del4, p.R501X and p.R2247X mutations independently (this was not possible for p.S3247X since it was too rare). The maternal effect and the increased frequency of mutations in mothers were found for all 3 mutations in both study populations (S4 Table and S5 Table). This suggests that the maternal effect is not specific to a given variant but a general characteristic of FLG loss-of-function mutations.
In the populations studied, mothers typically have a more prominent role than fathers in children’s health care [24]. We hypothesized that preferential ascertainment of AD-affected mothers carrying FLG mutations may be the cause of the observed maternal effect. Indeed, we observed a higher frequency of AD-affected mothers than fathers, which could be due to a genuine maternal effect or to ascertainment bias (AD prevalence in Central European mothers = 0.23 and fathers = 0.12; Northern European mothers = 0.35 and fathers = 0.19). In order to avoid this potential bias we repeated our analysis including only families in which both parents had a negative history of AD. Importantly, the maternal genotype effect remained strong and significant in the remaining population (Meta-S1 = 1.38; P = 0.003; S6 Table).
We also tested the potential effect of the paternal FLG genotype on the children. Since this option is not available in PREMIM/EMIM, we performed the analysis after exchanging the paternal and maternal genotypes on our genotype files. This analysis revealed no significant effect of the paternal FLG status (S7 Table).
A large proportion of the families included in the present study (60%) were recruited through an affected sib pair. Aiming to maximize power, all previous analyses were performed considering all affected siblings as independent individuals which may lead to biased risk parameter estimates. We therefore repeated the analysis including only one affected child per family and found that the magnitude of the maternal effect remained constant in this set of independent trios (S1meta-analysis = 1.45; P = 1.1 x 10−4; S8 Table).
Potential influence of maternal immunity
Filaggrin has a major role in cutaneous barrier function [1]. According to publicly available datasets [25–27] FLG expression is highest in skin and absent in tissues relevant for mother-child interactions such as uterus, placenta, or mammary gland (S1 Fig. and S2 Fig.). However, recent studies demonstrated that FLG mutations result in increased antigen penetration through the skin and the production of allergen-specific antibodies (IgE, specific sensitization) [6,7,28]. Thus, we hypothesized that systemic inflammatory responses in FLG carrier mothers may influence AD risk in children via feto-maternal immune crosstalk.
All available maternal plasma samples from the Central European study were therefore tested for the presence of allergen-specific IgE, which is a well-characterized biochemical marker of allergy [29]. We performed a stratified analysis in 253 families with and 311 families without maternal specific sensitization. In the families with maternal specific sensitization (+Mat_sens) we observed a strong maternal genotype (S1 = 1.63; P MCG vs CG = 0.005) and a weak child genotype effect (R1 = 1.37; ; Pnull = 0.03; Table 5). This was consistent with a marginal over-transmission of FLG mutations from parents to affected offspring in a transmission disequilibrium test (TDT, transmitted (T): untransmitted (U) = 97:65, P = 0.04; Table 6). In contrast, the opposite pattern was observed in the group of families with non-allergic mothers (−Mat_sens). Here, the maternal genotype effect was not significant (S1 = 1.18; P MCG vs CG = 0.3) while the child genotype effect was strong (R1 = 2.3; Pnull = 9.9 x 10−11). This observation was confirmed by a striking over-transmission of FLG mutations in the TDT (T:U = 169:68, P = 5.3 x 10−8). In concordance with the different rates of mutation transmission observed in both groups, the frequency of FLG mutations was significantly higher in AD-children of non-allergic mothers (mutation frequency 0.16 and 0.11 in affected children of −Mat_sens and +Mat_sens families, respectively; OR = 1.48; P = 0.005; Table 6).
Table 5. Parent-of-origin-analysis analysis stratified by maternal specific sensitization.
| Children Genotype model (CG) | ||||||
| Group | n a | R1 (CI) | R2 (CI) | S1 (CI) | P null b | P MCG vs CG c |
| +Mat_sens | 253 | 1.37 (0.97–1.94) | 2.98 (1.19–7.45) | - | 0.03 | - |
| −Mat_sens | 311 | 2.30 (1.64–3.22) | 7.19 (3.77–13.7) | - | 9.9 x 10−11 | - |
| Maternal Child Genotype model (MCG) | ||||||
| Group | n a | R1 (CI) | R2 (CI) | S1 (CI) | P null b | P MCG vs CG c |
| +Mat_sens | 253 | 1.37 (0.97–1.93) | 2.97 (1.19–7.43) | 1.63 (1.15–2.31) | 0.002 | 0.005 |
| −Mat_sens | 311 | 2.30 (1.64–3.22) | 7.19 (3.77–13.69) | 1.18 (0.86–1.61) | 3.2 x10−10 | 0.30 |
a Number of families.
b P value for the comparison with the null model with no effects.
c P value for the comparison with the Child Genotype model.
CI indicates 95% confidence interval. All results correspond to the combined FLG mutations. Note that all unrelated cases and population-based controls were excluded from this analysis due to missing maternal sensitization data.
Table 6. TDT analysis stratified by maternal specific sensitization.
| Transmission Disequilibrium Test (TDT) | Allelic test in children d | |||||
|---|---|---|---|---|---|---|
| Group | n a | T / U b | OR c | P | Mut freq | OR (P) |
| +Mat_sens | 253 | 97 / 65 | 1.49 | 0.04 | 0.11 | 1 |
| −Mat_sens | 311 | 169 / 68 | 2.49 | 5.3 x10−8 | 0.16 | 1.48 (0.005) |
a Number of families.
b Number of transmitted (T) and un-transmitted (U) alleles. Allele counts are referred to the combined FLG mutations.
c Odds ratio calculated as the ratio of transmitted versus un-transmitted alleles.
d Comparison of allelic frequency between AD-affected children of the −Mat_sens versus the +Mat_sens families.
Discussion
We report here that maternal loss-of-function mutations in FLG directly influence AD risk in the offspring independently of the child’s own genotype. Importantly, this maternal effect was observed consistently for all different FLG mutations tested and in 2 independent populations. Given that genomic imprinting and maternal genotype effects can lead to similar patterns of parent-of-origin effects [14], we specifically modelled both scenarios. Although we cannot completely exclude an imprinting effect, our data supports a direct maternal genotype effect of FLG mutations. This is consistent with the lack of known imprinted genes in the 1q21.3 genomic region containing FLG [25].
It is not obvious how maternal mutations in a skin-barrier gene can influence the child’s phenotype. However, given that filaggrin deficiency promotes specific sensitization, we hypothesized that systemic immune responses may play a role in the FLG maternal effect. Our observation that the FLG maternal effect is significant only in the group of sensitized mothers supports this hypothesis and indicates that FLG mutations in allergic mothers act as a strong environmental risk factor for AD in the child.
The importance of prenatal mother-child interaction in shaping the child’s immune phenotype is highlighted by studies in pregnant mice: while the induction of Th2 immune responses increased susceptibility to allergic asthma in the offspring, a protective effect was observed upon induction of Th1 responses, emphasizing the importance of maternal immune status during pregnancy [30–32]. Supporting evidence in humans arise from epidemiological studies showing that exposure to a microbial-rich farm environment during pregnancy protects children from the development of allergic diseases [33]. It is unknown how this “immunological imprinting” may be transmitted from mother to child, but animal and human studies suggest the induction of epigenetic modifications in relevant immune cells in the offspring [31,34]. Future large population-based studies with parental DNA, data on maternal allergic sensitization and biological material available for epigenetic analysis will be required to further explore this interesting hypothesis.
A recent mouse study found that parent-of-origin effects are widespread and account for an unexpectedly large proportion of complex trait heritability [35]. This is supported by human studies demonstrating that the parental origin of an allele inherited by the offspring can affect disease susceptibility to complex diseases [12,13,36–38]. However, evidence for the existence of maternal genotype effects, which occur without transmission of the risk allele to the offspring, comes mainly from animal studies analysing the effect of maternal gene knockouts in wild-type offspring [39]. Examples of such effects in humans are scarce and refer to rare congenital malformations [40,41]. The present work is, to our knowledge, the first report of a large maternal genotype effect in a common human disease. Notably, the magnitude of the maternal effect (RR = 1.5) was consistent in both data sets and exceeded that reported for most AD genetic risk factors identified to date.
Interestingly, AD is commonly the first manifestation of allergic disease and filaggrin deficiency is a risk factor for the transition from AD to other atopic diseases such as food allergy, hay fever, and asthma [5,16]. Thus, it is tempting to speculate that maternal FLG mutations may influence the risk of a much wider range of allergic disorders.
This and other studies provide proof-of-principle that associations originally discovered by case-control analysis can arise as a consequence of parent-of-origin effects, although with an underestimation of the effect size due to inaccurate genetic modelling [12]. Family-based studies re-evaluating previously identified susceptibility loci will enable the identification of parent-of-origin effects and help characterize part of the missing heritability in complex traits.
Methods
Ethics statement
The study was carried out in accordance to the approval of the ethics commission of the Charité—Universitätsmedizin Berlin (ref EA2/054/10) and following the guidelines of the declaration of Helsinki. Informed consent was obtained from all probands or their legal guardians.
Subjects
We investigated samples originating from European family-based and population-based studies. All samples were divided, according to the country of origin, into a Central and a Northern European study population (Table 1).
The GENUFAD study (Genetic Analysis of Nuclear Families with Atopic Dermatitis) recruited complete nuclear families with at least two children affected with early-onset (<2 years of age) and moderate to severe AD as previously described [20]. A doctor’s diagnosis of AD was made according to standard criteria [42]. The GENUFAD study contributed 522 complete German nuclear families to the Central European study and 32 Swedish families to the Northern European data set. A large proportion of these families have been reported in previous studies [16,43].
The MAS (Multicenter Allergy Study) is a previously described population-based birth cohort in which 1314 German infants were followed since 1990 to investigate the epidemiology of allergic diseases [44]. The diagnosis of AD was made as previously described [16]. 112 German MAS trios, consisting of a child with AD and both parents, were included in the Central European study.
The ETAC (Early Treatment of the Atopic Child) is a European study which recruited infants diagnosed with AD in their first year of life into a randomized, double blind, placebo controlled trial on the efficacy of cetirizine in the prevention of asthma [45]. Children with early onset and moderate to severe disease were selected for the present study when parental DNA was available. The ETAC study contributed 21 Swedish trios to the Northern European study. Additionally, 125 ETAC trios from different European countries were included in the Central European study (48 from the Netherlands, 23 from Italy, 20 from the UK, 15 from France, 13 from the Czech Republic, and 6 from Germany).
A previous study from Sweden contributed 397 families to the Northern European study group [17]. This included 272 complete affected sib pair families with AD diagnosed according to the U.K. Working Party’s Diagnostic Criteria [46]. The remaining 125 families were incomplete nuclear families including mother-child or father-child pairs.
In all family-based studies, information regarding the parental history of AD was obtained by a questionnaire at the time of family recruitment. The analysis shown in S6 Table was performed in families in which both parents had a negative history of AD. Families in which one or both parents had a positive or unknown disease history were excluded.
As described below, the analytical methods used allowed the incorporation of unrelated individuals to increase power. Thus, the Central European study also included previously published FLG genotypes of 772 unrelated German AD cases and 373 German controls from a previous GWAS [43]. In addition, we genotyped FLG mutations in 375 German children with AD diagnosed at a tertiary care center for pediatric allergy at Charité Universitätsmedizin Berlin. Also, previously published FLG genotype counts of 2,963 population-based German individuals from the International Study of Asthma and Allergies in Childhood II Study [47] were included. Likewise, in our Northern European study population we included genotypes of the 3 most prevalent FLG mutations (c.2282del4, p.R501X and p.R2447X) previously reported in the Swedish population-based BAMSE cohort [22]. The rarest FLG mutation (p.S3247X) was not available in the BAMSE dataset.
Data on specific allergic sensitization was available in a large proportion of mothers from the GENUFAD and MAS studies. Plasma levels of specific IgE against grass and birch pollen, ribwort, cat and dog dander, mold (Cladosporidium herbarum, Alternaria tenuis), hen’s egg, cow’s milk, fish, peanut, and house dust mite were determined using CAP-RAST-FEIA (Pharmacia). A mother was defined as sensitized if specific IgE ≥0.7 kU/l (CAP2) to at least one allergen was detected.
Genotyping of FLG mutations
Genomic DNA was prepared from whole blood by standard methods. The FLG c.2282del4 variant was analyzed with fluorescence-based semiautomated genotyping [16] and the FLG p.R501X, p.R2447X and p.S3247X mutations with Taqman allelic discrimination (Applied Biosystems, Foster City, California, USA) as previously described [4]. Genotyping of p.R2447X in the Northern European families was performed using a fluorescent Kaspar assay (KASP-By-Design genotyping assays, LGC group, Teddington, UK). Genotyping with Taqman and Kaspar was performed using a ViiA 7 Real-Time PCR System (Applied Biosystems, Foster City, California, USA). When analyzing the rare p.R2447X and p.S3247X mutations, a sample known to be a mutation carrier was included on each genotyping plate as a positive control.
The FLG mutations are named according to the nomenclature recommendations by den Dunnen and Antonarakis [48]. The positions of the mutations in the cDNA refer to the A of the ATG-translation initiation codon of NM_002016.1.
Statistical analysis
The PREMIM and EMIM tools were used to test for imprinting and maternal genotype effects of the FLG mutations [18,19]. First, the PREMIM tool was used to classify each trio according to the number of copies of FLG mutations carried by mother, father, and affected child. Incomplete nuclear families, unrelated AD cases, and population-based samples were also included in the analysis to increase the power to detect parent-of-origin effects [18,19]. Since the EMIM analysis is based on the assumption that the genotype frequencies in controls correspond to those in the general population, all population-based controls were included in the analysis irrespectively of disease status. Starting values for allele frequencies of the FLG mutations in the study population (including controls) were estimated with PREMIM (–a option).
The trios were analyzed using the EMIM tool, which uses a multinomial modelling approach to estimate genotype relative risk parameters on the basis of observed counts of genotype combinations in case-parent trios. The following parameters influencing the disease risk in the child were modelled with EMIM:
R1 (R2), the factor by which an individual’s disease risk is multiplied if they carry one (two) risk alleles at a given locus.
S1 (S2), the factor by which an individual’s disease risk is multiplied if the mother carries one (two) risk alleles at that locus.
Im (Ip) the factor by which an individual’s disease risk is multiplied if inheriting a risk allele from the mother (or father).
γ11 (interaction term), the factor by which an individual’s disease risk is multiplied if both mother and child have 1 copy of the risk allele.
Previous data indicated that FLG mutations do not fit an additive genetic model, since the risk of AD in the homozygous carriers is too high. Thus, instead of using the default EMIM settings assuming an additive model we choose to independently estimate the R1 and R2 parameters. Modelling of maternal genotype effects was done using the default additive model. All analyses were performed under the “conditional on exchangeable parental genotype” (CEPG) assumption. This assumption should protect from potential biases in parameter estimation due to the inclusion of families recruited through multiple affected individuals, at the cost of reduced power to detect parent-of-origin effects compared to assuming only Hardy-Weinberg equilibrium [18,19]. A step-by-step analysis was performed by including additional risk parameters in the model as indicated in S9 Table.
Maximum likelihood estimates were obtained from each model and a likelihood ratio test was performed to assess the significance among nested models. Note that it is not possible to directly compare the MCG and Im models in a likelihood ratio test, since they are not nested. However, they can be compared indirectly by comparison to the full model (Table 4).
In order to test for genetic interaction between the child and the maternal genotypes we included and interaction term in the model (γ11). This parameter estimates the factor by which an individual’s disease risk is multiplied if both mother and child have one copy of the risk allele. A likelihood ratio test comparing the Maternal Child Genotype (MCG) and the MCG-Interaction model was then performed (see S10 Table).
A meta-analysis of the results from the Central and Northern European populations was performed using METAL [23]. The inverse variance method was used and the corresponding betas and standard errors were obtained from the EMIM summary file. The meta-analysis was performed on the single risk parameters estimates (R1, R2, S1 or Im) from each model. An analysis of heterogeneity was also performed with METAL in order to evaluate if the observed effect sizes were homogeneous across datasets.
In order to increase power in the main analysis we allowed the inclusion of all affected offspring available in each family which were considered as independent trios (-xa option in PREMIM). This may lead to biased results when using large pedigrees but it is unlikely to have a large effect in our study since most families had only 1 or 2 affected children. In order to exclude this potential bias we repeated the analysis including one affected child per family (omitting the −xa option in PREMIM; S8 Table).
The Transmission Disequilibrium Test (TDT) was performed with PLINK [49]. In order to account for multiple affected offspring within families, empirical p-values were calculated with the —tdt —perm option, which flips the allele transmitted from parent to offspring with 50:50 probability. Allelic effects were calculated with PLINK —assoc using the offspring of the +Mat_Sens as controls and those from the −Mat_Sens as cases.
Haplotype frequencies on the central European Study were calculated with FAMHAP [50], which computes maximum-likelihood estimates obtained with the expectation-maximization algorithm.
Supporting Information
Data from the BioGPS Portal where mRNA levels were quantified with expression arrays in human tissues. [25,26].
(TIF)
Data from the GTEx Consortium where mRNA was quantified by Next generation Sequencing. [27]
(TIF)
Only high quality mutations were considered (filter = pass). a Human genome build GRCh37.p13 (chromosome 1). b Alias used in this manuscript. c Allele frequency in the combined set of 61,486 unrelated individuals. d Alternative allele count of genotypes in the combined population e Total number of called genotypes in the combined population. Data was accessed on December 12, 2014.
(XLSB)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOC)
(DOCX)
(DOCX)
Acknowledgments
We thank all individuals and families for their participation in this study. We thank all physicians and nurses involved in patient recruitment for their valuable contribution to the study. We are grateful to C. Flachmeier and S. Kolberg for excellent technical support.
The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by Deutsche Forschungsgemeinschaft (http://www.dfg.de/). HJC is supported by a Senior Research Fellowship in Basic Biomedical Science from the Wellcome Trust (Grant Reference 087436; http://www.wellcome.ac.uk/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brown SJ, McLean WH (2012) One remarkable molecule: filaggrin. J Invest Dermatol 132: 751–762. 10.1038/jid.2011.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE et al. (2006) Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet 38: 337–342. [DOI] [PubMed] [Google Scholar]
- 3. Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H et al. (2006) Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet 38: 441–446. [DOI] [PubMed] [Google Scholar]
- 4. Sandilands A, Terron-Kwiatkowski A, Hull PR, O'Regan GM, Clayton TH et al. (2007) Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet 39: 650–654. [DOI] [PubMed] [Google Scholar]
- 5. Brown SJ, Asai Y, Cordell HJ, Campbell LE, Zhao Y et al. (2011) Loss-of-function variants in the filaggrin gene are a significant risk factor for peanut allergy. J Allergy Clin Immunol 127: 661–667. 10.1016/j.jaci.2011.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fallon PG, Sasaki T, Sandilands A, Campbell LE, Saunders SP et al. (2009) A homozygous frameshift mutation in the mouse Flg gene facilitates enhanced percutaneous allergen priming. Nat Genet 41: 602–608. 10.1038/ng.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawasaki H, Nagao K, Kubo A, Hata T, Shimizu A et al. (2012) Altered stratum corneum barrier and enhanced percutaneous immune responses in filaggrin-null mice. J Allergy Clin Immunol 129: 1538–1546. 10.1016/j.jaci.2012.01.068 [DOI] [PubMed] [Google Scholar]
- 8. Lim RH, Kobzik L, Dahl M (2010) Risk for asthma in offspring of asthmatic mothers versus fathers: a meta-analysis. PLoS One 5: e10134 10.1371/journal.pone.0010134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldberg M, Eisenberg E, Elizur A, Rajuan N, Rachmiel M et al. (2013) Role of parental atopy in cow's milk allergy: a population-based study. Ann Allergy Asthma Immunol 110: 279–283. 10.1016/j.anai.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 10. Bisgaard H, Halkjaer LB, Hinge R, Giwercman C, Palmer C et al. (2009) Risk analysis of early childhood eczema. J Allergy Clin Immunol 123: 1355–1360. 10.1016/j.jaci.2009.03.046 [DOI] [PubMed] [Google Scholar]
- 11. Wadonda-Kabondo N, Sterne JA, Golding J, Kennedy CT, Archer CB et al. (2004) Association of parental eczema, hayfever, and asthma with atopic dermatitis in infancy: birth cohort study. Arch Dis Child 89: 917–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P et al. (2009) Parental origin of sequence variants associated with complex diseases. Nature 462: 868–874. 10.1038/nature08625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lawson HA, Cheverud JM, Wolf JB (2013) Genomic imprinting and parent-of-origin effects on complex traits. Nat Rev Genet 14: 609–617. 10.1038/nrg3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hager R, Cheverud JM, Wolf JB (2008) Maternal effects as the cause of parent-of-origin effects that mimic genomic imprinting. Genetics 178: 1755–1762. 10.1534/genetics.107.080697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Exome Aggregation Consortium (ExAC), Cambridge, MA (URL: http://exac.broadinstitute.org) [date (December, 2014) accessed].
- 16. Marenholz I, Nickel R, Ruschendorf F, Schulz F, Esparza-Gordillo J et al. (2006) Filaggrin loss-of-function mutations predispose to phenotypes involved in the atopic march. J Allergy Clin Immunol 118: 866–871. [DOI] [PubMed] [Google Scholar]
- 17. Ekelund E, Lieden A, Link J, Lee SP, d'Amato M et al. (2008) Loss-of-function variants of the filaggrin gene are associated with atopic eczema and associated phenotypes in Swedish families. Acta Derm Venereol 88: 15–19. 10.2340/00015555-0383 [DOI] [PubMed] [Google Scholar]
- 18. Howey R, Cordell HJ (2012) PREMIM and EMIM: tools for estimation of maternal, imprinting and interaction effects using multinomial modelling. BMC Bioinformatics 13: 149 10.1186/1471-2105-13-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ainsworth HF, Unwin J, Jamison DL, Cordell HJ (2011) Investigation of maternal effects, maternal-fetal interactions and parent-of-origin effects (imprinting), using mothers and their offspring. Genet Epidemiol 35: 19–45. 10.1002/gepi.20547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee YA, Wahn U, Kehrt R, Tarani L, Businco L et al. (2000) A major susceptibility locus for atopic dermatitis maps to chromosome 3q21. Nat Genet 26: 470–473. [DOI] [PubMed] [Google Scholar]
- 21. Muller S, Marenholz I, Lee YA, Sengler C, Zitnik SE et al. (2009) Association of Filaggrin loss-of-function-mutations with atopic dermatitis and asthma in the Early Treatment of the Atopic Child (ETAC) population. Pediatr Allergy Immunol 20: 358–361. 10.1111/j.1399-3038.2008.00808.x [DOI] [PubMed] [Google Scholar]
- 22. Ballardini N, Kull I, Soderhall C, Lilja G, Wickman M et al. (2013) Eczema severity in preadolescent children and its relation to sex, filaggrin mutations, asthma, rhinitis, aggravating factors and topical treatment: a report from the BAMSE birth cohort. Br J Dermatol 168: 588–594. 10.1111/bjd.12196 [DOI] [PubMed] [Google Scholar]
- 23. Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Case A, Paxson C (2001) Mothers and others: who invests in children's health? J Health Econ 20: 301–328. [DOI] [PubMed] [Google Scholar]
- 25. Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA et al. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A 101: 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu C, Orozco C, Boyer J, Leglise M, Goodale J et al. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10: R130 10.1186/gb-2009-10-11-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. GTEx Consortium. (2013) The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van den Oord RA, Sheikh A (2009) Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: systematic review and meta-analysis. BMJ 339: b2433 10.1136/bmj.b2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu FT, Goodarzi H, Chen HY (2011) IgE, mast cells, and eosinophils in atopic dermatitis. Clin Rev Allergy Immunol 41: 298–310. 10.1007/s12016-011-8252-4 [DOI] [PubMed] [Google Scholar]
- 30. Hamada K, Suzaki Y, Goldman A, Ning YY, Goldsmith C et al. (2003) Allergen-independent maternal transmission of asthma susceptibility. J Immunol 170: 1683–1689. [DOI] [PubMed] [Google Scholar]
- 31. Fedulov AV, Kobzik L (2011) Allergy risk is mediated by dendritic cells with congenital epigenetic changes. Am J Respir Cell Mol Biol 44: 285–292. 10.1165/rcmb.2009-0400OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Straubinger K, Paul S, Prazeres da CO, Ritter M, Buch T et al. (2014) Maternal immune response to helminth infection during pregnancy determines offspring susceptibility to allergic airway inflammation. J Allergy Clin Immunol. [DOI] [PubMed]
- 33. von ME, Vercelli D (2010) Farm living: effects on childhood asthma and allergy. Nat Rev Immunol 10: 861–868. 10.1038/nri2871 [DOI] [PubMed] [Google Scholar]
- 34. Brand S, Teich R, Dicke T, Harb H, Yildirim AO et al. (2011) Epigenetic regulation in murine offspring as a novel mechanism for transmaternal asthma protection induced by microbes. J Allergy Clin Immunol 128: 618–625. 10.1016/j.jaci.2011.04.035 [DOI] [PubMed] [Google Scholar]
- 35. Mott R, Yuan W, Kaisaki P, Gan X, Cleak J et al. (2014) The architecture of parent-of-origin effects in mice. Cell 156: 332–342. 10.1016/j.cell.2013.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shirakawa T, Li A, Dubowitz M, Dekker JW, Shaw AE et al. (1994) Association between atopy and variants of the beta subunit of the high- affinity immunoglobulin E receptor. Nat Genet 7: 125–129. [DOI] [PubMed] [Google Scholar]
- 37. Walley AJ, Chavanas S, Moffatt MF, Esnouf RM, Ubhi B et al. (2001) Gene polymorphism in Netherton and common atopic disease. Nat Genet 29: 175–178. [DOI] [PubMed] [Google Scholar]
- 38. Soderhall C, Marenholz I, Kerscher T, Ruschendorf F, Esparza-Gordillo J et al. (2007) Variants in a novel epidermal collagen gene (COL29A1) are associated with atopic dermatitis. PLoS Biol 5: e242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gleason G, Liu B, Bruening S, Zupan B, Auerbach A et al. (2010) The serotonin1A receptor gene as a genetic and prenatal maternal environmental factor in anxiety. Proc Natl Acad Sci U S A 107: 7592–7597. 10.1073/pnas.0914805107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yan L, Zhao L, Long Y, Zou P, Ji G et al. (2012) Association of the maternal MTHFR C677T polymorphism with susceptibility to neural tube defects in offsprings: evidence from 25 case-control studies. PLoS One 7: e41689 10.1371/journal.pone.0041689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordeeva LA, Voronina EN, Sokolova EA, Ermolenko NA, Gareeva JV et al. (2013) Association GSTT1, GSTM1 and GSTP1 (Ile105Val) genetic polymorphisms in mothers with risk of congenital malformations in their children in Western Siberia: a case-control study. Prenat Diagn 1–7. [DOI] [PubMed]
- 42. Hanifin JM, Rajka G (1980) Diagnostic Features of Atopic Dermatitis. Acta Derm (Stockholm) 92 (Suppl.): 44–47. [Google Scholar]
- 43. Esparza-Gordillo J, Weidinger S, Folster-Holst R, Bauerfeind A, Ruschendorf F et al. (2009) A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet 41: 596–601. 10.1038/ng.347 [DOI] [PubMed] [Google Scholar]
- 44. Lau S, Illi S, Sommerfeld C, Niggemann B, Bergmann R et al. (2000) Early exposure to house-dust mite and cat allergens and development of childhood asthma: a cohort study. Multicentre Allergy Study Group. Lancet 356: 1392–1397. [DOI] [PubMed] [Google Scholar]
- 45. Warner JO (2001) A double-blinded, randomized, placebo-controlled trial of cetirizine in preventing the onset of asthma in children with atopic dermatitis: 18 months' treatment and 18 months' posttreatment follow-up. J Allergy Clin Immunol 108: 929–937. [DOI] [PubMed] [Google Scholar]
- 46. Williams HC, Burney PG, Hay RJ, Archer CB, Shipley MJ et al. (1994) The U.K. Working Party's Diagnostic Criteria for Atopic Dermatitis. I. Derivation of a minimum set of discriminators for atopic dermatitis. Br J Dermatol 131: 383–396. [DOI] [PubMed] [Google Scholar]
- 47. Weidinger S, O'Sullivan M, Illig T, Baurecht H, Depner M et al. (2008) Filaggrin mutations, atopic eczema, hay fever, and asthma in children. J Allergy Clin Immunol 121: 1203–1209. 10.1016/j.jaci.2008.02.014 [DOI] [PubMed] [Google Scholar]
- 48. Den Dunnen JT, Antonarakis SE (2001) Nomenclature for the description of human sequence variations. Hum Genet 109: 121–124. [DOI] [PubMed] [Google Scholar]
- 49. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Herold C, Becker T (2009) Genetic association analysis with FAMHAP: a major program update. Bioinformatics 25: 134–136. 10.1093/bioinformatics/btn581 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data from the BioGPS Portal where mRNA levels were quantified with expression arrays in human tissues. [25,26].
(TIF)
Data from the GTEx Consortium where mRNA was quantified by Next generation Sequencing. [27]
(TIF)
Only high quality mutations were considered (filter = pass). a Human genome build GRCh37.p13 (chromosome 1). b Alias used in this manuscript. c Allele frequency in the combined set of 61,486 unrelated individuals. d Alternative allele count of genotypes in the combined population e Total number of called genotypes in the combined population. Data was accessed on December 12, 2014.
(XLSB)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOC)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.