Abstract
AIM: To evaluate the association between poly-morphisms XRCC1 Arg194Trp and Arg399Gln and XRCC3 Thr241Met and the risk for chronic gastritis and gastric cancer, in a Southeastern Brazilian population.
METHODS: Genotyping by PCR-RFLP was carried out on 202 patients with chronic gastritis (CG) and 160 patients with gastric cancer (GC), matched to 202 (C1) and 150 (C2) controls, respectively.
RESULTS: No differences were observed among the studied groups with regard to the genotype distribution of XRCC1 codons 194 and 399 and of XRCC3 codon 241. However, the combined analyses of the three variant alleles (194Trp, 399Gln and 241Met) showed an increased risk for chronic gastritis when compared to the GC group. Moreover, an interaction between the polymorphic alleles and demographic and environmental factors was observed in the CG and GC groups. XRCC1 194Trp was associated with smoking in the CG group, while the variant alleles XRCC1 399Gln and XRCC3 241Met were related with gender, smoking, drinking and H pylori infection in the CG and GC groups.
CONCLUSION: Our results showed no evidence of a rela-tionship between the polymorphisms XRCC1 Arg194Trp and Arg399Gln and XRCC3 Thr241Met and the risk of chronic gastritis and gastric cancer in the Brazilian population, but the combined effect of these variants may interact to increase the risk for chronic gastritis, considered a premalignant lesion. Our data also indicate a gene-environment interaction in the susceptibility to chronic gastritis and gastric cancer.
Keywords: Gastric cancer, Gastritis, XRCC1, XRCC3, Polymorphism, Environmental exposure
INTRODUCTION
DNA repair pathways are responsible for maintaining the integrity of the genome in face of environmental insults and general DNA replication errors, playing a role in protecting it against mutations that lead to cancer[1]. So, polymorphisms of DNA repair enzymes, which may alter the function or efficiency of the DNA repair, may contribute to an increased risk of environmental carcinogenesis[2]. These low-penetrance susceptibility genes have common variants and interact with environmental factors, contributing as a major factor to the populational incidence of cancer[3]. Several polymorphisms in genes that participate in different DNA repair pathways, such as XPD, XPF, ERCC1, XRCC1, XRCC3[4], hOGG1[5], XPA, XPB[6] and XPC[7], have been identified and related to cancer susceptibility.
The XRCC1 gene is responsible for a scaffolding protein that directly associates with other proteins such as DNA polymerase β, PARP (ADP-ribose polymerase) and DNA ligase III in a complex, to facilitate the processes of base excision repair (BER) or single-strand break repair[8]. The BER pathway repairs DNA damage caused by a variety of endogenous and exogenous factors, including oxidation, alkylating agents and ionizing radiation[1,9]. The XRCC1 protein can bind directly to both gapped and nicked DNA, as well as to gapped DNA associated with DNA polymerase β, suggesting that this protein might be independently involved in DNA damage recognition[10]. Two polymorphisms, more often found in XRCC1’ conserved sites, lead to a C→T substitution at codon 194 in exon 6 and to a G→A substitution at codon 399 in exon 10 of the gene, leading to the amino acid alterations arginine (Arg) to tryptophan (Trp) and arginine (Arg) to glutamine (Gln), respectively. These changes in conserved protein sites may alter the BER capacity, increasing the chances of DNA damage[4].
The Arg399Gln variant is more frequent and has been associated mainly with head and neck[11], colorectal[12], gastric[13], esophageal[14,15], breast[16] and lung[17,18] cancers. The Arg194Trp polymorphism has been related to colorectal[12], gastric[13], head and neck[19] and skin[20] cancers.
Protein XRCC3 functions in the DNA double-strand break (DSB) and cross-link repair[21] and interacts and stabilizes Rad51[22], one of the key components of the homologous repair (HR) pathway. The HR pathway uses a second intact copy of a homologous chromosome as a template to copy the information lost at the DSB site, resulting in a high-fidelity process and preventing chromosomal aberrations[9]. The main polymorphism in this gene involves the change of threonine (Thr) to methionine (Met) at codon 241 in exon 7[4]. Little is known about the functional consequences of this variation, although some studies observed a positive relation between the Thr241Met polymorphism and an increased risk for skin[23], bladder[24], breast[25] and lung[26] cancers.
So far, the investigations about interactions between XRCC1 and XRCC3 polymorphisms and environmental carcinogenesis have produced scarce and conflicting results[27,28,29,30], showing the functional complexity of these variants, that can include their interaction with environmental factors, thus modulating the susceptibility to cancer. Regarding gastric cancer, only a few studies were conducted to investigate its association with XRCC1 and XRCC3 variants[13,27,29,31].
In Brazil, gastric cancer is still one of the most frequent types of cancer. The estimate for 2005 points to the fourth place in incidence and mortality, with about 23 000 new cases and 12 000 deaths[32]. However, multiple factors are thought to play a role in gastric carcinogenesis, including diet[33], lifestyle[34], pathological changes in the stomach such as chronic gastritis[35], and genetic alterations[36,37], besides the infection by Helicobacter pylori, the first bacterium to be termed as a definitive cause of cancer[38].
Thus, we conducted a study to evaluate the association between the polymorphisms XRCC1 Arg194Trp and Arg399Gln and XRCC3 Thr241Met and the risk of chronic gastritis and gastric cancer in a Brazilian population, as well as the interaction between these polymorphisms and environmental factors involved in gastric carcinogenesis.
MATERIALS AND METHODS
Subjects
This was a case-control study on chronic gastritis and gastric cancer. The case groups comprised 202 patients with a histopathologically confirmed diagnosis of chronic gastritis (100 men and 102 women), with a mean age of 52 years (range 19-86 years), and 160 patients with a histopathologically confirmed diagnosis of gastric adenocarcinoma (118 men and 42 women), with a mean age of 61 years (range 28-93 years). All subjects were recruited from the Hospital de Base in São José do Rio Preto, SP, and from the Pio XII Foundation in Barretos, SP, Brazil. Gastric adenocarcinomas were classified as diffuse or intestinal types, according to the classification proposed by Lauren[39], and the chronic gastritis cases according to the Sidney System[40]. H pylori infection was histologically established by the Giemsa staining technique. Two cancer-free control groups with no previous history of gastric disease were matched to the case groups with respect to age, gender and ethnicity. The control group for chronic gastritis (C1) was composed of 202 healthy individuals (100 men and 102 women) with a mean age of 51 years (range 20-85 years), and the control group for gastric cancer (C2) consisted of 150 healthy volunteers (108 men and 42 women) with a mean age of 59 years (range 22-93 years). Epidemiological data on the study population were collected using a standard interviewer-administered questionnaire, with questions about current and past occupation, smoking habits, alcohol intake and family history of cancer. This work was approved by the National Research Ethics Committee and written informed consent was obtained from all individuals.
DNA extraction and genotype analyses
About 5 mL of whole blood were collected from all study participants in sterile EDTA-coated vacutainers. The samples were assigned a unique identifier code. DNA was extracted according to Abdel-Rahman et al[41] and stored at -20 ºC until used for genotyping.
Genotypic analyses of the XRCC1 gene were carried out by multiplex PCR-RFLP, using primers for codons 399 (F 5’-TTGTGCTTTCTCTGTGTCCA-3’ and R 5’-TCCTCCAGCCTTTTCTGATA-3’) and 194 (F 5’-GCCCCGTCCCAGGTA-3’ and R 5’-AGCCCCAAGACCCTTTCACT-3’), which generate a fragment of 615 and 491 bp, respectively, as previously described[12] with modifications. Briefly, PCR was performed in 25 μL reaction buffer containing 12.5 pmol each primer, 0.2 mmol/L of dNTPs, 3 mmol/L of MgCl2, about 100 ng DNA and 1 U of Taq DNA polymerase. The PCR products were digested overnight with 10 U of MspI at 37 ºC. The wild-type Arg allele for codon 194 is identified by the presence of a 293 bp band, and the mutant Trp allele by the presence of a 313 bp band (indicative of the absence of the MspI cutting site). For codon 399, the presence of two bands of 375 and 240 bp, respectively, identifies the wild-type Arg allele, while the uncut 615 bp band identifies the mutant Gln allele (indicative of the absence of the MspI cutting site). A 178 bp band, resulting from an additional invariant MspI cutting site in the 491 bp amplified fragment, is always present and serves as an internal control for complete enzyme digestion.
Polymorphism of the XRCC3 gene was deter-mined by PCR-RFLP, using codon 241 primers (F- 5’ GCCTGGTGGTCATCGACTC 3’ e R- 5’ ACAGGGCTCTGGAAGGCACT GCTCAGCTCACGCACC 3’), as previously described by David-Beabes et al[42]. The 25 μL PCR mixture con-tained about 100 ng of DNA, 12.5 pmol of each primer, 0.2 mmol/L of dNTPs, 2 mmol/L of MgCl2 and 1 U of Taq DNA polymerase. The 552 bp amplified product was digested overnight with 5 U of NlaIII at 37 ºC. The wild-type allele Thr was identified by the presence of two 239 and 313 bp bands, while the mutant allele Met was represented by 105, 208, and 239 bp bands.
Statistical analysis
Chi-square or Fisher’s exact tests were utilized to compare the groups with regard to genotype frequencies and putative risk factors such as age, gender, ethnicity, smoking, drinking, H pylori infection and histological type of adenocarcinoma. To investigate the gene-environment interactions, the odds ratios (OR) and their 95% confidence intervals (95% CI) were calculated, according to a combination of the XRCC1 and XRCC3 polymorphisms with putative risk factors. The statistical analyses were performed using Statidisk and GraphPad InStat computer software programs. A probability level (P) of less than 0.05 was used as criterion of significance.
RESULTS
Cases and controls did not show any statistically significant difference with regard to age, gender and ethnicity, indicating a well-matched study population. Gastric cancer (GC) patients were more likely to be cigarette smokers or alcohol drinkers than chronic gastritis (CG) patients and controls (C2), and this difference was statistically significant (P<0.05), the same occurring with CG patients when compared to controls (C1).
The XRCC1 and XRCC3 genotypes and the allele frequency distributions among cases and controls are presented in Table 1. The allele frequencies of polymorphisms 194Trp, 399Gln and 241Met were similar in cases and controls, not showing any statistically significant differences (P>0.05). However, a combined analysis of the XRCC1 Arg194Trp and Arg399Gln polymorphisms and the XRCC3 Thr241Met polymorphism (Table 2), and the assessment of the inter- and intra-gene interactions of these three polymorphisms revealed a statistically significant (P = 0.0372) association when the three variant alleles interacted in the chronic gastritis group, as compared to the gastric cancer group. Other combinations did not show any significant difference. The banding patterns of XRCC1 Arg194Trp and Arg399Gln and of XRCC3 Thr241Met are represented in Figure 1.
Table 1.
XRCC1 and XRCC3 allele frequencies in patients with chronic gastritis (CG) and gastric cancer (GC) and in the respective control groups C1 and C2
| Genotypes | CG (n = 202) | C1 (n = 202) | P | GC (n = 160) | C2 (n = 150) | P | |
| n (%) | n (%) | n (%) | n (%) | ||||
| XRCC1 | Arg/Arg | 176 (87.1) | 183 (90.6) | 140 (87.5) | 130 (86.7) | ||
| Codon 194 | Arg/Trp | 24 (11.9) | 19 (9.4) | 20 (12.5) | 19 (12.7) | ||
| Trp/Trp | 2 (1.0) | 0 (0.0) | 0 (0.0) | 1 (0.6) | |||
| Arg/Trp+Trp/Trp | 26 (12.9) | 19 (9.4) | 0.2683 | 20 (12.5) | 20 (13.3) | 0.8269 | |
| Allele frequency | 0.07 | 0.05 | 0.06 | 0.07 | |||
| XRCC1 | Arg/Arg | 98 (48.5) | 95 (47) | 73 (45.6) | 70 (46.7) | ||
| Codon 399 | Arg/Gln | 91 (45.0) | 82 (40.6) | 67 (41.9) | 57 (38.0) | ||
| Gln/Gln | 13 (6.5) | 25 (12.4) | 0.7651 | 20 (12.5) | 23 (15.3) | 0.5035 | |
| Arg/Gln+Gln/Gln | 104 (51.5) | 107 (53.0) | 87 (54.4) | 80 (53.3) | |||
| Allele frequency | 0.29 | 0.33 | 0.33 | 0.34 | |||
| XRCC3 | Thr/Thr | 92 (45.5) | 84 (41.6) | 84 (52.5) | 67 (44.7) | ||
| Codon 241 | Thr/Met | 81 (40.1) | 89 (44.1) | 53 (33.1) | 60 (40.0) | ||
| Met/Met | 29 (14.4) | 29 (14.3) | 0.4221 | 23 (14.4) | 23 (15.3) | 0.1679 | |
| Thr/Met+Met/Met | 110 (54.5) | 118 (58.4) | 76 (47.5) | 83 (55.3) | |||
| Allele frequency | 0.34 | 0.36 | 0.31 | 0.35 |
Table 2.
Association between XRCC1 and XRCC3 genotype profiles and risk for chronic gastritis (CG) and gastric cancer (GC)
| XRCC1 | XRCC3 | Groups | |||||||||
| Codon 194 | Codon 399 | Codon 241 | CG | GC | OR (95%CI) P | CG | C1 | OR (95%CI) P | GC | C2 | OR (95%CI) P |
| All wide-type genotypes | |||||||||||
| Arg | Arg | Thr | 33 | 32 | 1.0 (reference) | 33 | 34 | 1.0 (reference) | 32 | 23 | 1.0 (reference) |
| One variant polymorphism | |||||||||||
| Trp | Arg | Thr | 5 | 8 | 0.6 (0.17-2.04) 0.5469 | 5 | 6 | 1.2 (0.32-4.16) 1.0000 | 8 | 6 | 1.1 (0.32-3.44) 1.0000 |
| Arg | Gln | Thr | 51 | 38 | 1.4 (0.68-2.50) 0.5124 | 51 | 42 | 0.8 (0.42-1.51) 0.5234 | 38 | 34 | 1.2 (0.61-2.56) 0.5918 |
| Arg | Arg | Met | 50 | 27 | 2.0 (0.91-3.57) 0.1237 | 50 | 49 | 1.0 (0.51-1.75) 1.0000 | 27 | 34 | 2,0 (0.83-3.70) 0.1426 |
| Two variant polymorphisms | |||||||||||
| Trp | Gln | Thr | 3 | 5 | 0.6 (0.13-2.63) 0.7106 | 3 | 2 | 0.6 (0.1-4.16) 1.0000 | 5 | 4 | 1.1 (0.16-4.54) 1.0000 |
| Arg | Gln | Met | 43 | 43 | 1.0 (0.50-1.85) 1.0000 | 43 | 58 | 1.4 (0.69-2.43) 0.4309 | 43 | 39 | 1.2 (0.63-2.50) 0.5389 |
| Trp | Arg | Met | 10 | 6 | 1.6 (0.52-5.00) 0.5771 | 10 | 6 | 0.6 (0.19-1.78) 0.4106 | 6 | 7 | 1.6 (0.48-5.55) 0.5999 |
| Three variant polymorphisms | |||||||||||
| Trp | Gln | Met | 8 | 1 | 8.3 (0.91-100) 0.0372 | 8 | 5 | 0.6 (0.17-2.04) 0.5400 | 1 | 3 | 5.0 (0.40-50.0) 0.3113 |
Figure 1.
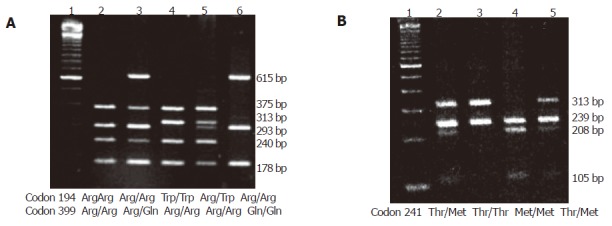
PCR-RFLP of XRCC1 and XRCC3 genes. A: XRCC1 gene. Lane 1: molecular weight marker. Lane 2: wild-type homozygous codons 194 and 399. Lane 3: wild-type homozygous codon 194 and heterozygous codon 399. Lane 4: mutant homozygous codon 194 and wild-type homozygous codon 399. Lane 5: heterozygous codon 194 and wild type-homozygous codon 399. Lane 6: wild-type homozygous codon 194 and mutant homozygous codon 399; B: XRCC3 gene. Lane 1: molecular weight marker. Lanes 2 and 5: heterozygous codon 241. Lane 3: wild-type homozygous codon 241. Lane 4: mutant homozygous codon 241.
Table 3 shows the associations of the different genotypes with the variables gender, smoking, drinking and H pylori infection, among the groups. For this analysis, we combined the heterozygous and mutant homozygous genotypes. Allele 194Trp was associated with smoking, with an increased OR for chronic gastritis (4.16, 95%CI = 1.16-16.66), when we compared the CG and C1 groups. The comparison between the CG and GC groups revealed that in men who were smokers and drinkers there was an association with increased OR’s for gastric cancer in the individuals with the alleles XRCC1 399Gln (3.03, 95%CI = 1.58-5.55; 2.27, 95%CI = 1.22-4.16 and 3.22, 95%CI = 1.75-5.88, respectively) and XRCC3 241Met (3.22, 95%CI = 1.66-6.25; 1.88, 95%CI = 1.01-3.57 and 2.63, 95%CI = 1.41-4.76, respectively). Comparing the CG and GC groups further, we found increased OR’s for gastric cancer in the association between H pylori-negative subjects and the alleles XRCC1 399Gln (0.26, 95%CI = 0.11-0.63) and XRCC3 241Met (0.36, 95%CI = 0.15-0.85). An association between smoking and drinking and the polymorphisms 399Gln (2.04, 95%CI = 1.17-3.57 and 2.63, 95%CI = 1.35-5.26, respectively) and 241Met (2.44, 95%CI = 1.42-4.16 and 2.13, 95%CI = 1.10-4.00, respectively) was observed when we compared the CG and C1 groups, with increased OR’s for chronic gastritis. Likewise, when we compared GC and C2, we observed an association between smoking and drinking, with an increased OR for stomach cancer, and alleles 399Gln (3.57, 95%CI = 1.85-6.66 and 5.55, 95%CI = 2.77-10.99) and 241Met (2.70, 95%CI = 1.38-5.26 and 4.34, 95%CI = 2.17-9.09). The other evaluated parameters as age and ethnicity, presented no association with the studied polymorphisms.
Table 3.
Association of heterozygous and mutant homozygous to the polymorphisms of codons 194 and 399 of the XRCC1 gene and to the codon 241 of the XRCC3 gene with demographic and environmental risk factors in chronic gastritis (CG) and gastric cancer (GC) patients and the respective control groups C1 and C2
| Groups |
Variable |
||||||||
|
Gender |
Smoke |
Drink |
H pylori infection |
||||||
| Male | Female | No | Yes | No | Yes | Positive | Negative | ||
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | ||
| XRCC1 | CG | 11 (42.3) | 15 (57.7) | 9 (34.6) | 17 (65.4) | 20 (77) | 6 (23) | ||
| Arg194Trp | |||||||||
| + | C1 | 10 (52.6) | 9 (47.4) | 13 (68.4) | 6 (31.6) | 17 (89.5) | 2 (10.5) | ||
| Trp194Trp | P = 0.4929 | P = 0.0250 | P = 0.4355 | ||||||
| XRCC1 | CG | 55 (53) | 49 (47) | 48 (46.2) | 56 (53.8) | 71 (68.3) | 33 (31.7) | 45 (53.6) | 39 (46.4) |
| Arg399Gln | GC | 67 (77) | 20 (23) | 24 (27.6) | 63 (72.4) | 35 (40.2) | 52 (59.8) | 9 (23.7) | 29 (76.3) |
| + | P = 0.0005 | P = 0.0084 | P = 0.0001 | P = 0.0021 | |||||
| Gln399Gln | CG | 55 (52.9) | 49 (47.1) | 48 (46.2) | 56 (53.8) | 71 (68.3) | 33 (31.7) | ||
| C1 | 52 (48.5) | 55 (51.4) | 68 (63.6) | 39 (36.4) | 91 (85) | 16 (15) | |||
| P = 0.5335 | P = 0.0111 | P = 0.0039 | |||||||
| GC | 67 (77) | 20 (23) | 24 (27.6) | 63 (72.4) | 35 (40.2) | 52 (59.8) | |||
| C2 | 56 (70) | 24 (30) | 46 (57.5) | 34 (42.5) | 63 (78.8) | 17 (21.2) | |||
| P = 0.3042 | P = 0.0001 | P = 0.0000 | |||||||
| XRCC3 | CG | 57 (51.8) | 53 (48.2) | 46 (41.8) | 64 (58.2) | 77 (70) | 33 (30) | 44 (52.4) | 40 (47.6) |
| Thr241Met | GC | 59 (77.6) | 17 (22.4) | 21 (27.6) | 55 (72.4) | 36 (47.4) | 40 (52.6) | 10 (28.6) | 25 (71.4) |
| + | P = 0.0004 | P = 0.0494 | P = 0.0019 | P = 0.0175 | |||||
| Met241Met | CG | 57 (51.8) | 53 (48.2) | 46 (41.8) | 64 (58.2) | 77 (47.4) | 33 (52.6) | ||
| C1 | 59 (50) | 59 (50) | 75 (63.6) | 43 (36.4) | 98 (79.6) | 20 (20.4) | |||
| P = 0.7838 | P = 0.0010 | P = 0.0110 | |||||||
| GC | 59 (77.6) | 17 (22.4) | 21 (27.6) | 55 (72.4) | 36 (47.4) | 40 (52.6) | |||
| C2 | 59 (71.1) | 24 (28.9) | 42 (50.6) | 41 (49.4) | 66 (79.6) | 17 (20.5) | |||
| P = 0.3458 | P = 0.0031 | P = 0.0000 | |||||||
DISCUSSION
There is increasing evidence that genetic variation leads to different DNA repair capacities in the human population. So, common polymorphisms can play a role in the individual genetic susceptibility to cancer[43]. Very few studies have investigated the role of polymorphisms of the DNA repair genes XRCC1 and XRCC3 in the risk of gastric cancer, and, to our knowledge, so far no study examined both gene polymorphisms in this type of cancer and in chronic gastritis. We conducted the first case-control study to investigate the relationship between the polymorphisms XRCC1 Arg194Trp and Arg399Gln and XRCC3 Thr241Met and the risk of chronic gastritis and gastric cancer, in a Southeastern Brazilian population.
Our data showed no association between the XRCC1 and XRCC3 polymorphisms and an increased risk for chronic gastritis and gastric cancer. To date, there are three published reports on investigations of the association between XRCC1 polymorphisms and gastric cancer risk[13,27,31], and only one that examined the influence of an XRCC3 polymorphism[29], with conflicting results. While Shen et al[13] found the wild-genotype Arg194Arg and the mutant-genotype Arg399Gln in the XRCC1 gene to be associated with an increased risk of gastric cardia cancer, Ratnasinghe et al[27] observed a significant reduction in the risk of this type of cancer, both in Chinese populations. Similarly, Lee et al[31] did not find any association with the risk of gastric cancer in a Korean population, but suggested that the 194Trp allele might be a protective allele with regard to gastric antral cancer. The only study that investigated the role of the XRCC3 polymorphism in gastric cancer in a Chinese population found no evidence of an association between this polymorphism and an increased risk of gastric cancer[29].
The differences observed in these reports may be due to the different types of gastric cancer studied (cardia and antrum), which may have a distinct pathogenesis and ethnical differences[31]. In the Brazilian population, most gastric cancers are located in the antral region, as in the Korean population, and are commonly associated with H. pylori infection. We also found that in the gastric cancer group the variant allele frequency of 194Trp was lower (0.06) and those of 399Gln and 241Met were higher (0.33 and 0.31, respectively) than those reported in Asian populations[13,27,29,31].
These differences can yet be due to the presence of variants of the common susceptibility polymorphisms, not just a single one, but DNA repair genes or activation and detoxification genes may jointly contribute to the susceptibility of gastric and other cancers. Thus, it is important to include more gene polymorphisms for the same or other DNA repair pathways to verify the gene-gene interactions, as well as the gene-environment interactions that may be important in the etiology of the disease.
When we assessed the association of XRCC1 Arg194trp and Arg399Gln and XRCC3 Thr241Met with the risk of chronic gastritis and gastric cancer, we found an increased risk for chronic gastritis when all three variant alleles were present at the same time, supporting the hypothesis of an additive effect of these three polymorphisms. There are no studies on chronic gastritis regarding its association with DNA repair gene polymorphisms, as there are for several metabolizing genes, such as GSTM1, GSTT1 and CYP2E1, in Brazilian[44] and Chinese[45] populations. Chronic gastritis, a frequent inflammation of the stomach[46] is considered a premalignant lesion[47]. Gastritis may start after an H pylori infection and progress over time from an initially superficial form to more severe forms, including severe atrophic gastritis with intestinal metaplasia[48]. About 10% of patients with gastric atrophy develop gastric cancer within a time period of 15 years[46]. Therefore, a reduced DNA repair capacity due to variant alleles may allow mutations to accumulate in the DNA of the epithelial cells of the stomach, resulting from the inflammatory process caused by H pylori or from environmental factors such as dietary habits and lifestyle, increasing the risk of gastric cancer.
Gastric cancer has a complex etiology in which genetic and environmental factors play an important role. In this study, we observed a statistically significant association between variant alleles and demographic and environmental factors such as gender, smoking, drinking and H pylori infection.
It is known that H pylori infection has a very important role in the development of chronic gastritis and in its development into gastric cancer[48]. Various mechanisms were proposed for H pylori-associated carcinogenesis, such as the formation of DNA adducts, the generation of free radicals, and a dysregulation of the gastric epithelial cell cycle[49,50]. So, these factors, associated with a decreased DNA repair capacity, may increase the risk for gastric cancer. Differently from these findings, we found a high frequency of variants 399Gln and 241Met in H pylori-negative gastric cancer patients, as compared to chronic gastritis patients. However, these data must have been influenced by the great number of H pylori-negative individuals, as compared to the H. pylori-positive individuals found in the gastric cancer group, probably due to an underestimate of the histological diagnosis in these patients.
In smokers, the presence of XRCC1 194Trp was more frequent in chronic gastritis cases, while polymorphisms XRCC1 399Gln and XRCC3 241Met were more frequent in both the chronic gastritis and the gastric cancer groups, compared with healthy controls. Whether the mechanisms of tobacco carcinogens act in human gastric cancer is currently uncertain. The main carcinogens contained in tobacco smoke include polyaromatic hydrocarbons (PAH), N-nitrosamines and aromatic amines. Cigarette smoking increases the number of single-strand breaks and DNA adducts, which, if left unrepaired, can lead to gene mutation[51]. These DNA damages can be repaired by BER, in which the XRCC1 protein has an important role. Functional studies of XRCC1 variants observed a significantly elevated level of sister chromatid exchange (SCE) in peripheral blood lymphocytes after in vitro exposure to the tobacco-specific NNK in carriers of the 399Gln allele, but the same was not observed for the 194Trp polymorphism[52]. Duell et al[53] reported higher frequencies of SCE for current smokers with the 399Gln polymorphism than for smokers with the Arg/Arg genotype.
Protein XRCC3 participates in the DSB repair by the homologous repair pathway and the 241Met variant may lead to biological implications for the enzyme’s function and/or the interaction with other proteins involved in DNA damage repair. Matullo et al[54] associated the 241Met polymorphism with 32P-DNA adduct levels, indicating a possible role of the XRCC3 gene in the repair of bulky DNA adducts. Thus, variations in DNA repair capacity caused by polymorphisms of DNA repair genes may modulate the genotoxic effect of tobacco smoking.
Excessive alcohol consumption can also lead to DNA damage through the production of free radical intermediates, such as reactive oxygen species, which are produced during the ethanol metabolism[55]. The frequency of DNA single-strand breaks also increases with chronic exposure to alcohol[56]. We observed an association with alcohol consumption in the patients with chronic gastritis and gastric cancer, and found an increased risk for these diseases when polymorphisms XRCC1 399Gln and XRCC3 241Met were present.
In conclusion, in the Brazilian population studied, we did not find evidence of a relationship between the polymorphisms XRCC1 Arg194Trp and Arg399Gln and XRCC3 Thr241Met and the development of chronic gastritis and gastric cancer. However, intra- and inter-gene interactions may contribute to the development of chronic gastritis, a precursor lesion of stomach cancer. We also verified a gene-environment interaction between the XRCC1 and XRCC3 polymorphisms, mainly with the habits of smoking and drinking, in the chronic gastritis and gastric cancer patients. Our study is an important addition to the small number of previously published reports on DNA repair gene variants in gastric cancer and shows the need for further studies in different populations, to elucidate the role of these polymorphisms in carcinogenesis.
ACKNOWLEDGMENTS
We thank Prof. Dr. Antonio José Manzato for assistance with the statistical analysis.
Footnotes
Supported by Brazilian Agency CAPES
Science Editor Guo SY Language Editor Elsevier HK
References
- 1.Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat Res. 2000;462:129–135. doi: 10.1016/s1383-5742(00)00024-7. [DOI] [PubMed] [Google Scholar]
- 2.Mohrenweiser HW, Jones IM. Variation in DNA repair is a factor in cancer susceptibility: a paradigm for the promises and perils of individual and population risk estimation? Mutat Res. 1998;400:15–24. doi: 10.1016/s0027-5107(98)00059-1. [DOI] [PubMed] [Google Scholar]
- 3.Shields PG, Harris CC. Cancer risk and low-penetrance susceptibility genes in gene-environment interactions. J Clin Oncol. 2000;18:2309–2315. doi: 10.1200/JCO.2000.18.11.2309. [DOI] [PubMed] [Google Scholar]
- 4.Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–608. [PubMed] [Google Scholar]
- 5.Ishida T, Takashima R, Fukayama M, Hamada C, Hippo Y, Fujii T, Moriyama S, Matsuba C, Nakahori Y, Morita H, et al. New DNA polymorphisms of human MMH/OGG1 gene: prevalence of one polymorphism among lung-adenocarcinoma patients in Japanese. Int J Cancer. 1999;80:18–21. doi: 10.1002/(sici)1097-0215(19990105)80:1<18::aid-ijc4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 6.Butkiewicz D, Rusin M, Harris CC, Chorazy M. Identification of four single nucleotide polymorphisms in DNA repair genes: XPA and XPB (ERCC3) in Polish population. Hum Mutat. 2000;15:577–578. doi: 10.1002/1098-1004(200006)15:6<577::AID-HUMU11>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Chavanne F, Broughton BC, Pietra D, Nardo T, Browitt A, Lehmann AR, Stefanini M. Mutations in the XPC gene in families with xeroderma pigmentosum and consequences at the cell, protein, and transcript levels. Cancer Res. 2000;60:1974–1982. [PubMed] [Google Scholar]
- 8.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular 'nick-sensor' in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: an update. Toxicology. 2003;193:3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 10.Marintchev A, Mullen MA, Maciejewski MW, Pan B, Gryk MR, Mullen GP. Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nat Struct Biol. 1999;6:884–893. doi: 10.1038/12347. [DOI] [PubMed] [Google Scholar]
- 11.Sturgis EM, Castillo EJ, Li L, Zheng R, Eicher SA, Clayman GL, Strom SS, Spitz MR, Wei Q. Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and neck. Carcinogenesis. 1999;20:2125–2129. doi: 10.1093/carcin/20.11.2125. [DOI] [PubMed] [Google Scholar]
- 12.Abdel-Rahman SZ, Soliman AS, Bondy ML, Omar S, El-Badawy SA, Khaled HM, Seifeldin IA, Levin B. Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett. 2000;159:79–86. doi: 10.1016/s0304-3835(00)00537-1. [DOI] [PubMed] [Google Scholar]
- 13.Shen H, Xu Y, Qian Y, Yu R, Qin Y, Zhou L, Wang X, Spitz MR, Wei Q. Polymorphisms of the DNA repair gene XRCC1 and risk of gastric cancer in a Chinese population. Int J Cancer. 2000;88:601–606. doi: 10.1002/1097-0215(20001115)88:4<601::aid-ijc13>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 14.Lee JM, Lee YC, Yang SY, Yang PW, Luh SP, Lee CJ, Chen CJ, Wu MT. Genetic polymorphisms of XRCC1 and risk of the esophageal cancer. Int J Cancer. 2001;95:240–246. doi: 10.1002/1097-0215(20010720)95:4<240::aid-ijc1041>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 15.Yu HP, Zhang XY, Wang XL, Shi LY, Li YY, Li F, Su YH, Wang YJ, Lu B, Sun X, et al. DNA repair gene XRCC1 polymorphisms, smoking, and esophageal cancer risk. Cancer Detect Prev. 2004;28:194–199. doi: 10.1016/j.cdp.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Duell EJ, Millikan RC, Pittman GS, Winkel S, Lunn RM, Tse CK, Eaton A, Mohrenweiser HW, Newman B, Bell DA. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:217–222. [PubMed] [Google Scholar]
- 17.Divine KK, Gilliland FD, Crowell RE, Stidley CA, Bocklage TJ, Cook DL, Belinsky SA. The XRCC1 399 glutamine allele is a risk factor for adenocarcinoma of the lung. Mutat Res. 2001;461:273–278. doi: 10.1016/s0921-8777(00)00059-8. [DOI] [PubMed] [Google Scholar]
- 18.Zhou W, Liu G, Miller DP, Thurston SW, Xu LL, Wain JC, Lynch TJ, Su L, Christiani DC. Polymorphisms in the DNA repair genes XRCC1 and ERCC2, smoking, and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:359–365. [PubMed] [Google Scholar]
- 19.Olshan AF, Watson MA, Weissler MC, Bell DA. XRCC1 polymorphisms and head and neck cancer. Cancer Lett. 2002;178:181–186. doi: 10.1016/s0304-3835(01)00822-9. [DOI] [PubMed] [Google Scholar]
- 20.Han J, Hankinson SE, Colditz GA, Hunter DJ. Genetic variation in XRCC1, sun exposure, and risk of skin cancer. Br J Cancer. 2004;91:1604–1609. doi: 10.1038/sj.bjc.6602174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 22.Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ. Evidence for simultaneous protein interactions between human Rad51 paralogs. J Biol Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- 23.Winsey SL, Haldar NA, Marsh HP, Bunce M, Marshall SE, Harris AL, Wojnarowska F, Welsh KI. A variant within the DNA repair gene XRCC3 is associated with the development of melanoma skin cancer. Cancer Res. 2000;60:5612–5616. [PubMed] [Google Scholar]
- 24.Matullo G, Guarrera S, Carturan S, Peluso M, Malaveille C, Davico L, Piazza A, Vineis P. DNA repair gene polymorphisms, bulky DNA adducts in white blood cells and bladder cancer in a case-control study. Int J Cancer. 2001;92:562–567. doi: 10.1002/ijc.1228. [DOI] [PubMed] [Google Scholar]
- 25.Smith TR, Miller MS, Lohman K, Lange EM, Case LD, Mohrenweiser HW, Hu JJ. Polymorphisms of XRCC1 and XRCC3 genes and susceptibility to breast cancer. Cancer Lett. 2003;190:183–190. doi: 10.1016/s0304-3835(02)00595-5. [DOI] [PubMed] [Google Scholar]
- 26.Jacobsen NR, Raaschou-Nielsen O, Nexø B, Wallin H, Overvad K, Tjønneland A, Vogel U. XRCC3 polymorphisms and risk of lung cancer. Cancer Lett. 2004;213:67–72. doi: 10.1016/j.canlet.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 27.Ratnasinghe LD, Abnet C, Qiao YL, Modali R, Stolzenberg-Solomon R, Dong ZW, Dawsey SM, Mark SD, Taylor PR. Polymorphisms of XRCC1 and risk of esophageal and gastric cardia cancer. Cancer Lett. 2004;216:157–164. doi: 10.1016/j.canlet.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Duan Z, Shen H, Lee JE, Gershenwald JE, Ross MI, Mansfield PF, Duvic M, Strom SS, Spitz MR, Wei Q. DNA repair gene XRCC3 241Met variant is not associated with risk of cutaneous malignant melanoma. Cancer Epidemiol Biomarkers Prev. 2002;11:1142–1143. [PubMed] [Google Scholar]
- 29.Shen H, Wang X, Hu Z, Zhang Z, Xu Y, Hu X, Guo J, Wei Q. Polymorphisms of DNA repair gene XRCC3 Thr241Met and risk of gastric cancer in a Chinese population. Cancer Lett. 2004;206:51–58. doi: 10.1016/j.canlet.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Sanyal S, Festa F, Sakano S, Zhang Z, Steineck G, Norming U, Wijkström H, Larsson P, Kumar R, Hemminki K. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- 31.Lee SG, Kim B, Choi J, Kim C, Lee I, Song K. Genetic polymorphisms of XRCC1 and risk of gastric cancer. Cancer Lett. 2002;187:53–60. doi: 10.1016/s0304-3835(02)00381-6. [DOI] [PubMed] [Google Scholar]
- 32.INCA-Instituto Nacional do Cancer, Ministerio da Saude. Estimativa 2005-Estimativas de incidencia por cancer no Brasil, 2004. Disponivel em. 2002. Available from: http: //www.inca.org.br.
- 33.Kobayashi M, Tsubono Y, Sasazuki S, Sasaki S, Tsugane S. Vegetables, fruit and risk of gastric cancer in Japan: a 10-year follow-up of the JPHC Study Cohort I. Int J Cancer. 2002;102:39–44. doi: 10.1002/ijc.10659. [DOI] [PubMed] [Google Scholar]
- 34.Sasazuki S, Sasaki S, Tsugane S. Cigarette smoking, alcohol consumption and subsequent gastric cancer risk by subsite and histologic type. Int J Cancer. 2002;101:560–566. doi: 10.1002/ijc.10649. [DOI] [PubMed] [Google Scholar]
- 35.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 36.Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–3215. doi: 10.1091/mbc.E02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tahara E. Genetic pathways of two types of gastric cancer. IARC Sci Publ. 2004;157:327–349. [PubMed] [Google Scholar]
- 38.Schistosomes , liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241. [Google Scholar]
- 39.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 40.Price AB. The Sydney System: histological division. J Gastroenterol Hepatol. 1991;6:209–222. doi: 10.1111/j.1440-1746.1991.tb01468.x. [DOI] [PubMed] [Google Scholar]
- 41.Abdel-Rahman SZ, Nouraldeen AM, Ahmed AE. Molecular interaction of [2,3-14C] acrylonitrile with DNA in gastric tissue of rat. J Biochem Toxicol. 1994;9:191–198. doi: 10.1002/jbt.2570090404. [DOI] [PubMed] [Google Scholar]
- 42.David-Beabes GL, Lunn RM, London SJ. No association between the XPD (Lys751G1n) polymorphism or the XRCC3 (Thr241Met) polymorphism and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:911–912. [PubMed] [Google Scholar]
- 43.Hu JJ, Mohrenweiser HW, Bell DA, Leadon SA, Miller MS. Symposium overview: genetic polymorphisms in DNA repair and cancer risk. Toxicol Appl Pharmacol. 2002;185:64–73. doi: 10.1006/taap.2002.9518. [DOI] [PubMed] [Google Scholar]
- 44.Colombo J, Rossit AR, Caetano A, Borim AA, Wornrath D, Silva AE. GSTT1, GSTM1 and CYP2E1 genetic polymorphisms in gastric cancer and chronic gastritis in a Brazilian population. World J Gastroenterol. 2004;10:1240–1245. doi: 10.3748/wjg.v10.i9.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Setiawan VW, Zhang ZF, Yu GP, Li YL, Lu ML, Tsai CJ, Cordova D, Wang MR, Guo CH, Yu SZ, et al. GSTT1 and GSTM1 null genotypes and the risk of gastric cancer: a case-control study in a Chinese population. Cancer Epidemiol Biomarkers Prev. 2000;9:73–80. [PubMed] [Google Scholar]
- 46.Cheli R, Giacosa A. Chronic atrophic gastritis and gastric mucosal atrophy--one and the same. Gastrointest Endosc. 1983;29:23–25. doi: 10.1016/s0016-5107(83)72493-4. [DOI] [PubMed] [Google Scholar]
- 47.Genta RM. Review article: Gastric atrophy and atrophic gastritis--nebulous concepts in search of a definition. Aliment Pharmacol Ther. 1998;12 Suppl 1:17–23. doi: 10.1111/j.1365-2036.1998.00003.x. [DOI] [PubMed] [Google Scholar]
- 48.Correa P. Helicobacter pylori and gastric carcinogenesis. Am J Surg Pathol. 1995;19 Suppl 1:S37–S43. [PubMed] [Google Scholar]
- 49.Moss SF. The carcinogenic effect of H. pylori on the gastric epithelial cell. J Physiol Pharmacol. 1999;50:847–856. [PubMed] [Google Scholar]
- 50.Zhang ZW, Patchett SE, Farthing MJ. Role of Helicobacter pylori and p53 in regulation of gastric epithelial cell cycle phase progression. Dig Dis Sci. 2002;47:987–995. doi: 10.1023/a:1015069519610. [DOI] [PubMed] [Google Scholar]
- 51.Newcomb PA, Carbone PP. The health consequences of smoking. Cancer. Med Clin North Am. 1992;76:305–331. doi: 10.1016/s0025-7125(16)30355-8. [DOI] [PubMed] [Google Scholar]
- 52.Abdel-Rahman SZ, El-Zein RA. The 399Gln polymorphism in the DNA repair gene XRCC1 modulates the genotoxic response induced in human lymphocytes by the tobacco-specific nitrosamine NNK. Cancer Lett. 2000;159:63–71. doi: 10.1016/s0304-3835(00)00532-2. [DOI] [PubMed] [Google Scholar]
- 53.Duell EJ, Wiencke JK, Cheng TJ, Varkonyi A, Zuo ZF, Ashok TD, Mark EJ, Wain JC, Christiani DC, Kelsey KT. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cells. Carcinogenesis. 2000;21:965–971. doi: 10.1093/carcin/21.5.965. [DOI] [PubMed] [Google Scholar]
- 54.Matullo G, Palli D, Peluso M, Guarrera S, Carturan S, Celentano E, Krogh V, Munnia A, Tumino R, Polidoro S, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 55.Brooks PJ. DNA damage, DNA repair, and alcohol toxicity--a review. Alcohol Clin Exp Res. 1997;21:1073–1082. [PubMed] [Google Scholar]
- 56.Daiker DH, Shipp BK, Schoenfeld HA, Klimpel GR, Witz G, Moslen MT, Ward JB. Effect of CYP2E1 induction by ethanol on the immunotoxicity and genotoxicity of extended low-level benzene exposure. J Toxicol Environ Health A. 2000;59:181–196. doi: 10.1080/009841000156961. [DOI] [PubMed] [Google Scholar]


