Abstract
Background
The methylenetetrahydrofolate reductase (MTHFR) is thought to be involved in the development of nonsyndromic cleft lip with or without cleft palate (NSCL/P). However, conflicting results have been obtained when evaluating the association between maternal MTHFR C677T and A1298C polymorphisms and the risk of NSCL/P. In light of this gap, a meta-analysis of all eligible case-control studies was conducted in the present study.
Materials and Methods
A total of 15 case-control studies were ultimately identified after a comprehensive literature search and Hardy-Weinberg equilibrium (HWE) examination. Cochrane’s Q test and index of heterogeneity (I2) indicated no obvious heterogeneity among studies.
Results
Fixed or random-effects models were used to calculate the pooled odds ratios (ORs). The results showed that the TT genotype in mothers increased the likelihood of having NSCL/P offspring 1.25 times (95% CI: 1.047-1.494) more than the CC homozygotes. Meanwhile, maternal TT genotype increased the risk of producing NSCL/P offspring in recessive model (OR=1.325, 95% CI: 1.124-1.562). However, the CT heterozygote and the CT+TT dominant models had no association with NSCL/P offspring compared with the CC wild-type homozygote model. Subgroup analyses based on ethnicity indicated that maternal TT genotype increased the likelihood of having NSCL/P offspring in Whites (OR=1.308, 95% CI: 1.059-1.617) and Asians (OR=1.726, 95% CI: 1.090-2.733) in recessive model. Also, subgroup analyses based on source of control showed that mothers with the 677TT genotype had a significantly increased susceptibility of having NSCL/P children in hospital based population (HB) when compared with CC homozygotes (OR=1.248, 95% CI: 1.024-1.520) and un- der the recessive model (OR=1.324, 95% CI: 1.104-1.588). Furthermore, maternal A1298C polymorphism had no significant association with producing NSCL/P offspring (dominant model OR=0.952, 95% CI: 0.816-1.111, recessive model OR=0.766, 95% CI: 0.567-1.036).
Conclusion
MTHFR C677T polymorphism is associated with the risk of generating NSCL/P offspring, and being a 677TT homozygote is a risk factor. MTHFR A1298C polymorphism was not associated with generating NSCL/P offspring. However, further work should be performed to confirm these findings.
Keywords: Methylenetetrahydrofolate Reductase, Cleft Lip, Meta-Analysis
Introduction
Cleft lip and palate is one of the most common congenital defects in humans (1), which is divided into two groups in genetics as syndromic and nonsyndromic (2). Of all patients with cleft lip and palate, only small portion are syndromic and most are nonsyndromic (3, 4). Based on clinical manifestations, nonsyndromic cleft lip and palate can be divided into nonsyndromic cleft lip with or without cleft palate (NSCL/P,OMIM 119530) and the palate only (CPO, OMIM 119540) (2). NSCL/P is a congenital facial malformation without any other structural or developmental abnormalities (1), and is different from CPO in embryologic origin and recurrence risks (5).
Although the etiology of NSCL/P is complex (6), numerous studies have reported that NSCL/P is associated with folate metabolism (7-10), and genes which encode key proteins of folate and methionine metabolism play a role in the susceptibility of NSCL/P (11). Thus, the gene encoding the methylenetetrahydrofolate reductase (MTHFR) enzyme is particularly attractive, because this enzyme is responsible for folate-dependent metabolism of homocysteine, which catalyses the reduction of 5, 10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, the predominant circulatory form of folate and the carbon donor for the remethylation of homocysteine to methionine (12).
The gene encoding the MTHFR enzyme is known to have at least two functional polymorphisms namely, C677T (rs1801133) and A1298C (rs1801131) for which their roles in the mechanisms of folate enzyme have been extensively investigated (12-16). The 677T allele results in an alanine to valine substitution at codon 222 (A222V), resulting in a thermolabile enzyme with 70% reduction in specific catalytic activity (13, 14). Similarly, the 1298C allele results in a glutamic acid to alanine substitution at codon 429 (G429A), resulting in a 40% reduction of MTHFR activity in vivo (15, 16). The low MTHFR activity caused by MTHFR polymorphisms maybe results in higher homocysteine or lower plasma folate levels, which both are associated with many diseases such as Down’s syndrome and neural tube defect (15, 17).
It has been hypothesized that NSCL/P may be associated with MTHFR which encodes a key protein in folate and methionine metabolism (11). Thus, MTHFR has been wildly studied to examine the relationship between its polymorphisms and the risk of NSCL/P but conflicting results were reported. Some studies found that the MTHFR C677T variant is associated with NSCL/P (18, 19). However, other studies, carried out in various populations around the world, found no or variable association (7, 20-24). Some studies indicated that the genotype of infants at C677T made a major contribution to the occurrence of NSCL/P (25-27) but others did not (28-31). Some investigations showed that the maternal genotype for the MTHFR C677T polymorphism had a significant impact on the occurrence of NSCL/P in their offspring (25, 32-37), but, other studies did not support this finding (29, 38-41).
Similarly, studies on the MTHFR A1298C polymorphism also yielded inconsistent results. The effect of MTHFR A1298C polymorphism was diverse from being a risk factor (18, 33) to no risk at all (7, 19, 23). Neither infant (26, 29, 42) nor maternal (40) MTHFR A1298C polymorphism obtained positive association with NSCL/P risk. In light of this gap, a meta-analysis on infant MTHFR polymorphisms and NSCL/P susceptibility was performed by Pan et al. (43), which suggested that infant MTHFR C677T polymorphism was involved in the development of NSCL/P. However, whether maternal MTHFR polymorphisms related to having NSCL/P children is yet to be confirmed. To resolve this confusion, we did a meta-analysis focusing on maternal MTHFR polymorphisms and the risk of having offspring with NSCL/P.
Materials and Methods
Study question
Are maternal MTHFR polymorphisms (C677T or A1298C) risk factors for having a child with NSCL/P?
Criteria for included studies
1. Explored the association of maternal MTHFR polymorphisms and NSCL/P children. 2. Casecontrol study design; considering the heterogeneity in different study designs, we only focused on casecontrol studies. Cross-sectional studies, case-parent triads, and transmission disequilibrium tests (TDT) designed studies were not included. 3. Cases were mothers who have children with NSCL/P. Control group were mothers without NSCL/P children and selected from the general population or hospital based population. 4. Provided distributions of the maternal MTHFR C677T and/or A1298C genotypes. 5. Control groups in studies did not deviate from Hardy-Weinberg equilibrium (HWE).
We did not consider the genotyping method. We only included information available from the publications and did not seek additional information by contacting primary authors. Studies were excluded if the disease was defined as either familial NSCL/P or orofacial clefts. Case reports, letters and review articles were excluded from the study.
Strategies for identified studies
A comprehensive literature search was performed using PubMed, Springer, Elsevier Digital Dissertations Databases, Scopus, and ISI web of knowledge with MeSH terms retrieval and free words retrieval for relevant articles published in English up to 15th March 2013. MeSH terms included "cleft lip" and "methylenetetrahydrofolate reductase (nadph2)". Free words included "cleft lip", "cleft", "lip", "methylenetetrahydrofolate" and "MTHFR". We extended our search to review the reference lists of retrieved articles and performed manual searching as a supplement. When a study had duplicate publication, only the most inclusive publication was considered. The full texts of candidate articles were examined by two investigators independently.
Data extraction
Data from each eligible study were extracted on custom-made data collection forms by two authors independently. For inconsistent evaluations, agreements were reached following discussion in our study group. For each study, the following characteristics were collected: first author, publication year, country, source of controls, characteristics of study population (ethnicity, sample size of case and control), and distributions of the maternal MTHFR C677T or A1298C genotypes among cases and controls. For studies with multiple gene polymorphisms, only data concerning MTHFR polymorphisms were included in the analysis. No modification of original data was performed. In addition, HWE was calculated based on the genotypes among controls. The data were extracted from published manuscripts, thus no research ethics board approval was necessary.
Statistical analysis
Stata 8.0 (Stata Corporation, College Station, TX) was used to perform all statistical analyses. Goodness-of-fit chi-squared test was used to assess the frequencies of MTHFR C677T and A1298C polymorphisms from expectation under HWE in controls. The strength of the association between the polymorphisms and NSCL/P was measured by ORs with 95% confidence intervals (CIs). The statistical significance of the observed OR was tested by Z-test. For the C677T polymorphism, we firstly used wild-type CC genotype as the reference group to evaluate the effects of the CT and TT genotypes on NSCL/P susceptibility. Then we estimated the effects of CT/TT versus CC and TT versus CT/CC assuming the dominant and recessive models of the T allele. Same procedures were also performed on the A1298C polymorphism. Furthermore, subgroup analyses were performed based on source of controls and ethnicity. Heterogeneity assumption between studies was examined by the Chi-square based on Q-test and I-square value (44). A fixedeffect model using the Mantel–Haenszel method was selected to pool data if there was homogeneity (p>0.05), otherwise, the random-effects model, using the Der Simonian and Laird method, was conducted.
Sensitivity analyses using the step by step exclusion method were conducted to assess whether each individual study affected the final results. Publication bias was evaluated by Begg’s test, the Egger’s asymmetry test and visual inspection of funnel plots. All the P values were for a twosided test and p<0.05 was considered statistically significant.
Sensitivity analyses using the step by step exclusion method were conducted to assess whether each individual study affected the final results. Publication bias was evaluated by Begg’s test, the Egger’s asymmetry test and visual inspection of funnel plots. All the P values were for a twosided test and p<0.05 was considered statistically significant.
Results
Description of studies
Fifteen case-control studies were identified via our search strategy, for which 8 were concerned with C677T exclusively, while 7 studies had analyzed both variants (Fig 1).
Fig 1.
A flow diagram for selection of studies and specific reasons for exclusion in this meta-analysis. NSCL/P; Nonsyndromic cleft lip with or without cleft palate, HWE; Hardy-Weinberg equilibrium and MTHFR; Methylenetetrahydrofolate reductase.
The characteristic information of the included studies such as first author, publication year, country, ethnicity, source of controls, sample size (case/control), genotype distributions (case/ control) are presented in table 1 and table 2. Little’s study was excluded because the genotype distribution among the control group was not in HWE (p<0.05) (45).
Table 1.
Main characteristics of MTHFR C677T polymorphism studies included in the meta-analysis
| Case | Control | Genotype (case/control) | Allele (case/control) | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First author | Year | Country | Ethnicity | Source of | n | n | CC | CT | TT | C n (%) | T n (%) | (P)## |
| Brandalizeet al. (21) | 2007 | Brazil | White | HB | 110 | 100 | 44/38 | 45/52 | 21/10 | 133(60)/128(64) | 87(40)/72(36) | 0.20 |
| Littleet al. (45)a | 2008 | England | White | PB | 96 | 226 | 46/86 | 42/119 | 8/21 | 134(70)/291(64)# | 58(30)/161(36) | 0.03 |
| Guo et al.(22) | 2009 | China | Asian | HB | 97 | 102 | 26/20 | 49/57 | 22/25 | 101(52)/97(48) | 93(48)/107(52) | 0.22 |
| Wanget al. (35) | 2012 | China | Asian | HB | 89 | 64 | 10/15 | 41/39 | 38/10 | 61(34)/69(54) | 117(66)/59(46) | 0.07 |
| Chornaet al. (25) | 2011 | Ukraine | European | PB | 27 | 50 | 19/34 | 2/2 | 6/14 | 40(74)/70(70) | 14(26)/30(30) | 0.05 |
| Ali et al.(19) | 2009 | India | Asian | PB | 116 | 214 | 78/176 | 37/36 | 1/2 | 193(83)/388(91) | 39(17)/40(9) | 0.92 |
| Bufalinoet al. (36) | 2010 | Brazil | Mixed | HB | 106 | 184 | 49/95 | 50/72 | 7/17 | 148(70)/262(71) | 64(30)/106(29) | 0.53 |
| Gasparet al. (11) | 2004 | Brazil | Mixed | HB | 336 | 644 | 174/327 | 131/269 | 31/48 | 479(71)/923(72) | 193(29)/365(28) | 0.47 |
| Whites | HB | 235 | 474 | 126/235 | 88/202 | 21/37 | 340(72)/672(71)# | 130(28)/276(29)# | 0.48 | |||
| Nonwhites | HB | 77 | 90 | 40/43 | 31/39 | 6/8 | 111(72)/125(69)# | 43(28)/55(31)# | 0.84 | |||
| Unclassified | HB | 24 | 80 | 8/49 | 12/28 | 4/3 | 28(58)/126(79)# | 20(42)/34(21)# | 0.68 | |||
| Gasparet al. (46) | 1999 | Brazil | White | HB | 59 | 90 | 30/37 | 19/40 | 10/13 | 79 (67)/114 (63) | 39 (33)/66 (37) | 0.68 |
| Millset al. (37) | 2008 | Ireland | White | HB | 465 | 1599 | 205/715 | 212/721 | 48/163 | 622(67)/2151(67)# | 308(33)/1047(33)# | 0.34 |
| Mostowskaet al. (47) | 2006 | Poland | European | PB | 121 | 81 | 60/42* | 46/33* | 15/6* | 166(69)/117(72) | 76 (31)/45 (28) | 0.89 |
| Pezzettiet al. (34) | 2004 | Italy | White | HB | 104 | 289 | 27/95 | 47/151 | 30/43 | 101(49)/341(59) | 107(51)/237(41) | 0.17 |
| Shotelersuket al. (33) | 2003 | Thailand | Asian | PB | 67 | 202 | 46/154 | 19/46 | 2/2 | 111(83)/354(88) | 23(17)/50(12) | 0.48 |
| Sozenet al. (23) | 2009 | Venezuela | Mixed | PB | 168 | 138 | 109/66 | 49/65 | 10/7 | 267(79)/197(71) | 69(21)/79(29) | 0.07 |
| Van Rooijet al. (7) | 2003 | Netherlands | White | PB | 148 | 170 | 78/84 | 55/74 | 15/12 | 211(71)/242(71)# | 85(28)/98(29)# | 0.43 |
| Tolarovaet al. (48) | 1998 | Argentina | White | PB | 93 | 84 | 39/39 | 37/33 | 17/12 | 115(62)/111(66)# | 71(38)/57(34)# | 0.26 |
PB; Population based, HB; Hospital based, HWE; Hardye Weinberg equilibrium, NA; Not available, a; Not enter final analysis because not fit HWE, *; Numbers calculated by text describe, #; Numbers calculated by the distribution of genotype and ##; P value of HWE were calculated by original data.
Table 2.
Main characteristics of MTHFR A1298C polymorphism studies included in the meta-analysis
| Case | Control | Genotype (case/control) | Allele (case/control) | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Firstauthor | Year | Country | Ethnicity | Source ofcontrols | n | n | AA | AC | CC | A n (%) | C n (%) | (P)## |
| Van Rooijet al. (7) | 2003 | Netherlands | White | PB | 125 | 159 | 57/76 | 52/67 | 16/16 | 166(66)/219(69)# | 84(34)/99(31)# | 0.83 |
| Shotelersuket al. (33) | 2003 | Thailand | Asian | PB | 67 | 202 | 30/108 | 33/80 | 4/14 | 93(69)/296(73) | 41(31)/108(27) | 0.88 |
| Pezzettiet al. (34) | 2004 | Italy | White | HB | 104 | 254 | 57/121 | 36/130 | 11/38 | 150(72)/372(64) | 58(28)/206(36) | 0.74 |
| Mills et al.(37) | 2008 | Ireland | White | HB | 366 | 1050 | 179/519 | 164/439 | 23/92 | 522(71)/1477(70)# | 210(29)/623(30)# | 0.95 |
| Ali et al.(19) | 2009 | India | Asian | PB | 116 | 214 | 64/99 | 47/97 | 5/18 | 175(75)/295(69) | 57(25)/133(31) | 0.4 |
| Sozen et al.(23) | 2009 | Venezuela | Mixed | PB | 168 | 138 | 119/101 | 47 /33 | 2 /4 | 285(85)/235(85) | 51(15)/41(15) | 0.52 |
| Tolarovaet al. (48) | 1998 | Argentina | White | PB | 86 | 78 | 56/50 | 27/25 | 3/3 | 114(78)/125(80)# | 33(22)/31(20)# | 0.95 |
PB; Population based, HB; Hospital based, HWE; Hardye Weinberg equilibrium, NA; Not available, #; Numbers calculated by the distribution of genotype and ##; P value of HWE were calculated by original data.
Heterogeneity test
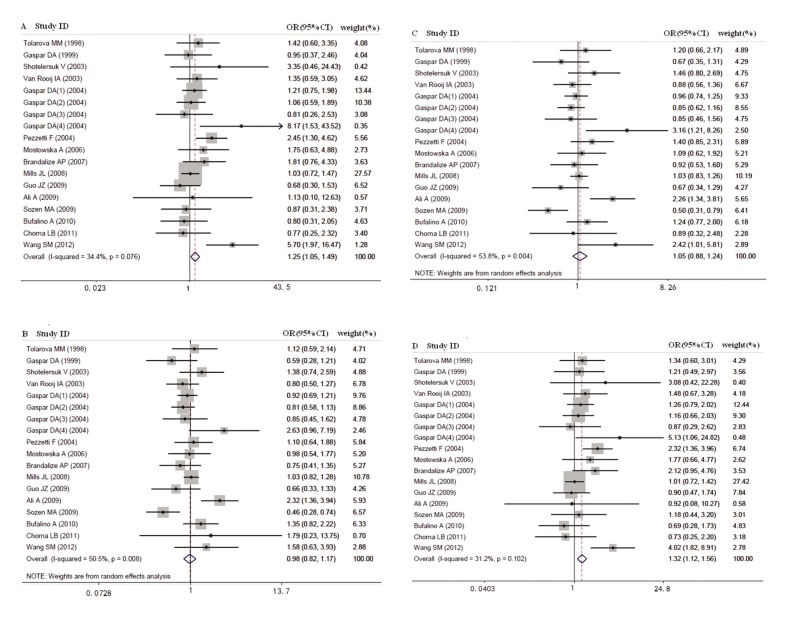
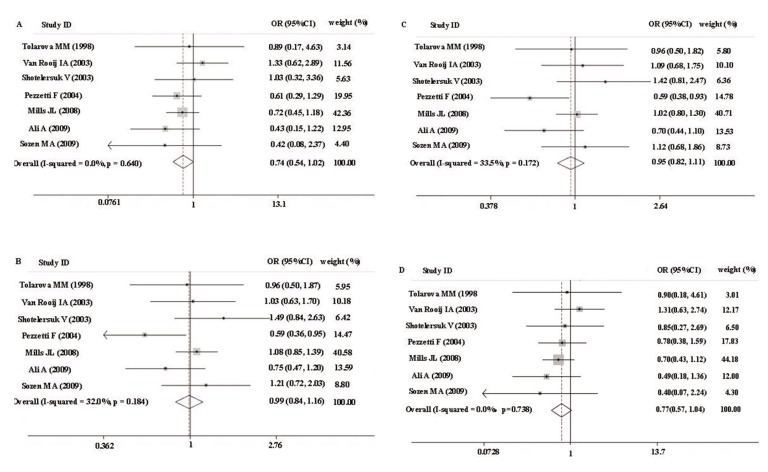
Overall, for maternal MTHFR C677T polymorphism, there were significant heterogeneity for heterozygote comparison (CT versus CC: Ph=0.008) and dominant model comparison (CT+TT versus CC: Ph=0.004), but not for the homozygote comparison (TT versus CC: Ph=0.076) and the recessive model comparison (TT versus CT+CC: Ph=0.102) (Table 3, Fig 2). Thus, we performed subgroup analysis stratified by source of controls and ethnicity to assess the cause of heterogeneity. Results suggested that ethnicity and source of controls were contributing to substantial heterogeneity. In A1298C polymorphism studies, there were no significant heterogeneity for all comparisons (all Ph>0.05, Table 4, Fig 3).
Table 3.
Results of Meta-analysis of MTHFR C677T polymorphism for studies on NSCL/P
| Groups | Studynumber | Sample size(case/control) | TT versus CC | CT versus CC | CT+TT versus CC (dominant) | TT versus CT+CC (recessive) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95% CI) | I2(%) | Ph | OR (95% CI) | I2(%) | Ph | OR (95% CI) | I2(%) | Ph | OR(95% CI) | I2(%) | Ph | ||||
| Overall | 15 | 2442/4655 | 1.251(1.047,1.494) | 34.4 | 0.076 | 0.982(0.824,1.171) | 50.5 | 0.008# | 1.048(0.884, 1.244) | 53.8 | 0.004# | 1.325(1.124, 1.562) | 31.2 | 0.102 | |
| Ethnicity | |||||||||||||||
| Asians | 4 | 369/582 | 1.565(0.887,2.761) | 71.1 | 0.016# | 1.378(0.796, 2.385) | 62.2 | 0.047# | 1.505(0.849, 2.667) | 67.5 | 0.026# | 1.726(1.090, 2.733) | 65.5 | 0.034# | |
| Whites | 8* | 1269/2853 | 1.243(0.992,1.558) | 13.2 | 0.327 | 0.920(0.794, 1.066) | 0.0 | 0.704 | 0.973(0.847, 1.118) | 0 | 0.712 | 1.308(1.059, 1.617) | 27.7 | 0.207 | |
| Others | 6* | 804/1220 | 1.172(0.839,1.639) | 26.1 | 0.238 | 0.969(0.666, 1.410) | 66.6 | 0.010# | 1.010(0.702, 1.452) | 68 | 0.008# | 1.194(0.865, 1.648) | 1.9 | 0.404 | |
| Source of control | |||||||||||||||
| HB | 8 | 1702/3716 | 1.248(1.024,1.520) | 56.9 | 0.01# | 0.957(0.821, 1.114) | 17.3 | 0.279 | 1.026(0.861, 1.223) | 38.4 | 0.093 | 1.324(1.104, 1.588) | 55.3 | 0.013# | |
| PB | 7 | 740/939 | 1.262(0.842,1.892) | 0 | 0.842 | 1.052(0.670, 1.650) | 73.0 | 0.001# | 1.074 (0.723,1.596) | 70.7 | 0.002# | 1.329(0.899, 1.965) | 0 | 0.885 | |
*; In Gaspar et al. (11) study, there have the data not only of whites but also of others, I2; Quantification of the heterogeneity, Ph; P values for heterogeneity from Q test, #; Random-effect model was used when p value for heterogeneity test <0.05, otherwise, fix-effect model was used, PB; Population based and HB; Hospital based.
Fig 2.
Forest plots of association between MTHFR C677T polymorphism and NSCLP risk.
A; TT vs. CC, B; CT vs. CC, C; CT+TT vs. CC and D; TT vs. CT+CC.
Table 4.
Results of Meta-analysis of MTHFR A1298C polymorphism for studies on NSCL/P
| Groups | Studynumber | Sample size(case/control) | TT versus CC | CT versus CC | CT+TT versus CC (dominant) | TT versus CT+CC (recessive) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95% CI) | I2(%) | Ph | OR (95% CI) | I2(%) | Ph | OR (95% CI) | I2(%) | Ph | OR(95% CI) | I2(%) | Ph | ||||
| Overall | 7 | 1032/2095 | 0.744 | 0 | 0.64 | 0.991 | 32 | 0.184 | 0.952 | 33.5 | 0.172 | 0.766 | 0 | 0.738 | |
| (0.545,1.016) | (0.844,1.164) | (0.816,1.111) | (0.567,1.036) | ||||||||||||
| Ethnicity | |||||||||||||||
| Asians | 2 | 183/416 | 0.611 | 15.7 | 0.276 | 0.986 | 69.5 | 0.070 | 0.929 | 73.1 | 0.054 | 0.618 | 0 | 0.479 | |
| (0.281,1.329) | (0.687,1.413) | (0.655,1.316) | (0.289,1.321) | ||||||||||||
| Whites | 4 | 681/1541 | 0.794 | 0 | 0.505 | 0.966 | 39.1 | 0.177 | 0.938 | 38 | 0.184 | 0.822 | 0 | 0.565 | |
| (0.561,1.125) | (0.797,1.170) | (0.780,1.126) | (0.588,1.150) | ||||||||||||
| Others | 1 | 168/138 | 0.424 | - | - | 1.209 | - | - | 1.124 | - | - | 0.404 | - | - | |
| (0.076,2.365) | (0.720,2.029) | (0.680,1.858) | (0.073,2.237) | ||||||||||||
| Source | |||||||||||||||
| of control | |||||||||||||||
| HB | 2 | 470/1304 | 0.690 | 0 | 0.715 | 0.828 | 79.3 | 0.028# | 0.807 | 76.9 | 0.037# | 0.722 | 0 | 0.797 | |
| (0.459,1.036) | (0.457,1.501) | (0.476,1.366) | (0.487,1.071) | ||||||||||||
| PB | 5 | 562/791 | 0.834 | 0 | 0.448 | 1.038 | 0 | 0.444 | 1.008 | 5.9 | 0.373 | 0.839 | 0 | 0.526 | |
| (0.513,1.358) | (0.818,1.316) | (0.802,1.266) | (0.523,1.345) | ||||||||||||
I2; Quantification of the heterogeneity, Ph; P values for heterogeneity from Q test, #; Random-effect model was used when p value for heterogeneity test <0.05, otherwise, fix-effect model was used, " -"; no I2 and Ph because only 1 study in this subgroup, PB; Population based and HB; Hospital based.
Fig 3.
Forest plots of association between MTHFR A1298C polymorphism and NSCLP risk.
A; CC vs. AA, B; AC vs. AA, C; AC+CC vs AA and D; CC vs. AC+AA.
The synthesis of effect size and subgroup analysis
Regarding maternal C677T polymorphism and NSCL/P offspring, 15 studies with a total of 2442 cases and 4655 controls were included. Maternal TT genotype contributed to elevated risks of having an NSCL/P child compared with the CC wild-type genotype (OR=1.251, 95% CI: 1.047- 1.494), and this effect appeared only in the HB population (OR=1.248, 95% CI: 1.024-1.520) after subgroup analysis by source of controls and ethnicity. Under the recessive model, 677TT conferred increased susceptibility to having a child with NSCL/P when using CT+CC as reference (OR=1.325, 95% CI: 1.124-1.562), and this effect appeared in Asians (OR=1.726, 95% CI: 1.090-2.733), Whites (OR=1.308, 95% CI: 1.059-1.617), and the HB population (OR=1.324, 95% CI: 1.104-1.588) after further subgroup analysis. However, the CT heterozygote and the CT+TT dominant model had no association with bearing NSCL/P offspring when compared with CC wild-type genotype (all 95% CI include 1) (Table 3, Fig 2).
With respect to the association between maternal MTHFR A1298C polymorphism and NSCL/P offspring, 7 studies with 1032 cases and 2095 controls were included. There was no association between the maternal MTHFR A1298C polymorphism and having NSCL/P offspring under all genetic models, since the null value of all ORs (null value=1) was well inside the 95% confidence intervals (Table 4, Fig 3).
Sensitivity analyses and publication bias
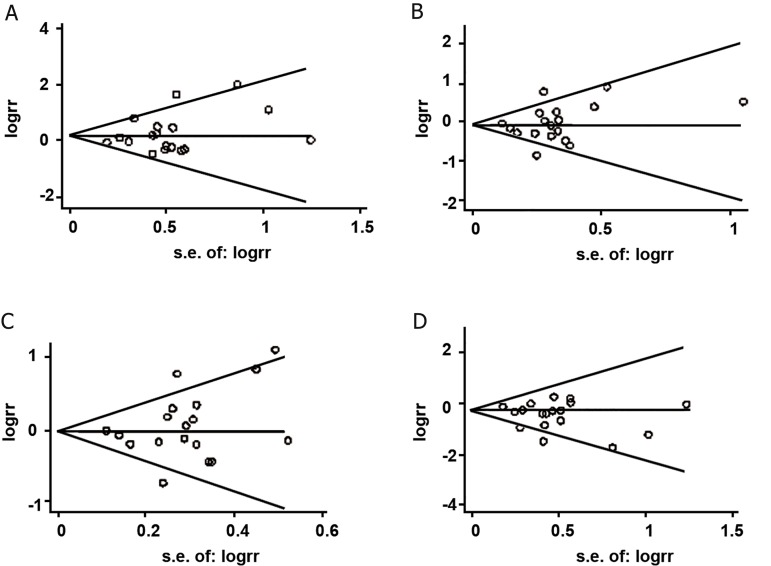
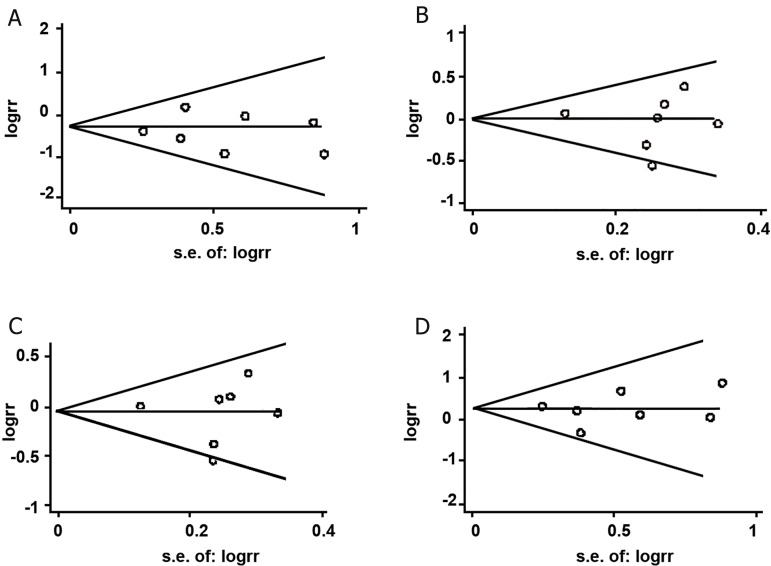
No individual study affected the final results according to sensitivity analyses (data not shown). Funnel plots of all studies revealed no asymmetrical distribution of ORs (Figes4-5), suggesting no significant publication bias in this study. Begg’s test and Egger’s test provided further statistical evidence of no significant publication bias in this meta-analysis (all p>0.05) (Table 5).
Fig 4.
Funnel plots for included studies of MTHFR C677T polymorphism and NSCLP risk. Begg’s funnel plot with pseudo 95% confidence limits. A; TT vs. CC, B; CT vs. CC, C; CT+TT vs. CC ad D; TT vs. CT+CC.
Fig 5.
Funnel plots for included studies of MTHFR A1298C polymorphism and NSCLP risk. Begg’s funnel plot with pseudo 95% confidence limits. A; CC vs. AA, B; AC vs. AA, C; AC+CC vs. AA and D; CC vs. AC+AA.
Table 5.
Results of Egger’s and Begg’s tests
| Comparison | Egger’s test | Begg’s test | |||
|---|---|---|---|---|---|
| t | P | 95% CI | Z | P | |
| C677T | |||||
| TT vs. CC | 1.27 | 0.221 | -0.566, 2.272 | 1.21 | 0.225 |
| CT vs. CC | 0.68 | 0.504 | -1.117, 2.180 | 0.53 | 0.596 |
| CT+TT vs. CC | 1.04 | 0.315 | -0.892, 2.603 | 0.83 | 0.405 |
| TT vs. CT+CC | -0.98 | 0.340 | -2.076, 0.760 | 0.23 | 0.820 |
| A1298C | |||||
| CC vs. AA | -0.24 | 0.821 | -2.444, 2.029 | 0.30 | 0.764 |
| AC vs. AA | -0.34 | 0.750 | -4.335, 3.332 | 0.30 | 0.764 |
| CC+AC vs. AA | -0.14 | 0.891 | -4.155, 3.716 | 0.60 | 0.548 |
| CC vs. AC+AA | -0.30 | 0.774 | -2.245, 1.771 | 0.00 | 1.000 |
Discussion
Principal findings
In the present meta-analysis, we focused on the relationship of maternal MTHFR polymorphisms (C677T and A1298C) and risk of having NSCL/P offspring. Results showed that mothers carrying the 677TT genotype had an increased susceptibility to have an NSCL/P offspring under both the recessive model and the CC homozygote model, but the CT heterozygote and the CT+TT dominant models had no association with bearing NSCL/P offspring when compared with CC genotype. This suggests that only the maternal 677TT genotype is a risk factor for having NSCL/P offspring. Subgroup analyses based on ethnicity and source of control showed that under the recessive model, maternal 677TT genotype increased the risk of having NSCL/P offspring in Whites, Asians, and the HB population, providing further evidence that the TT genotype is the risk factor. However, maternal MTHFR A1298C polymorphism had no association with having NSCL/P offspring under all genetic models, and results did not differ when subgroup analyses were undertaken.
In the present study, results from the dominant model were consistent with that reported by Verkleij-Hagoort et al. (49), which used single dominant model and also indicated that the maternal MTHFR C677T and A1298C polymorphisms were not independently associated with CLP with pooled ORs of 1.2 (95% CI: 0.9-1.5) and 1.0 (95% CI: 0.7-1.2). Results from the codominant model were similar to the study by Luo et al. (50), which only used the co-dominant model and also revealed that the maternal 677TT genotype elevated the risk of CL/P with a pooled OR of 1.32 (95% CI: 1.06-1.63) when compared with the normal 677CC genotype. However, the results of subgroup analyses were different. Luo et al. (50) demonstrated that maternal 677TT genotype increased the risk of having CL/P offspring in the white population with an OR of 1.36 (95% CI:1.05-1.76), while in present subgroup analyses, based on the same factors and the same genetic model, the effect was found in the hospital based population but not in Whites or Asians.
There are several reasons for the inconsistency between the present study and the other two meta-analyses. First, the applied genetic models are different. It is notable that besides the co-dominant and dominant models, the recessive model was also applied in our present meta-analyses and the results further supported the view that maternal 677TT genotype is a risk factor and is associated with having NSCL/P offspring. Second, types of study designs are different. Studies included in the two meta-analysis covered case-control and cohort studies (51) whereas our study was restricted to case-control studies. Third, the number of included studies varied. There were 10 and 12 studies included in the 2007 and 2012 metaanalyses respectively, but our meta-analysis comprised of 15 studies.
Furthermore, Verkleij-Hagoort et al. (49) and Luo et al. (50) had estimated infant MTHFR polymorphisms and CL/P susceptibility under the dominant and co-dominant models respectively in their meta-analysis, and all pooled results revealed no statistical association between infant C677T and A1298C variants and risk of CL/P. While meta-analysis performed by Pan et al. (43) which combined co-dominant, dominant and recessive models suggested that the infant 677TT genotype is associated with CL/P in recessive model. However, we did not examine infant MTHFR polymorphisms in the present study, so we can not conclude which one (maternal or infant MTHFR polymorphisms) is more related and more important with regards to NSCL/P.
Strength and weakness of the review
The associations between NSCL/P susceptibility and maternal MTHFR polymorphisms have been widely examined but provided inconsistent results. This absence of consensus among individual studies might be due to different ethnicities and study populations, different laboratory procedures used for genotyping, variable environment and diet, selection bias from both cases and controls subjects, and insufficient sample size (25, 33, 35, 36, 38, 39). However, meta-analyses have the advantage of increased statistical power by pooling the results from small individual studies and also can examination the variability between studies.
In the present study, to ensure the representativeness of the study subjects, we performed HWE test on the control groups in the final selected studies. Also, only NSCL/P was included to reduce etiologic heterogeneity. Furthermore, subgroup analyses based on ethnicity and source of control were performed to reduce ethnic heterogeneity and selection bias. However, certain limitations also need to be acknowledged. For one thing, the information of environmental interventions or maternal periconceptional behaviors was not available in our study, whereas these factors may influence the whole pregnancy outcome. For another thing, heterogeneity present in overall or subgroup analyses might be due to different regions, various genotyping methods and others mentioned above. But we only separated study populations into three groups: Asians, Whites, and others, and we did not perform any subgroup analysis based on genotyping method. Last, but not least, only case-control studies were selected, which are less powerful for genetic associations than transmission disequilibrium test in familybased designs.
Possible mechanism
It is not possible to explain the mechanism of the association between NSCL/P susceptibility and maternal MTHFR polymorphisms in the present study. However, the deficiency of nutritional folic acid during embryonic development has been proposed as a factor in the etiology of NSCL/P, and many studies suggest that maternal ingestion of folic acid during early pregnancy can reduce the risk of NSCL/P (7-10). According to this evidence, it has been hypothesized that NSCL/P might be associated with folate metabolism and polymorphic variants of the genes which encode key proteins in folate and methionine metabolism may play a role in the susceptibility of NSCL/P (11).
MTHFR is an important enzyme in folate metabolism. Normal MTHFR activity is crucial to maintain the pool of circulating folate and methionine and to prevent the accumulation of homocysteine (12). C677T and A1298C are two common functional single nucleotide polymorphisms localized in MTHFR, and their roles in the mechanisms of folate enzyme have been extensively researched. Compared to the CC and CT genotypes, the TT genotype is characterized by 70% enzymatic activity reduction (52). And it is the reason that the concentrations of folate in serum, plasma, and red blood cells decrease and plasma homocysteine concentrations increase mildly (12-14). Similarly, the A1298C polymorphism, resulting in a glutamic acid to alanine substitution at codon 429, also causes decreasing MTHFR enzyme activity, but is not associated with higher homocysteine or lower plasma folate levels (15, 16). This mechanism can explain all the results we gained in present study.
In addition, it is worth noting that linkage disequilibrium (LD) exists between MTHFR C677T and A1298C. Some studies had reported a stronger MTHFR activity loss in combined heterozygote (C677T/A1298C) in neural tube defect cases (15, 53), however, there are few studies that investigated the relationship between this compound genotype and NSCL/P risk, and quantitative analysis cannot be conducted owing to incomplete data.
Conclusion
The results of this study give supporting evidence for significant association between maternal MTHFR C677T polymorphism and risk of having an NSCL/P offspring, and confirm mothers with the 677TT genotype are susceptible to have a child with NSCL/P. Nevertheless, in view of the limitations of the present study, our findings should be interpreted prudently. Further work, especially those that address the combined effects of genetic and environmental factors on a specific phenotype of orofacial clefts should be performed to evaluate these findings.
Acknowledgments
This work was supported by the National Natural Science Foundation of China No. 81370779, 81170630 and U1204804. We declare that we have no conflict of interest.
References
- 1.Lidral AC, Moreno LM, Bullard SA. Genetic Factors and Orofacial Clefting. Semin Orthod. 2008;14(2):103–114. doi: 10.1053/j.sodo.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobourne MT. The complex genetics of cleft lip and palate. Eur J Orthod. 2004;26(1):7–16. doi: 10.1093/ejo/26.1.7. [DOI] [PubMed] [Google Scholar]
- 3.Sun H, Huang H. Study on genetic locus of nonsyndromic cleft lip with or without cleft palate. International Journal of Oral Medicine Volume. 2000;27(2):100–103. (in Chinese) [Google Scholar]
- 4.McDonnell R, Owens M, Delany C, Earley M, McGillivary A, Orr DJ, et al. Epidemiology of orofacial clefts in the east of Ireland in the 25-year period 1984-2008. Cleft Palate Craniofac J. 2014;51(4):e63–69. doi: 10.1597/11-299. [DOI] [PubMed] [Google Scholar]
- 5.Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: From genotypes to subphenotypes. Oral Dis. 2009;15(7):437–453. doi: 10.1111/j.1601-0825.2009.01577.x. [DOI] [PubMed] [Google Scholar]
- 6.Carinci F, Scapoli L, Palmieri A, Zollino I, Pezzetti F. Human genetic factors in nonsyndromic cleft lip and palate: an update. Int J Pediatr Otorhinolaryngol. 2007;71(10):1509–1519. doi: 10.1016/j.ijporl.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Van Rooij IA, Vermeij-Keers C, Kluijtmans LA, Ocke MC, Zielhuis GA, Goorhuis-Brouwer SM, et al. Does the interaction between maternal folate intake and the methylenetetrahydrofolate reductase polymorphisms affect the risk of cleft lip with or without cleft palate? Am J Epidemiol. 2003;157(7):583–591. doi: 10.1093/aje/kwg005. [DOI] [PubMed] [Google Scholar]
- 8.Badovinac RL, Werler MM, Williams PL, Kelsey KT, Hayes C. Folic acid-containing supplement consumption during pregnancy and risk for oral clefts: a meta-analysis. Birth Defects Res A Clin Mol Teratol. 2007;79(1):8–15. doi: 10.1002/bdra.20315. [DOI] [PubMed] [Google Scholar]
- 9.Wilcox AJ, Lie RT, Solvoll K, Taylor J, McConnaughey DR, Abyholm F, et al. Folic acid supplements and risk of facial clefts: national population based case-control study. BMJ. 2007;334(7591):464–464. doi: 10.1136/bmj.39079.618287.0B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson CY, Little J. Folate intake, markers of folate status and oral clefts: is the evidence converging? Int J Epidemiol. 2008;37(5):1041–1058. doi: 10.1093/ije/dyn098. [DOI] [PubMed] [Google Scholar]
- 11.Gaspar DA, Matioli SR, de Cassia Pavanello R, Araujo BC, Alonso N, Wyszynski D, et al. Maternal MTHFR interacts with the offspring’s BCL3 genotypes, but not with TGFA, in increasing risk to nonsyndromic cleft lip with or without cleft palate. Eur J Hum Genet. 2004;12(7):521–526. doi: 10.1038/sj.ejhg.5201187. [DOI] [PubMed] [Google Scholar]
- 12.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10(1):111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 13.Mills JL, Kirke PN, Molloy AM, Burke H, Conley MR, Lee YJ, et al. Methylenetetrahydrofolate reductase thermolabile variant and oral clefts. Am J Med Genet. 1999;86(1):71–74. [PubMed] [Google Scholar]
- 14.Molloy AM, Daly S, Mills JL, Kirke PN, Whitehead AS, Ramsbottom D, et al. Thermolabile variant of 5,10-methylenetetrahydrofolate reductase associated with low red-cell folates: implications for folate intake recommendations. Lancet. 1997;349(9065):1591–1593. doi: 10.1016/S0140-6736(96)12049-3. [DOI] [PubMed] [Google Scholar]
- 15.Van der Put NM, Gabreels F, Stevens EM, Smeitink JA, Trijbels FJ, Eskes TK, et al. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62(5):1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisberg I, Tran P, Christensen B, Sibani S, Rozen R. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab. 1998;64(3):169–172. doi: 10.1006/mgme.1998.2714. [DOI] [PubMed] [Google Scholar]
- 17.Rai AK, Singh S, Mehta S, Kumar A, Pandey LK, Raman R. MTHFR C677T and A1298C polymorphisms are risk factors for Down’s syndrome in Indian mothers. J Hum Genet. 2006;51(4):278–283. doi: 10.1007/s10038-005-0356-3. [DOI] [PubMed] [Google Scholar]
- 18.Wan WD, Wang LJ, Zhou XP, Zhou DL, Zhang QG, Huang JL, et al. Relationship between nonsyndromic cleft lip with or without cleft palate (NSCL/P) and genetic polymorphisms of MTHFR C677T and A1298C. Zhonghua Zheng Xing Wai Ke Za Zhi. 2006;22(1):8–11. [PubMed] [Google Scholar]
- 19.Ali A, Singh SK, Raman R. MTHFR 677TT alone and IRF6 820GG together with MTHFR 677CT, but not MTHFR A1298C, are risks for nonsyndromic cleft lip with or without cleft palate in an Indian population. Genet Test Mol Biomarkers. 2009;13(3):355–360. doi: 10.1089/gtmb.2008.0115. [DOI] [PubMed] [Google Scholar]
- 20.Zhu J, Ren A, Hao L, Pei L, Liu J, Zhu H, et al. Variable contribution of the MTHFR C677T polymorphism to non-syndromic cleft lip and palate risk in China. Am J Med Genet A. 2006;140(6):551–557. doi: 10.1002/ajmg.a.31115. [DOI] [PubMed] [Google Scholar]
- 21.Brandalize AP, Bandinelli E, Borba JB, Felix TM, Roisenberg I, Schuler-Faccini L. Polymorphisms in genes MTHFR, MTR and MTRR are not risk factors for cleft lip/palate in South Brazil. Braz J Med Biol Res. 2007;40(6):787–791. doi: 10.1590/s0100-879x2006005000112. [DOI] [PubMed] [Google Scholar]
- 22.Guo JZ, Song XM, Wang Y, Zhu WL, Li SQ, Li Y. Relationship between genetic polymorphisms of MTHFR C677T and nonsyndromic cleft lip with or without palate. Beijing Da Xue Xue Bao. 2009;41(4):432–436. [PubMed] [Google Scholar]
- 23.Sozen MA, Tolarova MM, Spritz RA. The common MTHFR C677T and A1298C variants are not associated with the risk of non-syndromic cleft lip/palate in northern Venezuela. J Genet Genomics. 2009;36(5):283–288. doi: 10.1016/S1673-8527(08)60116-2. [DOI] [PubMed] [Google Scholar]
- 24.Blanton SH, Kolle BS, Hecht JT, Mulliken JB, Martin ER. No evidence supporting MTHFR as a risk factor in the development of familial NSCLP. Am J Med Genet. 2000;92(5):370–371. doi: 10.1002/1096-8628(20000619)92:5<370::aid-ajmg17>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Chorna LB, Akopyan HR, Makukh HV, Fedoryk IM. Allelic polymorphism of MTHFR, MTR and MTRR genes in patients with cleft lip and/or palate and their mothers. Tsitol Genet. 2011;45(3):51–56. [PubMed] [Google Scholar]
- 26.Han Y, Pan Y, Du Y, Tong N, Wang M, Zhang Z, et al. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and nonsyndromic orofacial clefts susceptibility in a southern Chinese population. DNA Cell Biol. 2011;30(12):1063–1068. doi: 10.1089/dna.2010.1185. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J, Hao L, Li S, Bailey LB, Tian Y, Li Z. MTHFR, TGFB3, and TGFA polymorphisms and their association with the risk of non-syndromic cleft lip and cleft palate in China. Am J Med Genet A. 2010;152A(2):291–298. doi: 10.1002/ajmg.a.33113. [DOI] [PubMed] [Google Scholar]
- 28.Shaw GM, Rozen R, Finnell RH, Todoroff K, Lammer EJ. Infant C677T mutation in MTHFR, maternal periconceptional vitamin use, and cleft lip. Am J Med Genet. 1998;80(3):196–198. doi: 10.1002/(sici)1096-8628(19981116)80:3<196::aid-ajmg2>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 29.Jugessur A, Wilcox AJ, Lie RT, Murray JC, Taylor JA, Ulvik A, et al. Exploring the effects of methylenetetrahydrofolate reductase gene variants C677T and A1298C on the risk of orofacial clefts in 261 Norwegian case-parent triads. Am J Epidemiol. 2003;157(12):1083–1091. doi: 10.1093/aje/kwg097. [DOI] [PubMed] [Google Scholar]
- 30.Davalos-Rodriguez IP, Ramirez-Lizardo EJ, Mena JP, Ledezma-Rodriguez V, Omayra-Davalos N, Gonzalez-Mercado MG, et al. Non-syndromic cleft lip/cleft palate and C677T methylene-tetrahydrofolate reductase variant in Mexican children. Rev Med Inst Mex Seguro Soc. 2009;47(5):549–552. [PubMed] [Google Scholar]
- 31.Reutter H, Birnbaum S, Lacava AD, Mende M, Henschke H, Berge S, et al. Family-based association study of the MTHFR polymorphism C677T in patients with nonsyndromic cleft lip and palate from central Europe. Cleft Palate Craniofac J. 2008;45(3):267–271. doi: 10.1597/06-174. [DOI] [PubMed] [Google Scholar]
- 32.Martinelli M, Scapoli L, Pezzetti F, Carinci F, Carinci P, Stabellini G, et al. C677T variant form at the MTHFR gene and CL/P: a risk factor for mothers? Am J Med Genet. 2001;98(4):357–360. doi: 10.1002/1096-8628(20010201)98:4<357::aid-ajmg1108>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 33.Shotelersuk V, Ittiwut C, Siriwan P, Angspatt A. Maternal 677CT/1298AC genotype of the MTHFR gene as a risk factor for cleft lip. J Med Genet. 2003;40(5):e64–e64. doi: 10.1136/jmg.40.5.e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pezzetti F, Martinelli M, Scapoli L, Carinci F, Palmieri A, Marchesini J, et al. Maternal MTHFR variant forms increase the risk in offspring of isolated nonsyndromic cleft lip with or without cleft palate. Hum Mutat. 2004;24(1):104–105. doi: 10.1002/humu.9257. [DOI] [PubMed] [Google Scholar]
- 35.Wang SM, Wang JH, Yu JC, Wei B, Wang KH, Liu JY, et al. Association between parental MTHFR gene polymorphism 677C/T and nonsyndromic cleft lip and palate in offspring. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2012;29(4):464–467. doi: 10.3760/cma.j.issn.1003-9406.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 36.Bufalino A, Ribeiro Paranaiba LM, Nascimento de Aquino S, Martelli-Junior H, Oliveira Swerts MS, Coletta RD. Maternal polymorphisms in folic acid metabolic genes are associated with nonsyndromic cleft lip and/or palate in the Brazilian population. Birth Defects Res A Clin Mol Teratol. 2010;88(11):980–986. doi: 10.1002/bdra.20732. [DOI] [PubMed] [Google Scholar]
- 37.Mills JL, Molloy AM, Parle-McDermott A, Troendle JF, Brody LC, Conley MR, et al. Folate-related gene polymorphisms as risk factors for cleft lip and cleft palate. Birth Defects Res A Clin Mol Teratol. 2008;82(9):636–643. doi: 10.1002/bdra.20491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vujkovic M, Steegers EA, van Meurs J, Yazdanpanah N, van Rooij IA, Uitterlinden AG, et al. The maternal homocysteine pathway is influenced by riboflavin intake and MTHFR polymorphisms without affecting the risk of orofacial clefts in the offspring. Eur J Clin Nutr. 2010;64(3):266–273. doi: 10.1038/ejcn.2009.138. [DOI] [PubMed] [Google Scholar]
- 39.Mostowska A, Hozyasz KK, Jagodzinski PP. Maternal MTR genotype contributes to the risk of non-syndromic cleft lip and palate in the Polish population. Clin Genet. 2006;69(6):512–517. doi: 10.1111/j.1399-0004.2006.00618.x. [DOI] [PubMed] [Google Scholar]
- 40.Jugessur A, Shi M, Gjessing HK, Lie RT, Wilcox AJ, Weinberg CR, et al. Maternal genes and facial clefts in offspring: a comprehensive search for genetic associations in two population-based cleft studies from Scandinavia. PLoS One. 2010;5(7):e11493–e11493. doi: 10.1371/journal.pone.0011493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi M, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, et al. Genome wide study of maternal and parent-of-origin effects on the etiology of orofacial clefts. Am J Med Genet A. 2012;158A(4):784–794. doi: 10.1002/ajmg.a.35257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, Hou W, Chen EJ, Liu XH, Hou CL, Zhang XH. Association of methylenetetrahydrofolate reductase A1298C polymorphisms with non-syndromic cleft lip with or without cleft palate. Zhonghua Kou Qiang Yi Xue Za Zhi. 2011;46(7):394–397. [PubMed] [Google Scholar]
- 43.Pan Y, Zhang W, Ma J, Du Y, Li D, Cai Q, et al. Infants’ MTHFR polymorphisms and nonsyndromic orofacial clefts susceptibility: a meta-analysis based on 17 case-control studies. Am J Med Genet A. 2012;158A(9):2162–2169. doi: 10.1002/ajmg.a.35503. [DOI] [PubMed] [Google Scholar]
- 44.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Little J, Gilmour M, Mossey PA, Fitzpatrick D, Cardy A, Clayton-Smith J, et al. Folate and clefts of the lip and palate -a U.K.-based case-control study: Part II: Biochemical and genetic analysis. Cleft Palate Craniofac J. 2008;45(4):428–438. doi: 10.1597/06-151.1. [DOI] [PubMed] [Google Scholar]
- 46.Gaspar DA, Pavanello RC, Zatz M, Passos-Bueno MR, Andre M, Steman S, et al. Role of the C677T polymorphism at the MTHFR gene on risk to nonsyndromic cleft lip with/without cleft palate: results from a case-control study in Brazil. Am J Med Genet. 1999;87(2):197–199. doi: 10.1002/(sici)1096-8628(19991119)87:2<197::aid-ajmg15>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 47.Prescott NJ, Winter RM, Malcolm S. Maternal MTHFR genotype contributes to the risk of non-syndromic cleft lip and palate. J Med Genet. 2002;39(5):368–369. doi: 10.1136/jmg.39.5.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tolarova MM, van Rooij IALM, Pastor M, Van der Put NMJ, Goldberg AC, Hol F, et al. A common mutation in the MTHFR gene is a risk factor for nonsyndromic cleft lip and palate anomalies. Am J Hum Genet. 1998;63:A27–A27. [Google Scholar]
- 49.Verkleij-Hagoort A, Bliek J, Sayed-Tabatabaei F, Ursem N, Steegers E, Steegers-Theunissen R. Hyperhomocysteinemia and MTHFR polymorphisms in association with orofacial clefts and congenital heart defects: a meta-analysis. Am J Med Genet A. 2007;143A(9):952–960. doi: 10.1002/ajmg.a.31684. [DOI] [PubMed] [Google Scholar]
- 50.Luo YL, Cheng YL, Ye P, Wang W, Gao XH, Chen Q. Association between MTHFR polymorphisms and orofacial clefts risk: a meta-analysis. Birth Defects Res A Clin Mol Teratol. 2012;94(4):237–244. doi: 10.1002/bdra.23005. [DOI] [PubMed] [Google Scholar]
- 51.Nurk E, Tell GS, Refsum H, Ueland PM, Vollset SE. Associations between maternal methylenetetrahydrofolate reductase polymorphisms and adverse outcomes of pregnancy: the Hordaland Homocysteine Study. Am J Med. 2004;117(1):26–31. doi: 10.1016/j.amjmed.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 52.Rozen R. Genetic predisposition to hyperhomocysteinemia: deficiency of methylenetetrahydrofolate reductase (MTHFR) Thromb Haemost. 1997;78(1):523–526. [PubMed] [Google Scholar]
- 53.Richter B, Stegmann K, Roper B, Boddeker I, Ngo ET, Koch MC. Interaction of folate and homocysteine pathway genotypes evaluated in susceptibility to neural tube defects (NTD) in a German population. J Hum Genet. 2001;46(3):105–109. doi: 10.1007/s100380170096. [DOI] [PubMed] [Google Scholar]