Summary
Huntington's Disease (HD) is caused by a polyglutamine tract expansion in huntingtin (HTT). Despite HTTs ubiquitous expression, there is early and robust vulnerability in striatum, the cause of which is poorly understood. Here, we provide evidence that impaired striatal mTORC1 activity underlies varied metabolic and degenerative phenotypes in HD brain and show that introducing the constitutively active form of the mTORC1 regulator, Rheb, into HD mouse brain, alleviates mitochondrial dysfunction, aberrant cholesterol homeostasis, striatal atrophy, impaired dopamine signaling, and increases autophagy. We also find that the expression of Rhes, a striatum-enriched mTOR activator, is reduced in HD patient and mouse brain, and that exogenous addition of Rhes alleviates motor deficits and improves brain pathology in HD mice. Our combined work indicates that impaired Rhes/mTORC1 activity in HD brain may underlie the notable striatal susceptibility and thus presents a promising therapeutic target for HD therapy.
Huntington's Disease (HD) is a fatal autosomal-dominant neurodegenerative disease caused by CAG repeat expansion in exon 1 of huntingtin, which encodes the protein huntingtin (HTT)(1993). Despite HTT expression in all tissues and brain regions, the striatum demonstrates the most profound and early degeneration. Mutant HTT (mHTT) negatively affects multiple cellular pathways, including mitochondria biogenesis (Cui et al., 2006; Tsunemi et al., 2012), cholesterol homeostasis (Karasinska and Hayden, 2011; Valenza and Cattaneo, 2011; Valenza et al., 2005), axonal growth (Li et al., 2001), and synaptogenesis (Milnerwood and Raymond, 2010), all of which may contribute to neuronal dysfunction and loss. These varied phenotypes may result from disruption of a core component regulating a range of fundamental biological processes. Identifying such a primary pathogenic event would facilitate therapeutic development.
Mechanistic target of rapamycin (mTOR) is a serine-threonine kinase that integrates signals to regulate cell growth and metabolism (Laplante and Sabatini, 2012). mTOR forms 2 distinct complexes, mTORC1 and mTORC2. Under nutrient-rich conditions, mTORC1 is activated and promotes protein translation and cell growth. In contrast, nutrient withdrawal inactivates mTORC1 and initiates macroautophagy (hereafter referred to as autophagy) as a cell survival mechanism. mTORC1 positively controls mitochondrial biogenesis (Cunningham et al., 2007) and regulates lipid homeostasis by controlling cholesterol synthesis (Peterson et al., 2011; Porstmann et al., 2008). Moreover, in the brain, mTORC1 promotes myelination, axon growth, and regeneration (Kim et al., 2012; Park et al., 2008; Sun et al., 2011), and genetic deletion of the Ras homologue enriched in brain (Rheb), a key activator of mTORC1, causes decreased cortical thickness and defective myelination (Zou et al., 2011).
In the striatum Rhes (Ras homolog enriched in the striatum) serves as a key activator of mTORC1 (Subramaniam et al., 2012). Genetic knockout of Rhes reduces mTORC1 activity, and attenuates adverse responses to L-DOPA induced dyskinesia (Subramaniam et al., 2012). Additionally, Rhes facilitates SUMOylation (Subramaniam et al., 2009), a process implicated in HD pathogenesis (Steffan et al., 2004). In vitro, Rhes promotes SUMOylation of mHTT to induce cytotoxicity (Subramaniam et al., 2009), and reducing exogenously added Rhes with siRNAs increased survival of cells transfected with mHTT fragments (Lu and Palacino, 2013; Seredenina et al., 2011). In vivo, genetic ablation of Rhes protects against neurotoxin-induced striatal lesions (Mealer et al., 2013), and transiently delays motor symptom onset in R6/1 HD mice (Baiamonte et al., 2013). Because Rhes is highly expressed in the striatum (Spano et al., 2004), it has been proposed that Rhes-mHTT interactions may underlie the prominent striatal degeneration in HD.
However, other data call to question a pathogenic role for Rhes in HD. Rhes levels are reduced in HD patient caudate nucleus (Hodges et al., 2006), and Rhes ablation in R6/1 HD models does not prevent brain degeneration (Baiamonte et al., 2013). Furthermore, Rhes KO mice develop brain atrophy and behavioral abnormalities resembling those found in HD mice (Baiamonte et al., 2013; Spano et al., 2004). In addition, Rhes promotes autophagy (Mealer et al., 2014), a well-established protective mechanism in HD. Therefore, given the critical function of Rhes in mediating striatal mTORC1 signaling, we hypothesized that a concomitant loss of Rhes and mTORC1 activity contributes to the early striatal pathology in HD, and that enhancing mTORC1 function, through upregulation of Rheb or Rhes, would be neuroprotective.
To test this hypothesis we acutely modulated Rhes and mTORC1 activity in adult striata of HD transgenic mice. We found beneficial effects with mTORC1 activation, including improved mHTT-associated metabolic phenotypes and reversal of striatal atrophy. Consistently, restoring mTORC1 activity or Rhes levels in vivo was protective. Further, we showed that the neuroprotective property of Rhes is dependent on its GTPase activity, which is required for activating mTORC1 but independent of SUMOylation related activity. Collectively, these data suggest that impaired Rhes/mTORC1 activity is relevant to the notable striatal pathogenesis in HD and suggest that impaired mTORC1 function may represent a fundamental mechanism underlying the complex disease phenotypes in HD.
Results
mTORC1 activity is reduced in the striatum of HD
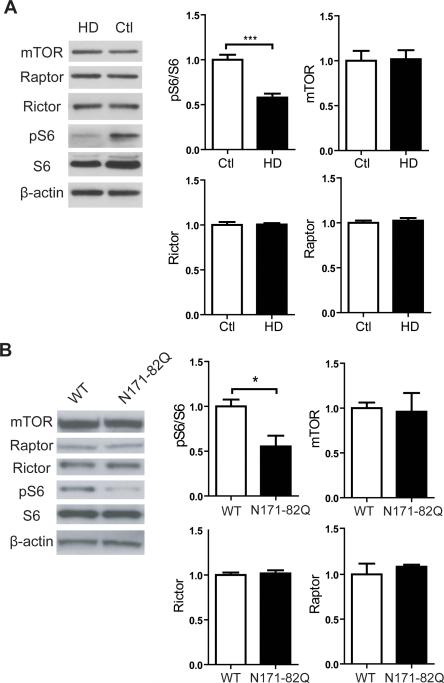
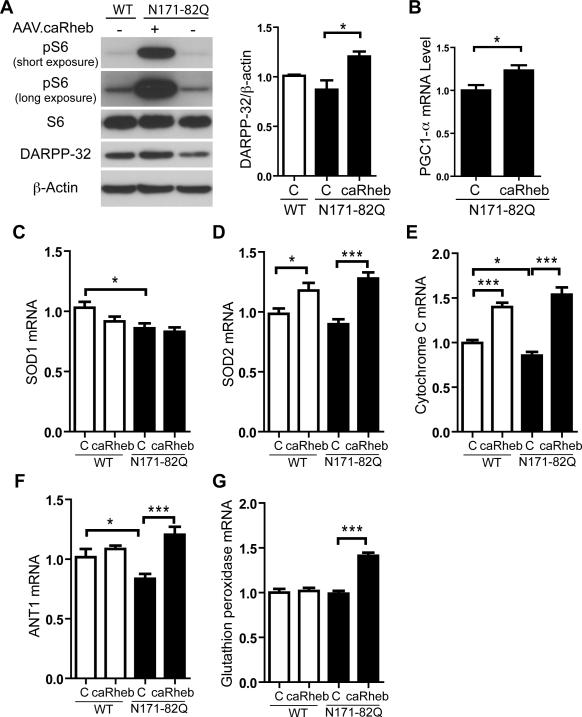
As a first test of our hypothesis, we examined mTORC1 activity in HD brain. We found that mTORC1 activity is reduced in striatal tissues from HD patients and N171-82Q mice as detected by reduced phosphorylation of ribosomal protein S6 (pS6), an established marker of mTORC1 activity (Figure 1A; Figures S1A,B). In HD mice, mTORC1 activity is impaired at 6 weeks of age, prior to the onset of neurological symptoms (Figure 1B). The reduced mTORC1 activity is not associated with reduced expression of mTOR, or components of the mTOR complex (e.g. Rictor and Raptor)(Figures 1A,B; Figure S1A). We next engineered adeno-associated viruses (AAVs), which transduce striatal neurons (Figure S2A) (McBride et al., 2008), to express Rheb, a well-established positive regulator of mTORC1 (Laplante and Sabatini, 2012). In this manner, we can acutely enhance mTORC1 activation in vivo. We used a constitutively active Rheb mutant (caRheb; S16H) previously shown to activate mTORC1 signaling in mouse brain (Kim et al., 2012; Zou et al., 2011). Unilateral injection of AAV.caRheb into N171-82Q mouse striata markedly induced mTORC1 activity as indicated by increased pS6 at 3 weeks post-injection (Figure 2A). Selective reduction of DARPP-32 levels, a fundamental component of dopamine signaling in medium spiny neurons (MSN), is an indicator of pathological progression in HD mouse and human brains (Bibb et al., 2000; Hodges et al., 2006). AAV.caRheb transduction promoted a 33% increase of DARPP-32 levels in N171-82Q mouse striata compared to contralateral uninjected tissue (Figure 2A). Moreover, AAV.caRheb upregulated expression of the mitochondrial-transcriptional regulator, PPARγ coactivator 1α (PGC1-α) (Figure 2B), a master regulator of mitochondrial biogenesis that when increased improves mitochondrial function and motor deficits in HD models (Cui et al., 2006; Tsunemi et al., 2012). Consistent with increased mitochondria activity, the levels of reactive oxygen species detoxifying (ROS) genes, SOD2, cyt-c, and ANT1 were also increased in AAV.caRheb treated brains (Figures 2C-F). Glutathione peroxidase, a recently identified neuroprotective antioxidant enzyme in HD model organisms (Mason et al., 2013), selectively increased in HD brains after caRheb transduction (Figure 2G). The improved profile could not be explained by an indirect effect of virus injection or downregulation of the mHTT transgene; similar results were obtained when AAV.caRheb injected brains were compared to AAV.eGFP treated brains, and caRheb did not affect mHTT transgene expression (Figures S2B-H).
Figure 1. mTORC1 activity is reduced in HD human and mouse striatum.
(A) Immunoblotting of mTOR, Rictor, Raptor, and mTORC1 activity (pS6) showed reduced mTORC1 activity in the striatum of patients with HD (N=10; see Fig S1A for expanded series) compared to unaffected individuals (N=6). β-actin was used as a loading control. Data represent mean ± SEM. ***P<0.001, Student's t-test. (B) Immunoblotting of mTOR, Rictor, Raptor, and pS6 from 6-week-old N171-82Q and WT mouse striatal lysates. β-actin was used as a loading control. Densitometry analyses for panels A and B revealed reduced pS6 levels in HD patient (N=10) vs. controls (N=6), and 6-week-old N171-82Q mice (N=4) compared to age- and sex- matched WT littermates (N=4). Data represent mean ± SEM.*P<0.05; ***P<0.001, Student's t-test.
Figure 2. Activation of mTORC1 pathway corrects metabolism-related deficits in HD mice.
(A) Western blot shows increased pS6 and DARPP-32 immunoreactivity in N171-82Q mice treated with unilateral injection of AAV.caRheb, compared to untreated. Mice were injected at 7 weeks of age and striatal lysates harvested at 10 weeks of age. Right, densitometry quantification of DARPP-32 immunoreactivity (N=4 mice per group; *P<0.05, One-way ANOVA with Tukey's post-hoc test). (B) RT-qPCR analysis of PGC1-α in striatal homogenates from 10-week-old N171-82Q mice treated with unilateral injection of AAV.caRheb at 7 weeks of age. Striatal lysates from uninjected contralateral hemispheres served as internal controls (N=8 per group). (C-G) RT-qPCR analysis of ROS detoxifying genes in striatal homogenates from 10-week-old N171-82Q and WT mice after unilateral injection of AAV.caRheb at 7 weeks of age. Lysates from uninjected contralateral hemispheres served as internal controls (N=8 per group). All genes were normalized to endogenous β-actin. Data represent mean ± SEM. C=Control. *P<0.05, **P<0.01, ***P<0.001, Student's t-test.
We next performed in vitro studies and treated striatal cells that express expanded (Q111) and normal (Q7) Htt (Trettel et al., 2000) with an ATP-competitive inhibitor of mTOR, Torin1. Torin1 potently inhibits mTOR functions including those that are resistant to inhibition by rapamycin (Thoreen et al., 2009). Torin1 treatment reduced pS6 and p-4E-BP1 levels as well as DARPP-32 expression in Q7 and Q111 cells (Figure S3A). Torin1 also repressed PGC1-α, the PGC1-α-regulated genes cAMP response element-binding protein (CREB), and transducer of regulated CREB 1 (TORC1) (Figures S3A, B). Together, our in vitro and in vivo results indicate that mTORC1 regulates PGC1-α and PGC1-α regulated metabolic genes in the setting of WT and mutant HTT alleles.
mTORC1 controls lipogenic gene expression in HD brains
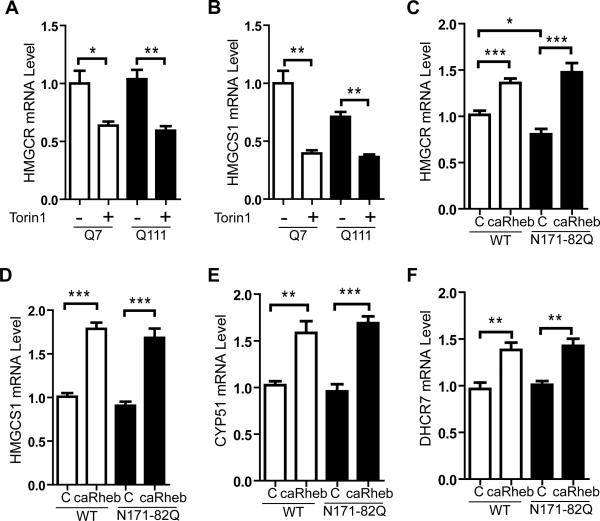
We next determined whether impaired mTORC1 activity underlies other HD-specific metabolic pathway alterations. Aberrant cholesterol biosynthesis has been observed in HD mouse and human brains (Karasinska and Hayden, 2011; Valenza and Cattaneo, 2011). The expression of genes required for lipid biosynthesis is controlled by nuclear transactivation of sterol regulatory element-binding proteins (SREBPs). mHTT has been reported to inhibit nuclear translocation of SREBP in cells and HD mice, which may contribute to dysfunctional cholesterol synthesis (Valenza et al., 2005). mTORC1 also regulates SREBP transactivation (Peterson et al., 2011; Porstmann et al., 2008). To determine if reducing mTORC1 activity alters cholesterol synthesis in the setting of HD, we applied Torin1 to cultured Q7 or Q111 striatal cells. Torin1 reduced expression of the SREBP target genes HMG-CoA reductase (HMGCR), the rate-limiting enzyme in cholesterol synthesis and HMGCS1 in both mutant and normal cell lines (Figures 3A, B).
Figure 3. mTOR regulates cholesterol biosynthetic pathway genes in HD mice striata.
(A and B) WT (Q7) and mutant (Q111) striatal cells grown in normal media were treated with 250 nM Torin1 or DMSO (control). Total cell extracts were obtained 24 hours after treatment. Gene expression levels of HMGCR and HMGCS1 were normalized to endogenous β-actin. (C-F) RT-qPCR analysis of lipogenic genes from striatal homogenates of 10-week-old N171-82Q and WT mice after unilateral injection of AAV.caRheb at 7 weeks of age. Lysates from uninjected contralateral hemispheres served as internal controls (N=8 per group). All genes were normalized to endogenous β-actin. Data represent mean ± SEM. C=Control. *P<0.05, **P<0.01, ***P<0.001, Student's t-test.
In N171-82Q mice, HMGCR is significantly reduced (Figure 3C) compared to controls, similar to that found in HD patient brains (Hodges et al., 2006). AAV.caRheb normalized HMGCR levels, and also increased expression sterol biosynthesic enzymes: HMG-CoA synthase 1 (HMGCS1), lanosterol 14α-demethylase (CYP51), and 7-dehydrocholesterol reductase (DHCR7) (Figures 3C-F; Figures S3 C-F). Similar to our in vitro findings, mTOR's regulation of striatal lipogenic gene expression is independent of the state of polyglutamine expansion in HTT, as AAV.caRheb also increased expression of these genes in WT striata as well (Figures 3C-3F). Although the reduction of HMGCR gene expression is mild in N171-82Q mice (transgene expressed from prion promoter) reduced expression of HMGCR, HMGCS1, DHCR7, and CYP51 are observed in HD human and other mouse models (Hodges et al., 2006; Valenza et al., 2005). Together, our results indicate that mTORC1 promotes lipogenic gene expression in the striatum, and suggest that impaired mTORC1 activity may contribute to reduced lipogenic gene expression in HD.
mTORC1 enhances pathways implicated in mHTT clearance
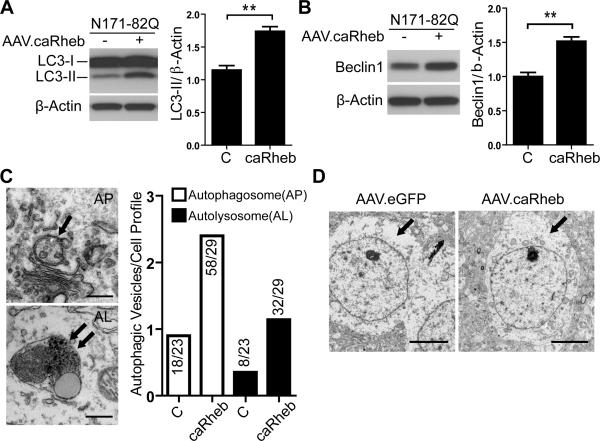
mTORC1 activity is generally associated with antagonizing autophagy induction, although autophagy can occur through mTORC1 dependent and independent pathways. With relevance to HD, basal autophagic activity is critical for maintaining neuronal survival and promoting aggregate clearance (Jeong et al., 2009; Ravikumar et al., 2004; Yamamoto et al., 2006). As such, we next examined the effects of mTORC1 activation on autophagy in AAV.caRheb treated HD mouse striata. We found increased basal levels of the autophagosome-membrane-associated protein, LC3-II in N171-82Q mice compared to WT mice (Figure S4). AAV.caRheb treatment increased levels of LC3-II (Figure 4A) and Beclin-1, a protein necessary for autophagy induction (Figure 4B). To distinguish between enhanced autophagy and impairment of autophagolysosomal maturation, electron microscopy (EM) was used to quantify the relative number of autophagosomes and autolysosomes in brains of mice treated with AAV.caRheb in one hemisphere or AAV.eGFP in the other hemisphere. While the autophagosomes/autolysosome ratio was similar between hemispheres, both autophagosomes and autolysosomes were increased with AAV.caRheb treatment compared to the control treated contralateral sides (Figure 4C), suggesting that autophagy is activated in the setting of increased mTORC1 activity in vivo. We also noted that control treated (AAV.eGFP) N171-82Q mice brain was depleted of electron-dense endoplasm granular mass and cytoplasmic organelles (Figure 4D), a phenotype similar to the striatal pathology induced by nucleolar stress (Kreiner et al., 2013). Conversely, cells in AAV.caRheb treated hemispheres showed enriched electron-dense cytoplasmic organelles.
Figure 4. AAV.caRheb increases basal autophagy in N171-82Q mice.
(A and B) Biochemical assessment of autophagy (LC3 and Beclin1) in striatal lysates from N171-82Q mice after unilateral injection of AAV.caRheb at 7 weeks of age. Striata were harvested 3 weeks post-injection. Uninjected contralateral striata serve as internal controls (N=6 mice per group). Representative western blots and densitometry analysis revealed increased LC3II and Beclin1 levels in AAV.caRheb-treated N171-82Q mice striata. β-actin was used as a loading control. Data are mean ± SEM.**P<0.01; Student's t-test. (C) Left panels: representative photomicrographs of an autophagosome (arrow) and an autolysosome (double arrow). Scale bar is 0.5 μm. Right panel: frequency of autophagosomes and autolysosomes detected in striatal neurons from AAV.eGFP and AAV.caRheb treated N171-82Q mice (3 mice examined per group; 7-10 cells/hemisphere/animal). Ratio indicates number of autophagosome or autolysosome over total number of cells counted. (D) Ultrastructural TEM analysis of striatal sections from N171-82Q mice after AAV.eGFP or AAV.caRheb injections into opposite hemispheres (N=3 mice per group). Left panel: a representative photomicrograph of striatal neurons from control treated striata shows electron-lucent cytoplasm, devoid of endoplasm granular mass and organelles. Right panel: AAV.caRheb treated neurons show normal cytoplasm enriched with organelles and endoplasm granular mass. Scale bar is 5 μm.
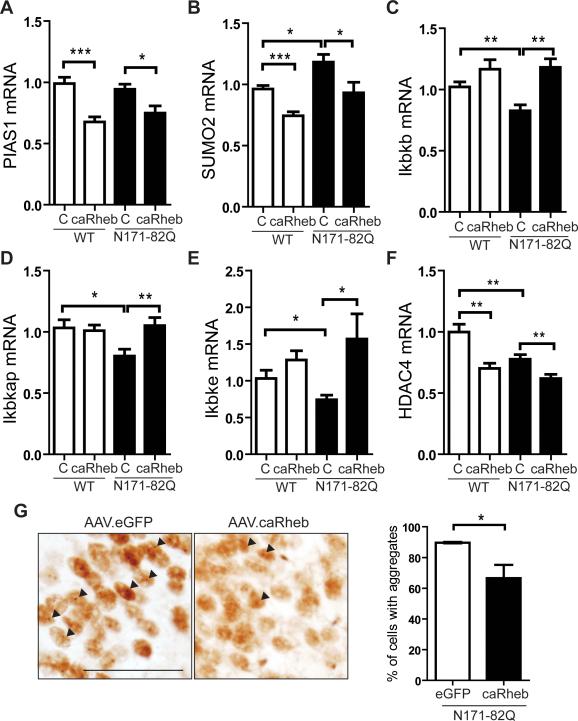
Recent work suggests that autophagic/proteasomal degradation of mHTT occurs through post-translational modifications (PTM) (Jeong et al., 2009; Jia et al., 2012; O'Rourke et al., 2013; Thompson et al., 2009). For example, the SUMO E3 ligase PIAS1 increases mHTT aggregate formation by promoting SUMO2 conjugation to mHTT in cell models, and reducing the PIAS1 ortholog in Drosophila expressing the mHTT protein is protective (O'Rourke et al., 2013). We found that SUMO2 levels were elevated in 14-week-old N171-82Q mouse striatum and that AAV.caRheb repressed PIAS1 and SUMO2 expression in both N171-82Q mice and WT control littermates (Figures 5A, B). We also examined IκB kinase (IKK), a kinase that induces mHTT degradation and targets mHTT for clearance by the proteasome and lysosome (Thompson et al., 2009). Endogenous IKK related genes (Ikbkb, Ikbkap, and Ikbke) were reduced in N171-82Q mouse striata, and AAV.caRheb restored their expression (Figures 5C-E). Additionally, HDAC4 is an important regulator of mHTT aggregate formation and reduction of HDAC4 reduces cytoplasmic aggregates, improves neuronal function, and extends the life span of HD mice (Mielcarek et al., 2013). Here, we found that AAV.caRheb transduction reduced HDAC4 expression in N171-82Q mouse striata (Figure 5F). HTT fragment-containing aggregates are few and inconsistent in striatum, but robust in hippocampus in N171-82Q mice. Therefore, we tested aggregate burden in the hippocampus. AAV efficiently transduces hippocampal neurons (Figure S5), and AAV.caRheb injected animals showed significantly reduced mHTT aggregate load compared to AAV.eGFP injected mice (Figure 5G), consistent with the observed effects of AAV.caRheb on genes involved in mHTT clearance.
Figure 5. mTORC1 regulates genes involved in mHTT aggregate clearance.
(A-F) RT-qPCR analysis of genes from striatal homogenates of 10-week-old N171-82Q and WT mice after unilateral injection of AAV.caRheb at 7 weeks of age. The uninjected contralateral side served as an internal control (N=8 per group). (A and B) Expression of genes implicated in the SUMO modification of mHTT. (C-E) Expression of IKK related genes (Ikbkb, Ikbkap, and Ikbke). (F) Expression of HDAC4. All genes were normalized to endogenous β-actin. Data represent mean ± SEM. C=Control. *P<0.05, **P<0.01, ***P<0.001, Student's t-test.(G) Left panels, immunohistochemical staining of N171-82Q mouse striatum with EM48 2 weeks after a single unilateral injection of AAV.caRheb and AAV.eGFP into the hippocampus of 10-week-old N171-82Q mice (N=4 per treatment group; Scale bar: 50 μm). Right panel, AAV.caRheb reduced EM48-positive aggregates. Arrows: Em48 positive aggregates. Data represent mean ± SEM. *P<0.05. Student's t-test.
Enhanced mTORC1 activity rescues striatal atrophy and improves motor phenotypes in HD mice
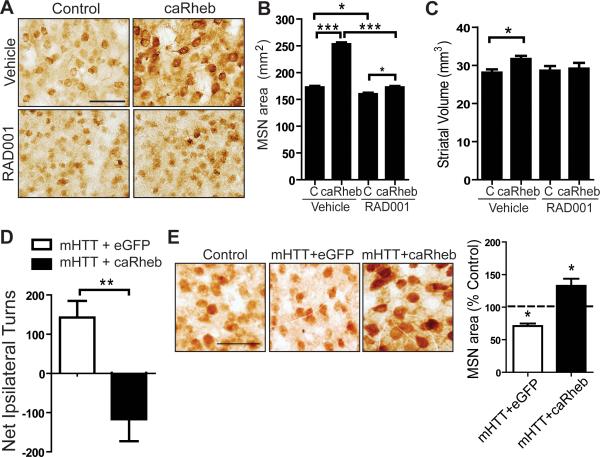
N171-82Q mice display pronounced striatal atrophy with preserved MSN numbers (Cheng et al., 2011; Schilling et al., 1999). To examine how mTORC1 activity impacts striatal volume in diseased brain, we performed unilateral AAV.caRheb injection into the striata of 11-week-old N171-82Q mice, an age by which there is measurable striatal atrophy (Cheng et al., 2011), and compared the treated hemisphere with the untreated side. AAV.caRheb positively impacted MSN cell size and striatal volume compared to untreated contralateral hemispheres (Figures 6A-C). To confirm that caRheb acts through mTORC1, we administered RAD001, a known mTORC1 inhibitor (Fox et al., 2010). RAD001 treatment reversed the morphological effects of caRheb and attenuated S6 phosphorylation (Figures 6A-6C; Figures S6A, B). Additionally, we found that 2 weeks of RAD001 treatment in the absence of caRheb caused a mild but significant exacerbation of MSN atrophy in N171-82Q mice (Figures 6A, B). These results suggest that AAV.caRheb promotes MSN growth or preserves MSN size through the mTORC1 pathway, and that impairing mTORC1 in vivo exacerbates this phenotype.
Figure 6. mTORC1 promotes MSN growth and counteracts mHTT-induced motor phenotypes.
(A) Immunohistochemical staining of MSNs with anti-DARPP-32, 2 weeks after a single unilateral injection of AAV.caRheb into the striatum of 11-week-old N171-82Q mice. Mice were given vehicle or RAD001 for 2 weeks. (B and C) Quantification of MSN cell area and striatal volumes from tissues of animals treated as in (A) (N=4 per treatment group; mean ± SEM). (D) WT mice injected unilaterally with AAV.mHTT/AAV.eGFP or AAV.mHTT/AAV.caRheb at 6 weeks of age. When tested 4 weeks post-injection, AAV.mHTT/AAV.eGFP induced ipsilateral rotational behavior in response to amphetamine. Co-injection of AAV.caRheb altered AAV.mHTT-induced ipsilateral rotation following amphetamine administration, with mice displaying contralateral rotation (N=4 per treatment group). *P<0.05, **P<0.001, ***P<0.001, Student's t-test. Scale bars: 100 μm (A-D). (E) Left panels, Immunohistochemical staining of MSNs with anti-DARPP-32 of sections from AAV.mHTT/AAV.eGFP or AAV.mHTT/AAV.caRheb injected mice from (D). The contralateral uninjected hemisphere is shown as control. Right panels, quantification of MSN area expressed as percentage of uninjected contralateral striata.*P<0.05, Student's t-test. Scale bars: 50 μm.
Impaired striatal dopamine circuitry is associated with motor symptoms in HD mice (Bibb et al., 2000; Johnson et al., 2006). Therefore, to determine the functional impact of AAV.caRheb on dopamine pathways we unilaterally injected N171-82Q mice and subjected them to amphetamine induced rotational tests. Amphetamine triggers dopamine release and induces rotational behavior in mice if there is imbalanced dopamine signaling in one hemisphere versus the other. For this, we co-injected AAV expressing mHTT (AAV.mHTT) into 6-week-old WT mice with either AAV.caRheb or control virus (AAV.eGFP). Unilateral injection of AAV.mHTT/AAV.eGFP caused ipsilateral rotation toward the injected side, an indication of mHTT toxicity (Figure 6D). In contrast, co-injection of AAV.caRheb with AAV.mHTT did not induce ipsilateral rotation, instead mice displayed contralateral rotations (Figure 6D). Consistent with the behavioral results, injection of AAV.mHTT/eGFP caused MSN atrophy compared to untreated contralateral striatum whereas co-injection of AAV.caRheb with AAV.mHTT prevented MSN atrophy (Figure 6E). Thus, enhancing mTORC1 activity benefits mHTT-driven neuronal atrophy and dopamine pathway impairment.
Exogenous addition of Rhes, a striatum-enriched mTOR activator, rescues mHTT-associated disease phenotypes in HD transgenic mice
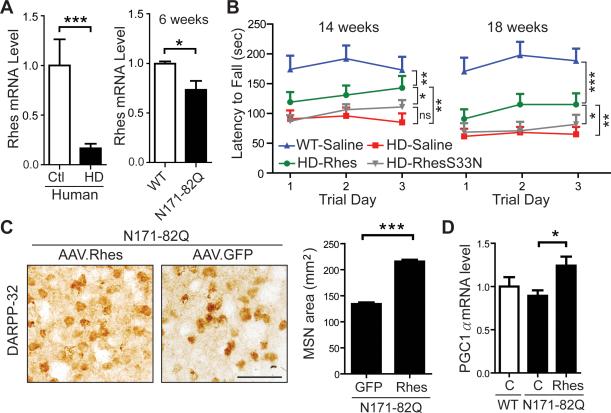
In the striatum, mTOR is predominantly regulated by a striatal-enriched GTPase protein, Rhes (Subramaniam et al., 2012). Prior in vitro studies imply a toxic role of Rhes in HD (Subramaniam et al., 2009), but its disease relevance in vivo is unclear. We found that Rhes levels were reduced in HD human and mouse striata (Figures 7A and S7A). Rheb levels are not reduced (Figure S7B). Notably, Rhes is significantly reduced prior to the onset of neurological symptoms; 6-week-old N171-82Q mice have 73% of wild type Rhes levels even though clinical symptoms are not evident until 12-14 weeks of age (Figures 7A and S7C). We therefore investigated how enhancing Rhes levels impacts disease phenotypes. As the N-terminal fragment of mHTT was previously shown to interact with Rhes and confer cytotoxicity in vitro, the N171-82Q model is ideal for testing this question in vivo (Subramaniam et al., 2009). If Rhes is a critical mediator of HD toxicity, acute overexpression of Rhes in N171-82Q mouse striata would be expected to exacerbate disease phenotypes. AAVs expressing Rhes (AAV.Rhes) were injected into 7-week-old N171-82Q mice and WT littermates. In contrast to what was expected from earlier in vitro work, but consistent with our findings on mTORC1 activation by Rheb, Rhes overexpression improved motor function in N171-82Q mice compared to saline-treated, disease littermates as determined by rotarod testing at 14 and 18 weeks of age (Figure 7B). In addition, after unilateral injection, AAV.Rhes significantly increased MSN cell size compared to control-treated contralateral hemispheres (Figures 7C). Similar to Rheb, AAV.Rhes upregulated expression of the mitochondrialtranscriptional regulator, PGC1-α (Figure 7D).
Figure 7. Intrastriatal Rhes overexpression improves disease phenotypes in N171-82Q mice.
(A) RT-qPCR analysis of endogenous Rhes levels from striatal lysates of unaffected individuals (Ctl, N=4) and HD patients (N=10), and 6-week-old N171-82Q (N=5) and WT (N=4) littermates. Data represent mean ± SEM. *P<0.05, ***P<0.001, Student's t-test. (B) Rotarod assessment of N171-82Q and WT mice after bilateral injection of AAV.Rhes, AAV.RhesS33N, or saline into the striatum at 7 weeks of age (N=10-14 mice per group at 14 weeks of age; N=6-13 mice per group at 18 weeks of age). Rotarod data from three consecutive days at 14 and 18 weeks of age are shown as latency to fall. Saline and AAV.RhesS33N-injected N171-82Q mice performed significantly worse compared to AAV.Rhes-injected N171-82Q mice on the rotarod at 14 and 18 weeks of age. NS=Not statistically significant. Data represent mean ± SEM. *P<0.05; **P<0.01; ***P<0.001, One-way ANOVA with Tukey's post-hoc test. (C) DARPP-32 staining and quantification of MSN area of N171-82Q mice striatal tissue sections after unilateral injection of AAV.Rhes and contralateral injection of AAV.GFP at 7 weeks of age. Tissues were harvested at 19 weeks of age (N=3 mice/group; GFP=245 cells; Rhes=251 cells). Data represent mean ± SEM. ***P<0.001, Student's t-test. Scale bars: 50 μm. (D) qPCR analysis of PGC-1α in striata from AAV.Rhes-or saline-treated N171-82Q and WT mice (n=6-9 per group) harvested at the end of the study (19 weeks of age). C=Control. *P<0.05, Student's t-test.
In cell based assays, Rhes enhances mHTT toxicity by promoting SUMOylation of mHTT (Subramaniam et al., 2009). To ascertain if the observed disease modifying effects of Rhes in vivo are via the mTORC1 or SUMOylation pathways, we generated AAVs that express RhesS33N (AAV.RhesS33N), a Rhes mutant with abolished GTPase activity and reduced mTORC1 activation but intact SUMOylation modulatory activity (Subramaniam et al., 2012; Subramaniam et al., 2009). Following striatal injections, AAV.RhesS33N-treated N171-82Q mice showed reduced mTORC1 activity compared to N171-82Q mice injected with AAV.Rhes (Figure S8). AAV.RhesS33N failed to modify motor deficits in N171-82Q mice, which performed similarly to control treated N171-82Q mice (Figure 7B). Notably, Rhes S33N does not cause further behavioral exacerbation, as one would predict based on prior in vitro observation (Subramaniam et al., 2009). Collectively, these data suggest that Rhes rescues HD behavioral deficits in part via mTORC1 activation.
Discussion
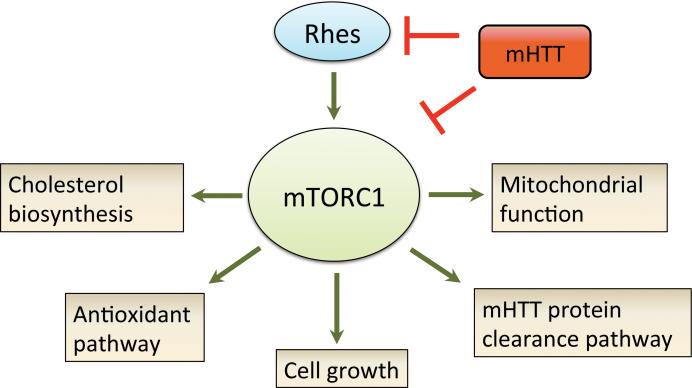
Cumulatively, we show that impaired mTORC1 activity is upstream of various phenotypic changes associated with HD, and may underlie the metabolic and degenerative phenotypes. We find that mTORC1 activation promoted energy metabolism, autophagy, and striatal cell function in HD mice. Consistent with a neuroprotective role of mTORC1, we find that Rhes, a striatum-enriched mTOR activator, is reduced in HD patient brains and HD mouse models prior to onset of neurological symptoms. Also, exogenous Rhes addition alleviated motor deficits and brain pathology in HD mice. Notably, the ability to promote striatal cell growth and function in symptomatic HD mice supports the notion that there is plasticity in mHTT-laden neurons (Yamamoto et al., 2000), and highlights the possibility of reverting striatal atrophy after disease onset through mTORC1 activation. Because mHTT has an increased propensity to bind Rhes (Subramaniam et al., 2009) and mTOR (Ravikumar et al., 2004), it is possible that a concomitant loss of Rhes and mTOR function by mHTT may render the striatum more vulnerable to early degeneration in HD (Figure 8). Given that striatal volume is a strong predictor of disease progression in HD patients (Tabrizi et al., 2013), therapies that restore striatal mTORC1 activity to normal levels may alleviate disease.
Figure 8. Proposed mechanism through which impaired mTOR contributes to complex disease phenotypes in HD.
Under normal physiological condition, mTOR regulates varied biological processes that are crucial for neuronal function and survival. In HD, mHTT disrupts mTORC1 function by direct interaction with mTOR, causing neuronal dysfunction by negatively impacting multiple pathways. In the striatum, mHTT further impairs mTORC1 activity by interfering with Rhes, a striatum enriched mTOR regulator. Thus, a concomitant loss of Rhes and mTORC1 activity may render striatal tissue vulnerable to early degeneration in HD.
A key disease mechanism by which mHTT interferes with energy production is repression of PGC-1α activity, whose expression is reduced in human caudate and HD transgenic mouse striata (Cui et al., 2006; Hodges et al., 2006; Tsunemi et al., 2012). Reduced PGC-1α levels may also sensitize HD brains to oxidative stress as dopaminergic neurons in PGC-1α KO mice are vulnerable to MPTP mediated oxidative damage (St-Pierre et al., 2006). Restoring brain PGC-1α expression ameliorates striatal atrophy in R6/2 HD mice (Cui et al., 2006), and improves motor phenotypes in the N171-82Q and R6/2 HD mouse models (Cui et al., 2006; Tsunemi and La Spada, 2011). Our finding that mTOR is a positive regulator of PGC-1α activity in HD mouse brains provides a therapeutic target for enhancing PGC-1α activity and a potential mechanism by which PGC-1α activity may be impaired.
At first blush, these data may appear to contradict the conventional view in the HD field indicating that mTORC1 inhibition is protective (Ravikumar et al., 2004; Roscic et al., 2011). Inhibition of mTORC1 by systemic delivery of rapamycin attenuates disease phenotypes in Drosophila and N171-82Q HD mice (Ravikumar et al., 2004). It is possible however, that the behavioral improvements induced by rapamycin may reflect the beneficial effects on skeletal muscle, rather than mTORC1 inhibition in the brain. In line with this, in R6/2 HD mice treated with rapamycin, rotarod improvement is transient and associated with accelerated weight loss and enhanced brain atrophy (Fox et al., 2010). Interestingly, mTORC1 activity is abnormally elevated in R6/2 mouse skeletal muscles (She et al., 2011). Together with our data, these findings support the notion that restoring balanced mTORC1 activity is critical and raise the interesting point that there can be tissue specific effects of mHTT on mTOR activity; unlike skeletal muscle, brain mTORC1 activity is already impaired in HD.
The finding that mTORC1 enhancement is neuroprotective also provides a mechanistic basis for earlier studies supporting a role for various agents shown to be therapeutic in HD models. For example, restoring BDNF-TrkB signaling has well-established neuroprotective effects in several preclinical HD models (Jiang et al., 2013; Simmons et al., 2013; Xie et al., 2010; Zuccato et al., 2001). A physiological consequence of this signaling cascade is mTORC1 activation (Troca-Marin et al., 2011; Zhou et al., 2010). Moreover, an RNAi screen identified Lkb-1 as a potent disease modifier in cell and drosophila HD models (Schulte et al., 2011). Lkb-1 is a tumor suppressor that negatively regulates mTORC1 (Shaw et al., 2004). Suppressing Lkb-1 improved mHTT-induced dysmorphic neurite growth in primary neurons, and rescued lethality and neurodegenerative phenotypes in drosophila expressing a mHTT allele (Schulte et al., 2011). Together, these results are consistent with our view that re-balancing mTORC1 activity is beneficial in HD.
Autophagy is essential for cell homeostasis and protein clearance (Wong and Cuervo, 2010). We find that mTORC1 activation stimulates autophagic activity in HD mouse brains and alters expression of genes implicated in promoting mHTT degradation (PIAS1, SUMO2, IKK, and HDAC4). These results may be surprising as mTORC1 is generally considered inhibitory to autophagy. However, recent evidence demonstrate that autophagy can occur under mTOR stimulated conditions (Narita et al., 2011), and under prolonged starvation conditions, reactivation of mTOR promotes formation of lysosomes, which are required for sustained autophagy (Yu et al., 2010). We suspect that enhanced basal or quality control autophagy is a necessary response to cell growth and metabolic activity, and unlike starvation-induced autophagy, is consistent with mTORC1 activation. Although the precise mechanism by which mTORC1 induces autophagy is unclear, we found mTORC1 enhancement increased beclin1 levels, an essential component involved in autophagic vesicle nucleation. Interestingly, overexpression of beclin 1 induces autophagic clearance of mHTT aggregates (Shoji-Kawata et al., 2013). Additionally, we find that mTORC1 upregulates expression of IKK, a known autophagy inducer that promotes mHTT degradation (Thompson et al., 2009). Further, stimulation of IGF1/Akt signaling, which in-turn activates mTORC1, induces mHTT phosphorylation and targets mHTT degradation (Humbert et al., 2002; Yamamoto et al., 2006). Cumulatively the data show that varied mechanisms for mHTT degradation are interrelated and controlled by mTORC1 activity.
The striatum-enriched protein, Rhes, is proposed to cause striatal degeneration by promoting SUMOylation of mHTT; siRNA knockdown of SUMO1 abolishes Rhes mediated cytotoxicity in vitro (Subramaniam et al., 2009). Contrary to the potential toxic role of Rhes, we find that Rhes addition improves neuropathological and motor phenotypes in HD mice, and that this neuroprotective property of Rhes requires its GTPase activity. This is in contrast to the in vitro condition, where the GTPase activity of Rhes is not involved with mHTT-mediated cytotoxicity. Interestingly, we find that reducing Rhes levels in vivo causes compensatory upregulation of SUMO1 in HD mouse striata (data not shown). This is consistent with the observation that SUMO1 and SUMO2 modified proteins accumulate in HD postmortem human brains (O'Rourke et al., 2013). Additionally, overexpressing the GTPase deficient but SUMOylation intact Rhes mutant does not exacerbate motor phenotypes in N171-82Q mice as one would predict based on prior in vitro results (Subramaniam et al., 2009), implying that the in vivo modulators of MSN toxicity is distinct from that in an isolated culture system.
Consistent with a cytoprotective role of Rhes, we recently reported that siRNA knock-down of Rhes exacerbates striatal atrophy and behavioral phenotypes in transgenic HD mice (Lee et al., 2014). On the other hand, other work showed that crossing Rhes KO mice with R6/1 mice delayed rotarod deficits (Baiamonte et al., 2013). However, genetic deletion of Rhes does not prevent striatal atrophy in the R6/1 mice; and Rhes KO mice demonstrate severe brain degeneration similar to HD mice (Baiamonte et al., 2013). Further, Rhes levels are reduced in HD patients and HD mouse model striata, with symptomatic HD patients showing a striking 83% reduction of striatal Rhes compared to unaffected individuals (Figure 7A). Rhes has recently been shown to promote autophagy (Mealer et al., 2014), and regulate intracellular iron homeostatsis (Choi et al., 2013). Therefore, restoring rather than reducing Rhes may be beneficial in HD brains.
The finding that normalizing mTORC1 activity improves HD phenotypes may have broad therapeutic implication for neurological diseases with deregulated mTOR activity. For instance, neurodevelopmental disorders such as fragile X mental retardation and autism are generally associated with hyperactive mTORC1 activity; suppressing mTORC1 in these conditions improves disease phenotypes (Bhattacharya et al., 2012; Tsai et al., 2012). In contrast, in the case of amyotrophic lateral sclerosis (ALS) where neuronal mTORC1 activity is reduced, stimulation of mTORC1 counteracts the degenerative process (Saxena et al., 2013). Thus, restoring the homeostatic level of mTORC1 function may broadly benefit neurological diseases.
In summary, our study provides new insight into the inter-connectivity of complex pathological phenotypes in HD, which converge on the mTORC1 pathway. And, together with prior work in other neurodegenerative disease fields, highlights the observation that the extent of mTORC1 activation must be carefully controlled for therapeutic utility. While reducing the expression of the mHTT in the brain would be a direct way to rescue mTORC1 activity in the setting of HD, a rebalance of mTORC1 activity or small molecules that disrupt RhesmHTT binding may be of therapeutic benefit.
Experimental Procedures
Animals
All animal protocols were approved by the University of Iowa Animal Care and Use Committee. N171-82Q mice were from Jackson Labs (Bar Harbor, ME) and maintained on B6C3F1/J background. Hemizygous and age-matched wild-type littermates were used for the indicated experiments. Mice were housed in a controlled temperature environment on a 12-hour light/dark cycle. Food and water were provided ad libitum. For pharmacological studies, RAD001 was obtained from LC Laboratories, diluted to 2mg/ml in 2% DMSO, and stored at − 20°C. Freshly thawed vehicle and RAD001 (30 μmol/kg) were given 3 times per week (Monday, Wednesday, and Friday) for two weeks by gavage as described previously(Fox et al., 2010). Mice were sacrificed 24 hours after the last dose.
Plasmids and AAVs
AAV vectors serotype 1 (AAV2/1) were used for this study. Rhes and Rheb cDNA sequences were amplified from a mouse striatum cDNA library. A Flag epitope was cloned 3’ for western blot assay. GTPase inactive Rhes mutant (RhesS33N) and hyperactive Rheb mutant (caRheb;S16H) were generated by QuikChange site-directed mutagenesis (Agilent Technology). Primers for mutagenesis used the QuikChange Primer Design Program (http://www.stratagene.com). Rhes, RhesS33N, and caRheb sequences were cloned into AAV vectors downstream of the chicken β-actin promoter. AAV.Rhes and AAV.RhesS33N vectors also express enhanced GFP (eGFP) under the control of an IRES sequence. N-terminal myc-tagged N171-82Q was PCR amplified from a pCMV-HD-N171-82Q plasmid (Harper et al., 2005), and cloned into AAV vectors to generate AAV.mHTT. AAVs were made by University of Iowa Vector Core. Titers (vector genomes/ml; vg/ml) were assessed by RT-PCR (Rhes: 3×1012 vg/ml, RhesS33N: 3×1012 vg/ml, caRheb: 3×1012 vg/ml, eGFP: 3×1012 vg/ml, and mHTT (N171-82Q): 3×1012 vg/ml).
Injections
The coordinates for striata were 0.86 mm rostral to bregma, ±1.8 mm lateral to midline, 3.5 mm ventral to the skull surface. N171-82Q mice and WT littermates were injected with AAV.Rhes, AAV.RhesS33N, or saline at 7 weeks of age. Bilaterally injections were with 5-μl of AAV. For unilateral injection studies, 5-μl of AAV was used. The coordinates for hippocampus were AP:-2.0 mm, ML:± 1.5 mm, DV:-2.3 mm relative to bregma; mice were injected unilaterally with 1-ul of AAV. For amphetamine induced rotation tests in WT mice, 3 μl AAV.mHTT was co-injected with either 2 μl AAV.caRheb or 2 μl AAV.eGFP into the striatum. Injection rates for all studies were 0.2 μl/min. Mice were sacrificed at indicated ages using standard approved methods (Harper et al., 2005; McBride et al., 2008).
Cell culture and transfections
Mutant (Q111) and WT (Q7) striatal cells (Trettel et al., 2000) with full-length HTT were kindly provided by Dr. Marcy MacDonald. The Q7 and Q111 cells were grown at 37°C in Dulbecco's modified Eagle's medium (DMEM; Sigma Chemical Co, St. Louis, MO) supplemented with 10% fetal bovine serum, 1% non-essential amino acids, 2 mM L-glutamine, and 400 μg/ml G418 (Geneticin; Invitrogen, Carlsbad, CA). For mTOR inhibition studies, cells were treated with DMSO or 250 nM Torin1 (Tocris, Bristol, UK) for 24 hours.
Mouse Brain Isolation
Mice used for histological analyses were anesthetized with a ketamine/xylazine mix and transcardially perfused with 20 ml of 0.9% cold saline, followed by 20 ml of 4% paraformaldehyde in 0.1 M PO4 buffer. Brains were removed and postfixed overnight, and 40-μm thick sections were collected. Mice used for molecular analyses were perfused with 20 ml of 0.9% cold saline, and brain was removed and blocked into 1-mm-thick coronal slices. Tissue punches of striatum were taken using a tissue corer (1.4 mm in diameter) and flash frozen in liquid nitrogen and stored at −80°C until used.
Immunohistochemistry and quantitation
Floating, 40-μm coronal brain sections were used for histochemistry. The following primary antibodies were used: DARPP-32, 1:200, Cell Signaling; Em48 1:200, gift from X. J. Li, Emory University School of Medicine; phospho-S6 antibody, S235/236, 1:300, Cell Signaling. Goat anti-rabbit or goat anti-mouse biotinylated IgG secondary antibodies were used (1:200; Vector laboratories) and developed with avidin-biotinylated horseradish peroxidase complexes (ABC; Vector Labs). In all procedures, deletion of the primary antibody served as control. Images were captured on an Olympus BX60 light microscope and DP70 camera with Olympus DP Controller software (Olympus, Melville, NY). For each brain, four representative sections were chosen and areas of 70 DARPP-32 positive medium spiny neurons (MSN) were manually traced under 40x objective from each hemisphere. The areas of DARPP-32 positive MSN were quantified by Olympus DP2-BSW software (version 2.1). The average MSN areas of ipsilateral injected sides were compared to contralateral control sides and expressed as means ± SEM. EM48 positive inclusions were counted by an experimenter blinded to treatment groups.
Stereology
Striatal volume was calculated using point counting methods with the Cavalieri estimator of volume (Michel and Cruz-Orive, 1988). Images of DARPP-32 stained coronal sections (40 μm thick) were captured using an Olympus BX60 light microscope and DP70 camera with Olympus DP Controller software (Olympus, Melville, NY). An acetate sheet with a grid pattern was placed on the image at random angles, and the number of grid intersections overlying the striatum counted. A random systematic set of serial sections at an interval of 480 μm apart was used. A minimum of 200 points were counted over 5 sections. Left and right striatal volumes were determined independently on the same sections from the point counts by using the formula , where , t=distance between sections, and Ap=area per point.
Electron microscopy
Mice were anesthetized and perfused intracardially with saline solution followed by 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer pH 7.4. Brains were removed and post-fixed in 2.5% glutaraldehyde, 0.1M sodium cacodylate buffer pH 7.4 for 24 hours. 100 μm coronal sections were cut with vibratome and fixed in 1% OsO4 in 0.1M cacodylate for 1h, stained with 1% uranyl acetate, dehydrated and embedded in Eponate 812. Ultrathin sections of striatum (80 nm) were examined with a JEOL EX 1200 transmission electron microscope.
Western blot analysis
Mouse striata were lysed in RIPA buffer with protease inhibitors (Complete Mini, Roche Applied Science, Mannheim, Germany) and phosphatase inhibitors (PhosSTOP Phosphatase Inhibitor Cocktail, Roche Applied Science, Mannheim, Germany). Protein concentration was determined by BCA assay (Pierce, Rockford, IL) and 20-30 μg of protein was reduced and ran on SDS-PAGE on 4%–10% acrylamide gels and transferred to 0.2 μm Immobilon PVDF membranes (Millipore, Billerica, MA). Primary antibodies were: DARPP-32, 1:1000, Cell Signaling; β-actin, Sigma-Aldrich, 1:5000; S6,1:1000, Cell Signaling; phospho-S6, S235/236, 1:1000, Cell Signaling; 4E-BP1, 1:1000, Cell Signaling; phospho-4E-BP1,T37/46, 1:1000, Cell Signaling; mTOR, 1:1000, Cell Signaling; Rictor, 1:1000, Cell Signaling; Raptor,1:1000, Cell Signaling; CREB, 1:1000, Cell Signaling; TORC1, 1:1000, Cell Signaling; PGC1-α,1:1000, abcam; LC3,1:1000, Novus Biologicals; and Beclin1,1:1000, Cell Signaling. Secondary antibodies were HRP-goat anti-mouse IgG and HRP-goat anti-rabbit IgG (Cell Signaling). Blots were developed using ECL Plus Western Blotting Detection System (GE Healthcare, Pittsburg, PA), and exposed to film for images. Protein quantification was performed using NIH ImageJ software or the VersaDocTM Imaging System (Biorad) and Quantity One R analysis software. Band densities were normalized to β-actin from the same samples and lanes.
Brain samples
Coronal sections of human autopsy brain tissues were obtained from unaffected individuals and patients with Vonsattel grade 2, 3 and 4 HD (Dr. Christopher Ross, Johns Hopkins University; New York Brain Bank at Columbia University, Alzheimer Disease Research Center, Taub Institute). Tissues were flash frozen with postmortem intervals (PMI) ranging from 13 to 49 hours. Caudate/putamen nuclei were dissected from the frozen tissues placed on an ice-cold metal stage and homogenized in RIPA buffer with protease inhibitors (Complete Mini, Roche Applied Science, Mannheim, Germany) and phosphatase inhibitors (PhosSTOP Phosphatase Inhibitor Cocktail, Roche Applied Science, Mannheim, Germany). Lysates were spun at 14,000g for 20 min. The supernatant was collected for western blot analysis. Additional frozen tissues were processed for RNA extraction using TRIzol.
Quantitative real-time PCR (RT-qPCR)
RNA was isolated from striatal punches using 1 ml of TRIzol. Random-primed first-strand complementary DNA (cDNA) synthesis was performed using 500 ng total RNA (High Capacity cDNA Reverse Transcription Kit; Applied Biosystems, Foster City, CA) per manufacturer's protocol. Real-time PCR was performed on a sequence detection system (Prism 7900HT, Applied Biosystems) using SYBR green PCR mix (Invitrogen) or TaqMan 2x Universal Master Mix (Applied Biosystems). The following Taqman primer/probe sets were obtained from Applied Biosystem: Rhes (Mm04209172_m1 and Hs04188091_mH), PGC1-α (Mm01208835_m1), and HMGCS1 (Mm01304569_m1), and HDAC4 (Mm01299557_m1). The primer sequences for SYBR green RT PCR are available upon request. Relative gene expression was determined using the ΔΔCT method, normalizing to β-actin.
Behavioral Tests
Amphetamine induced rotational assay
Mice were injected with intraperitoneal amphetamine (3 mg/kg; Sigma), and then placed individually in the center of a square plastic activity chamber (50 × 50 cm). Mice were allowed to habituate in the chamber for 20 min before amphetamine induction. Rotational behavior was then recorded for 40 min by a ceiling-mounted video camera. Net rotation was expressed as number of ipsilateral turns – number of contralateral turns in a 40 min session.
Accelerating Rotarod
Male N171-82Q mice were tested for baseline motor function at 6 weeks of age, and then at 10, 14, and 18 weeks of age. Before each test, mice were first habituated on the rotarod for 4 minutes and rested for one hour. The tests were conducted three trials per day (with 30 minutes of rest between trials) for four consecutive days. For each trial, mice were placed on the rod that accelerates from 4-40 rotations per min over 4 minutes, and then speed maintained at 40 rpm. Latency to fall (or if mice hung on for two consecutive rotations without running) was used as a rating of motor performance. The trials were stopped at 300 seconds, and mice remaining on the rotarod at that time were scored as 300 sec. Data from the three trials for each group on each day are presented as means ± SEM. Mice were always tested in the dark phase of the light/dark cycle. All behavioral experiments were conducted with the experimenter blind to mouse genotypes and treatments.
Statistical analyses
Data were analyzed using Student's t-test or One-way ANOVA analysis, followed by Tukey's post hoc analyses to assess for significant differences between individual groups. All statistical analyses were performed using GraphPad Prism version 5.0c. Data are expressed as mean ± SEM. In all cases, P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
Supported by NIH grants NS50210 (to BLD) and NS052789-S1 (to LMT and BLD) and the Roy J Carver Trust (BLD). We would like to thank Dr. Nandakumar Narayanan for assistance with the amphetamine induced rotational assay, and Dr. Christopher Ross from Johns Hopkins University for patient brain tissues.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Baiamonte BA, Lee FA, Brewer ST, Spano D, LaHoste GJ. Attenuation of Rhes activity significantly delays the appearance of behavioral symptoms in a mouse model of Huntington's disease. PLoS One. 2013;8:e53606. doi: 10.1371/journal.pone.0053606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Kaphzan H, Alvarez-Dieppa AC, Murphy JP, Pierre P, Klann E. Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron. 2012;76:325–337. doi: 10.1016/j.neuron.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb JA, Yan Z, Svenningsson P, Snyder GL, Pieribone VA, Horiuchi A, Nairn AC, Messer A, Greengard P. Severe deficiencies in dopamine signaling in presymptomatic Huntington's disease mice. Proc Natl Acad Sci U S A. 2000;97:6809–6814. doi: 10.1073/pnas.120166397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Peng Q, Hou Z, Aggarwal M, Zhang J, Mori S, Ross CA, Duan W. Structural MRI detects progressive regional brain atrophy and neuroprotective effects in N171-82Q Huntington's disease mouse model. Neuroimage. 2011;56:1027–1034. doi: 10.1016/j.neuroimage.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BR, Bang S, Chen Y, Cheah JH, Kim SF. PKA modulates iron trafficking in the striatum via small GTPase, Rhes. Neuroscience. 2013;253:214–220. doi: 10.1016/j.neuroscience.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Fox JH, Connor T, Chopra V, Dorsey K, Kama JA, Bleckmann D, Betschart C, Hoyer D, Frentzel S, Difiglia M, et al. The mTOR kinase inhibitor Everolimus decreases S6 kinase phosphorylation but fails to reduce mutant huntingtin levels in brain and is not neuroprotective in the R6/2 mouse model of Huntington's disease. Mol Neurodegener. 2010;5:26. doi: 10.1186/1750-1326-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci U S A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Humbert S, Bryson EA, Cordelieres FP, Connors NC, Datta SR, Finkbeiner S, Greenberg ME, Saudou F. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- Jeong H, Then F, Melia TJ, Jr., Mazzulli JR, Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Kast RJ, Steffan JS, Thomas EA. Selective histone deacetylase (HDAC) inhibition imparts beneficial effects in Huntington's disease mice: implications for the ubiquitin-proteasomal and autophagy systems. Hum Mol Genet. 2012;21:5280–5293. doi: 10.1093/hmg/dds379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Peng Q, Liu X, Jin J, Hou Z, Zhang J, Mori S, Ross CA, Ye K, Duan W. Small-molecule TrkB receptor agonists improve motor function and extend survival in a mouse model of Huntington's disease. Hum Mol Genet. 2013;22:2462–2470. doi: 10.1093/hmg/ddt098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Rajan V, Miller CE, Wightman RM. Dopamine release is severely compromised in the R6/2 mouse model of Huntington's disease. J Neurochem. 2006;97:737–746. doi: 10.1111/j.1471-4159.2006.03762.x. [DOI] [PubMed] [Google Scholar]
- Karasinska JM, Hayden MR. Cholesterol metabolism in Huntington disease. Nat Rev Neurol. 2011;7:561–572. doi: 10.1038/nrneurol.2011.132. [DOI] [PubMed] [Google Scholar]
- Kim SR, Kareva T, Yarygina O, Kholodilov N, Burke RE. AAV transduction of dopamine neurons with constitutively active Rheb protects from neurodegeneration and mediates axon regrowth. Mol Ther. 2012;20:275–286. doi: 10.1038/mt.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiner G, Bierhoff H, Armentano M, Rodriguez-Parkitna J, Sowodniok K, Naranjo JR, Bonfanti L, Liss B, Schutz G, Grummt I, et al. A neuroprotective phase precedes striatal degeneration upon nucleolar stress. Cell Death Differ. 2013 doi: 10.1038/cdd.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Sowada MJ, Boudreau RL, Aerts AM, Thedens DR, Nopoulos P, Davidson BL. Rhes Suppression Enhances Disease Phenotypes in Huntington's Disease Mice. J Huntingtons Dis. 2014;3:65–71. doi: 10.3233/JHD-140094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li SH, Yu ZX, Shelbourne P, Li XJ. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington's disease mice. J Neurosci. 2001;21:8473–8481. doi: 10.1523/JNEUROSCI.21-21-08473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Palacino J. A novel human embryonic stem cell-derived Huntington's disease neuronal model exhibits mutant huntingtin (mHTT) aggregates and soluble mHTT-dependent neurodegeneration. FASEB J. 2013;27:1820–1829. doi: 10.1096/fj.12-219220. [DOI] [PubMed] [Google Scholar]
- Mason RP, Casu M, Butler N, Breda C, Campesan S, Clapp J, Green EW, Dhulkhed D, Kyriacou CP, Giorgini F. Glutathione peroxidase activity is neuroprotective in models of Huntington's disease. Nat Genet. 2013;45:1249–1254. doi: 10.1038/ng.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealer RG, Murray AJ, Shahani N, Subramaniam S, Snyder SH. Rhes, a Striatal-selective Protein Implicated in Huntington Disease, Binds Beclin-1 and Activates Autophagy. J Biol Chem. 2014;289:3547–3554. doi: 10.1074/jbc.M113.536912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealer RG, Subramaniam S, Snyder SH. Rhes Deletion Is Neuroprotective in the 3-Nitropropionic Acid Model of Huntington's Disease. J Neurosci. 2013;33:4206–4210. doi: 10.1523/JNEUROSCI.3730-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel RP, Cruz-Orive LM. Application of the Cavalieri principle and vertical sections method to lung: estimation of volume and pleural surface area. J Microsc. 1988;150:117–136. doi: 10.1111/j.1365-2818.1988.tb04603.x. [DOI] [PubMed] [Google Scholar]
- Mielcarek M, Landles C, Weiss A, Bradaia A, Seredenina T, Inuabasi L, Osborne GF, Wadel K, Touller C, Butler R, et al. HDAC4 reduction: a novel therapeutic strategy to target cytoplasmic huntingtin and ameliorate neurodegeneration. PLoS Biol. 2013;11:e1001717. doi: 10.1371/journal.pbio.1001717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milnerwood AJ, Raymond LA. Early synaptic pathophysiology in neurodegeneration: insights from Huntington's disease. Trends Neurosci. 2010;33:513–523. doi: 10.1016/j.tins.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332:966–970. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke JG, Gareau JR, Ochaba J, Song W, Rasko T, Reverter D, Lee J, Monteys AM, Pallos J, Mee L, et al. SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep. 2013;4:362–375. doi: 10.1016/j.celrep.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Roscic A, Baldo B, Crochemore C, Marcellin D, Paganetti P. Induction of autophagy with catalytic mTOR inhibitors reduces huntingtin aggregates in a neuronal cell model. J Neurochem. 2011;119:398–407. doi: 10.1111/j.1471-4159.2011.07435.x. [DOI] [PubMed] [Google Scholar]
- Saxena S, Roselli F, Singh K, Leptien K, Julien JP, Gros-Louis F, Caroni P. Neuroprotection through Excitability and mTOR Required in ALS Motoneurons to Delay Disease and Extend Survival. Neuron. 2013;80:80–96. doi: 10.1016/j.neuron.2013.07.027. [DOI] [PubMed] [Google Scholar]
- Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- Schulte J, Sepp KJ, Wu C, Hong P, Littleton JT. High-content chemical and RNAi screens for suppressors of neurotoxicity in a Huntington's disease model. PLoS One. 2011;6:e23841. doi: 10.1371/journal.pone.0023841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seredenina T, Gokce O, Luthi-Carter R. Decreased striatal RGS2 expression is neuroprotective in Huntington's disease (HD) and exemplifies a compensatory aspect of HD-induced gene regulation. PLoS One. 2011;6:e22231. doi: 10.1371/journal.pone.0022231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- She P, Zhang Z, Marchionini D, Diaz WC, Jetton TJ, Kimball SR, Vary TC, Lang CH, Lynch CJ. Molecular characterization of skeletal muscle atrophy in the R6/2 mouse model of Huntington's disease. Am J Physiol Endocrinol Metab. 2011;301:E49–61. doi: 10.1152/ajpendo.00630.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, Macduff D, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013 doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DA, Belichenko NP, Yang T, Condon C, Monbureau M, Shamloo M, Jing D, Massa SM, Longo FM. A Small Molecule TrkB Ligand Reduces Motor Impairment and Neuropathology in R6/2 and BACHD Mouse Models of Huntington's Disease. J Neurosci. 2013;33:18712–18727. doi: 10.1523/JNEUROSCI.1310-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano D, Branchi I, Rosica A, Pirro MT, Riccio A, Mithbaokar P, Affuso A, Arra C, Campolongo P, Terracciano D, et al. Rhes is involved in striatal function. Mol Cell Biol. 2004;24:5788–5796. doi: 10.1128/MCB.24.13.5788-5796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Napolitano F, Mealer RG, Kim S, Errico F, Barrow R, Shahani N, Tyagi R, Snyder SH, Usiello A. Rhes, a striatal-enriched small G protein, mediates mTOR signaling and L-DOPA-induced dyskinesia. Nat Neurosci. 2012;15:191–193. doi: 10.1038/nn.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Park KK, Belin S, Wang D, Lu T, Chen G, Zhang K, Yeung C, Feng G, Yankner BA, et al. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi SJ, Scahill RI, Owen G, Durr A, Leavitt BR, Roos RA, Borowsky B, Landwehrmeyer B, Frost C, Johnson H, et al. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington's disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 2013 doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, Martinez-Vincente M, Arrasate M, O'Rourke JG, Khashwji H, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- Troca-Marin JA, Alves-Sampaio A, Montesinos ML. An increase in basal BDNF provokes hyperactivation of the Akt-mammalian target of rapamycin pathway and deregulation of local dendritic translation in a mouse model of Down's syndrome. J Neurosci. 2011;31:9445–9455. doi: 10.1523/JNEUROSCI.0011-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, Steinberg J, Crawley JN, Regehr WG, Sahin M. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, Lazarowski ER, Damian VA, Masliah E, La Spada AR. PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4:142ra197. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, La Spada AR. PGC-1alpha at the intersection of bioenergetics regulation and neuron function: From Huntington's disease to Parkinson's disease and beyond. Prog Neurobiol. 2011 doi: 10.1016/j.pneurobio.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenza M, Cattaneo E. Emerging roles for cholesterol in Huntington's disease. Trends Neurosci. 2011;34:474–486. doi: 10.1016/j.tins.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Valenza M, Rigamonti D, Goffredo D, Zuccato C, Fenu S, Jamot L, Strand A, Tarditi A, Woodman B, Racchi M, et al. Dysfunction of the cholesterol biosynthetic pathway in Huntington's disease. J Neurosci. 2005;25:9932–9939. doi: 10.1523/JNEUROSCI.3355-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Hayden MR, Xu B. BDNF overexpression in the forebrain rescues Huntington's disease phenotypes in YAC128 mice. J Neurosci. 2010;30:14708–14718. doi: 10.1523/JNEUROSCI.1637-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Lin DS, Zheng F, Sutton MA, Wang H. Intracellular calcium and calmodulin link brain-derived neurotrophic factor to p70S6 kinase phosphorylation and dendritic protein synthesis. J Neurosci Res. 2010;88:1420–1432. doi: 10.1002/jnr.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhou L, Du XX, Ji Y, Xu J, Tian J, Jiang W, Zou Y, Yu S, Gan L, et al. Rheb1 is required for mTORC1 and myelination in postnatal brain development. Dev Cell. 2011;20:97–108. doi: 10.1016/j.devcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.