Abstract
Background
Maternal metabolic demands change dramatically during the course of gestation and must be coordinated with the needs of the developing placenta and fetus. The liver is critically involved in metabolism and other important functions. However, maternal hepatic adjustments to pregnancy are poorly understood.
Aim
The aim of the study was to evaluate the influences of pregnancy on the maternal liver growth and gene expression profile.
Methods
Holtzman Sprague-Dawley rats were mated and sacrificed at various stages of gestation and postpartum. The maternal Livers were analyzed in gravimetric response, DNA content by PicoGreen dsDNA quantitation reagent, hepatocyte ploidy by flow cytometry, and hepatocyte proliferation by ki-67 immunostaining. Gene expression profiling of nonpregnant and gestation d18.5 maternal hepatic tissue was analyzed using a DNA microarray approach and partially verified by northern blot or quantitative real-time PCR analysis.
Results
During pregnancy, the liver exhibited approximately an 80% increase in size; proportional to the increase in body weight of the pregnant animals. The pregnancy-induced hepatomegaly was a physiological event of liver growth manifested by increases in maternal hepatic DNA content and hepatocyte proliferation. Pregnancy did not affect hepatocyte polyploidization. Pegnancy-dependent changes in hepatic expression were noted for a number of genes, including those associated with cell proliferation, cytokine signaling, liver regeneration, and metabolism.
Conclusions
The metabolic demands of pregnancy cause marked adjustments in maternal liver physiology. Central to these adjustments are an expansion in hepatic capacity and changes in hepatic gene expression. Our findings provide insights into pregnancy-dependent hepatic adaptations.
Keywords: Hepatomegaly, liver growth, hepatocyte proliferation, pregnancy, lactation
Introduction
During pregnancy, a series of coordinated physiological adaptations occur in many maternal organ systems, including hyperplasia of beta cell mass in pancreatic islets required for elevated production of insulin (1), splenic growth and development of the erythroid lineage (2), forebrain olfactory neurogenesis associated with maternal behaviors (3), immune system adjustments to protect the fetal-maternal interface from immunological attack (4), increased blood volume and cardiac output to meet the needs of fetal-maternal bio-exchange of gas, nutrients, and metabolic wastes (5). These widespread maternal physiological changes are highly coordinated with the development and growth of the placenta and fetus and are essential for successful establishment and maintenance of pregnancy.
Maternal metabolic demands change dramatically during the course of gestation. The liver is critically involved in metabolism and a variety of physiological functions involving the uptake, storage and distribution of both nutrients and vitamins, maintenance of blood sugar levels, regulation of circulating plasma lipids, the synthesis of circulating plasma proteins and metabolism of nutrients, toxic compounds, and drugs. During pregnancy, it is anticipated that the liver would increase its functional capacity to accommodate the nutritional and metabolic needs of developing placentas and fetuses. Several lines of evidence indicate the functional adjustments of the maternal liver to pregnancy, such as pregnancy-dependent changes in drug, lipid, cholesterol, and glucose metabolism (6-13). These adjustments are associated with the alterations of metabolic enzyme activity, nuclear receptor expression, and gonadal and pituitary hormone signaling in the maternal liver during pregnancy (13-17). Notably, it has been observed that maternal liver weight increases during pregnancy in the mouse and rat (7, 10, 14, 18). However, the impact of pregnancy on maternal liver weight has not been systematically evaluated. In this investigation, we examined hepatic adaptations to pregnancy at organ, cellular, and molecular levels.
Materials and methods
Animals and tissue preparation
Holtzman Sprague-Dawley rats (8 weeks of age) were obtained from Harlan Sprague Dawley Inc. (Indianapolis, IN). The presence of sperm in the vaginal smear of the rat was considered to be gestational d0.5. Animals were provided food and water ad libitum. Animals were placed on a 12 h light:12 h dark cycle and the temperature and relative humidity were maintained between 20-24°C and 40-60%, respectively. Liver tissue was collected from non-pregnant (n=7), gestational d4.5 (n=4), d8.5 (n=7), d11.5 (n=13), d13.5 (n=13), d15.5 (n=5), d18.5 (n=7), d20.5 (n=7), d21.5 (n=5) and postpartum d10 with and without pups (n=5 and n=7, respectively) rats and weighed. The body weights of non-pregnant rats, pregnant rats with conceptuses, and postpartum d10 rats were recorded. All collected tissues were then snap-frozen in liquid nitrogen and stored at -80°C. Animal care and experimental protocols were approved by The University of Kansas Medical Center Animal Care and Use Committee.
DNA content
Liver tissues collected from non-pregnant, pregnant (gestation d11.5 and d18.5) and postpartum d10 (with and without pups) rats were completely digested overnight at 55°C in a buffer containing 100 mM NaCl, 50 mM Tris-HCl (pH 8.0), 25 mM EDTA, 0.5% SDS and 400μg/ml of proteinase K (n=3 per group). The digested samples were mixed with PicoGreen dsDNA quantitation reagent (Molecular Probes, Eugene, OR). The mixed samples were excited at 485 nm and the fluorescence emission intensity was measured at 538 nm using a spectrofluorometer (Finstruments Fluorokan II, MTX Lab System, Inc., Vienna, VA). DNA concentrations were determined using a standard curve of fluorescence emission intensity plotted versus DNA concentration. The standard curve was generated with the reagents provided by the vendor.
Ki-67 immunostaining
Liver tissues were collected from nonpregnant, pregnant (gestational d4.5, d8.5, d11.5, d13.5, d15.5, d18.5, and d21.5), and postpartum d10 non-lactating and lactating rats. Formalin-fixed and paraffin-embedded liver tissue sections were prepared and subjected to Ki-67 immunostaining with a primary Ki-67 antibody (NeoMarkers, Fremont, CA) according to the manufacturer's instructions. This procedure was used to monitor hepatocyte proliferation in the liver.
Hepatocyte isolation and ploidy analysis
Primary rat hepatocytes were isolated using the procedure described by Kim and coworkers (19). Rats were anesthetized before in situ perfusion of the liver through the portal vein with Hanks Balanced Salt Solution followed by Gibco Liver Digestion Medium (Invitrogen, Carlsbad, CA). RPMI1640 with supplements (10% fetal bovine serum, FBS, 100 U/ml penicillin, 100 μg/ml streptomycin) was added to the dissociated hepatoctyes. Hepatocytes were passed through sterile gauze and isolated by centrifugation. Trypan blue was used to determine cell viability which exceeded 80%. Propidium iodide staining was performed using 2 × 106 cells/ml. Hepatocytes were resuspended in 1 ml of 0.9% sodium chloride and fixed by adding 2.5 ml of 90% cold ethanol. Following a 30 min incubation at room temperature, cells were centrifuged at 50 ×g for 2 min and the pellet was resuspended in 1 ml of propidium iodide (50 μg/ml in phosphate buffered saline, pH 7.2). In addition, 100 μl of Rnase A (1 mg/ml) was added to the cells and incubated for 30 min at 37°C. Hepatocytes were analyzed by flow cytometry using BD LSRII and FACS Diva (BD Biosciences, San Jose, CA).
Microarray analysis
Total RNA was prepared from liver tissue of non-pregnant (n=9) and gestation d18.5 (n=9) using TRIzol reagent according to the manufacturer's protocol (Invitrogen, Carlsbard, CA). RNA extractions were pooled to form three groups of three livers for each collection day in nuclease-free water at a concentration of 1.0 μg/μl. The Affymetrix GeneChip system was used to process GeneChip expression arrays (Affymetrix, Santa Clara, CA) and performed by the University of Kansas Medical Center Microarray Facility. Reverse transcription (RT) utilizing 5 μg total RNA was conducted using the SuperScript Choice System (Invitrogen). The RT was driven by the annealing of an oligo dT primer coupled with a T7 promoter sequence. The resulting cDNA was labeled with biotin using the GeneChip Expression 3′ Amplification reagent kit (Affymetrix). Biotin labeled cRNA was then fragmented with Mg++ and K+ at high temperature to a size range of ∼50-200 bp. Biotin-labeled fragmented cRNAs were hybridized to the Rat 230.20 GeneChip Expression array (Affymetrix) for 16 h at 45°C and 60 rpm in a GeneChip Hybridization Oven 640 (Affymetrix). Hybridized GeneChips underwent low and high stringency washing and R-phycoerythrin-streptavidin staining procedures using a GeneChip Fluidics Station (Affymetrix). GeneChips were scanned using a Genechip scanner 3000 7G with autoloader. Fluidics and scan functions were controlled by GCOS software version 1.4. Expression data sets were analyzed using the expression analysis software GeneSpring 7.0.
Cloning
cDNAs for the genes listed in Table 1 were isolated by RT-PCR from total RNA extracts prepared with TRIzol reagent from either non-pregnant or gestation d18.5 rat livers. Five μg of total RNA and 0.5 μg of oligo dT were used for reverse transcription reactions with Superscript II RT (Invitrogen). PCR was performed using Platinium Taq DNA High Fidelity polymerase (Invitrogen) with specific primers (Table 1). PCR was conducted for 30 cycles under the following conditions; preheat, 94°, denature 94° for 1 min, anneal, 60° for 1 min, extension, 72° for 1 min. PCR products were separated on 1% agarose gels and stained with ethidium bromide. Amplified products were subcloned into pCRII-TOPO vector with the TOPO TA Cloning kit (Invitrogen). cDNAs were sequenced by the Northwestern University DNA Sequencing Center (Chicago, IL).
Table 1. PCR Primer Sequences.
| Gene | Symbol | GenBank | Primer | Sequence (5′ to 3′) |
|---|---|---|---|---|
| Alpha-2 macroglobulin | A2m | NM_012488 | Fwd | GAAGAGTCCGTGGCTTTCTG |
| Rev | TAGCAGGCGTTGCTATCCTT | |||
| Patatin-like phospholipase domain containing 3 | Pnpla3 | XM_343302 | Fwd | TTCTACCACATCGGGGCTAC |
| Rev | CGGAAGGAAGGAGGGATTAG | |||
| Nephroblastoma overexpressed gene | Nov | NM_030868 | Fwd | ACCTGTGGCTCAGAGGAGAA |
| Rev | CCCTGGCAAACACTGAAACT | |||
| Early growth response 1 | Egr1 | NM_012551 | Fwd | CCACAACAACAGGGAGACCT |
| Rev | TCTTGCGTTCATCACTCCTG | |||
| Hepcidin antimicrobial peptide | Hamp | NM_053469 | Fwd | ACAGAAGGCAAGATGGCACT |
| Rev | TGCAACAGAGACCACAGGAG | |||
| Insulin-like growth factor binding protein 1 | Igfbp1 | NM_013144 | Fwd | CAGCAAACAGTGCGAGACAT |
| Rev | CTGCCCTTTCAAAGCAGAAC | |||
| Neurotrophic tyrosine kinase, receptor, type 1 | Ntrk1 | NM_021589 | Fwd | TACCTAGCCAGCCTGCACTT |
| Rev | TCCAGACTCCTAGCCCAGAA | |||
| Achaete-scute complex homolog-like 1 | Ascl1 | NM_022384 | Fwd | GGCTCAACTTCAGTGGCTTC |
| Rev | ACACAGGATCTCCTGCCATC | |||
| Prominin 1 | Prom1 | NM_021751 | Fwd | GCAAAAGAGGTCGAGGAGTG |
| Rev | GCAGAATCCAAAGAGGCAAG | |||
| Glyceraldehyde-3-phosphate dehydrogenase | G3pdh | NM_017008 | Fwd | ACCACAGTCCATGCCATCAC |
| Rev | TCCACCACCCTGTTGCTGTA |
Northern analysis
Northern blotting was conducted as previously described (20). RNA was extracted from tissues using TRIzol. Total RNA (20 μg) was separated on 1% formaldehyde-agarose gels and transferred to nylon membranes. Blots were probed with [32P]-labeled cDNAs indicated in Table 1. G3pdh cDNA was used as an internal control to demonstrate equal loading and RNA integrity.
Quantitative RT-PCR (qRT-PCR)
cDNAs were synthesized with total RNA (1 μg) from each sample using M-MLV reverse transcriptase (Invitrogen), diluted 5 times with water, and subjected to SYBR qRT-PCR to quantify mRNA levels of the genes listed in Table 2. Primers (Table 2) were designed using Primer Express 2.0 (Applied Biosystems, Foster City, CA). Real-time PCR amplification of cDNAs was carried out in a reaction mixture (10 μl) containing SYBR GREEN PCR Master Mix (Applied Biosystems) and primers (600 nM each). Amplification and fluorescence detection were carried out using the ABI Prism 7500 Real Time PCR System (Applied Biosystems). Cycling conditions included an initial hold step (95 °C for 10 min) and 40 cycles of a 2-step PCR (92 °C for 15 s, then 60 °C for 1 min), followed by a dissociation step (95 °C for 15 s, 60 °C for 15 s, and then 95 °C for 15 s). The comparative CT method was used for relative quantification of the amount of mRNA for each sample normalized to 18S RNA.
Table 2. qRT-PCR Primer Sequences.
| Gene | Symbol | GenBank | Primer | Sequence (5′ to 3′) |
|---|---|---|---|---|
| Tumor necrosis factor receptor superfamily, member 9 (CD137, ILA, 4-1BB) | Tnfrsf9 | AI_236084 | Fwd | ACTTTCTGCAGTAAATACCCTC |
| Rev | GAACCTGAAATAGCCTTGACAC | |||
| Tumor-associated calcium signal transducer 1 | Tacstd1 | BG_376410 | Fwd | GTCCTTGTTCCATTCATCTAAGAG |
| Rev | CCATCTCCTTTATCTCAGCC | |||
| Monocyte to macrophage differentiation-associated 2 | Mmd2 | BI_296275 | Fwd | GTGATTTACTTCTTCATTGCGG |
| Rev | AAGCTTGTACCGTTCATGGA | |||
| Protein phosphatase 1, regulatory subunit 3C | Ppp1r3c | BM_390827 | Fwd | GGTCCTTACAATGGTCTTCAG |
| Rev | CATTCCTTCTGTGATTTGGCT | |||
| 18s rRNA | 18s rRNA | M11188 | Fwd | GCAATTATTCCCCATGAACG |
| Rev | GGCCTCACTAAACCATCCAA |
Statistical analysis
The data were analyzed by analysis of variance and post hoc comparisons determined by Newman-Keuls Test. Data analysis of hepatocyte ploidy was analyzed by comparing means using the two-tailed Student's t-test.
Results
Pregnancy induces maternal hepatomegaly
Holtzman Sprague-Dawley rats were mated and sacrificed at various stages of gestation and postpartum. Gravimetric responses of maternal livers were assessed throughout pregnancy and postpartum (d10) in non-lactating and lactating conditions. Significant increases in maternal liver weight were observed during pregnancy (Fig. 1A&B). In comparison with pre-pregnancy liver sizes, pregnancy-dependent increases in liver mass were statistically significant at gestation d8.5 and sustained thereafter, culminating in about an 80% liver enlargement (hepatomegaly). Pregnant and nonpregnant rats did not significantly differ in liver-to-body weight ratios (Fig. 1C). The observation indicates that the maternal liver adjusts its size proportionally to the collective increases in maternal body weight and the weights of the placentas and fetuses. Following parturition, the maternal liver regressed at d10 postpartum in the non-lactating condition, whereas the maternal liver retained its pre-partum sizes in the lactating condition. In summary, we have identified an adaptive response of the maternal liver to pregnancy and lactation.
Fig. 1. Maternal liver weight responses to pregnancy and lactation in the rat.

Tissues were collected from non-pregnant (np), pregnant (gestational d4.5, d8.5, d11.5, d13.5, d15.5, d18.5, and d21.5), and postpartum d10 non-lactating (nlac) and lactating (lac) rats and weighed. Maternal liver weight responses are shown in Panel A. Representative gross morphology of nonpregnant (NP) and gestation d18.5 livers is shown in Panel B. Liver weight responses were also normalized to body weight as shown in Panel C. Asterisks indicate values that are significantly different from values for non-pregnant animals (P < 0.05; n = 5 to 7).
Pregnancy-induced hepatomegaly is associated with increases in hepatic DNA content and hepatocyte proliferation
In order to determine the cellular basis underlying pregnancy-dependent liver weight increases, we first measured the total DNA content in the livers of nonpregnant rats, pregnant rats (gestation d11.5 and d18.5), and at d10 postpartum in both non-lactating and lactating rats. During pregnancy, liver DNA content was significantly increased on gestation d18.5 as compared with nonpregnant rats (Fig. 2). Following parturition, lactating rats possessed higher liver DNA content than non-lactating rats (Fig. 2). Pregnancy and lactation associated-increases in liver size correlated with increases in DNA content.
Fig. 2. Measurement of liver DNA content from non-pregnant, pregnant, and post-partum rats.
Liver tissues collected from non-pregnant (np), pregnant (gestational d11.5 and d18.5) and postpartum d10 non-lactating (nlc) and lactating (lac) rats were completely digested overnight. DNA content was measured using Quant-iT PicoGreen dsDNA Kit. DNA concentrations were determined using a standard curve of fluorescence emission intensity plotted versus DNA concentration. Asterisks indicate values that are significantly different from values for non-pregnant animals (P < 0.05; n = 5 to 7).
A significant number of hepatocytes routinely undergo endoreduplication generating large polyploid cells (21). Hepatocyte polyploidization has been associated with many physiological and pathological processes (22). It is unknown whether pregnancy alters hepatocyte endoreduplication and whether this might impact liver DNA content and growth. Thus, we determined the impact of pregnancy on hepatocyte DNA content. Primary hepatocytes were isolated from the livers of nonpregnant and gestation d18.5 rats, stained with propidium iodide, and subjected to flow cytometry analysis. Livers from pregnant and nonpregnant rats did not differ in their contents of diploid (2N), tetraploid (4N), and octoploid (8N) hepatocytes (Fig. 3). The data indicate that pregnancy does not exert a significant affect on hepatocyte polyploidization.
Fig. 3. Hepatocyte ploidy analysis.

Primary rat hepatocytes were isolated from non-pregnant (np) and gestational d18.5 animals and stained with propidium iodide. Hepatocyte ploidy (2N, 4N, and 8N) was analyzed by flow cytometry using BD LSRII and FACS Diva. Histograms from the flow cytometry are shown in Panel A. Populations of hepatocytes with different DNA contents are shown in Panel B.
Ki-67 immunostaining on tissue sections prepared from livers of nonpregnant, pregnant, and postpartum rats were performed to further assess the impact of pregnancy and lactation on hepatocyte proliferation. A gestation-dependent increase in the number of Ki-67-positive hepatocytes was observed (Fig. 4). Maternal hepatocyte proliferation occurred before implantation (gestation d4.5), continued through midgestation, peaked at gestation d18.5, and subsequently arrested just prior to parturition. Postpartum liver samples did not exhibit evidence of hepatocyte proliferation (Fig. 4). The data indicate that maternal hepatocytes undergo replication throughout most of gestation.
Fig. 4. Ki-67 immunostaining in liver tissues.

Liver tissues were collected from nonpregnant (NP), pregnant (gestational d4.5, d8.5, d11.5, d13.5, d15.5, d18.5, and d21.5), and postpartum d10 non-lactating (nlac) and lactating (lac) rats. The tissues were fixed in formalin and embedded in paraffin. Liver tissue sections were prepared and Ki-67 immunostaining was performed. The nuclei of Ki67-positive cells stained dark brown. (A) Representative liver sections immunohistochemically stained for Ki-67 are shown for NP and gestation d18.5 rats. (B) Ki67-positive hepatocytes were counted (40 × optical field) and the results are shown as mean number of positive nuclei per field ± SD. Asterisks indicate values that are significantly different from values for non-pregnant animals (P < 0.05; n = 3).
Pregnancy alters maternal hepatic gene expression
To identify potential regulators of pregnancy-dependent liver growth, gene expression profiling of nonpregnant and gestation d18.5 maternal hepatic tissue was analyzed using a DNA microarray approach. Genes exhibiting at least a four-fold upregulation or downregulation in mRNA expression (pregnant versus nonpregnant) are listed in Tables 3 and 4. Verification of the changes in hepatic transcript levels of these genes was extended to the entire course of pregnancy and postpartum by northern blot or qRT-PCR analysis (Fig. 5). Genes exhibiting at least a 2.5-fold but less than 4-fold upregulation or downregulation in mRNA expression (pregnant versus nonpregnant) are listed in Supplemental Tables 1 and 2.
Table 3. Selected genes exhibiting increased hepatic expression in pregnant rats.
| Gene | Symbol | Ratio* | Functional group |
|---|---|---|---|
| Nephroblastoma overexpressed gene | Nov | 81.9 | Ligand-signaling |
| Alpha 2-macroglobulin | A2m | 32.7 | Acute phase protein |
| Early growth response 1 | Egr1 | 15.2 | Transcriptional regulation |
| Patatin-like phospholipase domain containing 3 | Pnpla3 | 6.4 | Lipid metabolism |
| Neurotrophic tyrosine kinase, receptor, type 1 | Ntrk1 | 4.8 | Ligand receptor-signaling (neural) |
| Achaete-scute complex homolog-like 1 | Ascl1 | 4.7 | Transcriptional regulation (neural) |
| Insulin-like growth factor binding protein 1 | Igfbp1 | 4.6 | Ligand transport |
| Similar to hypothetical protein – MGC13251 | RGD1308350 | 4.5 | Unknown |
| Tumor necrosis factor receptor superfamily, member 9 (CD137, ILA, 4-1BB) | Tnfrsf9 | 4.4 | Ligand receptor-signaling |
| Prominin 1 | Prom1 | 4.2 | Stem cell surface protein |
Pregnant:non-pregnant
Table 4. Selected genes exhibiting decreased hepatic expression in pregnant rats.
| Gene | Symbol | Ratio* | Functional group |
|---|---|---|---|
| Unknown Est | --------- | 0.05 | Unknown |
| Tumor-associated calcium signal transducer 1 | Tacstd1 | 0.14 | Epithelial cell adhesion molecule |
| Cytochrome P450, 3a18 | Cyp3a18 | 0.16 | Metabolic enzyme |
| Flavin containing monoxygenase 1 | Fmo1 | 0.19 | Metabolic enzyme |
| Hepcidin antimicrobial peptide | Hamp | 0.21 | iron metabolism and inflammation |
| X-prolyl aminopeptidase 2, membrane bound | Xpnpep2 | 0.21 | Prolidase |
| Sulfotransferase family, cytosolic, 1C, member 2 | Sult1c2 | 0.22 | Metabolic enzyme |
| Protein phosphatase 1, regulatory subunit 3C | Ppp1r3c | 0.22 | Glycogen metabolism |
| Monocyte to macrophage differentiation-associated 2 | Mmd2 | 0.23 | Membrane receptor |
| Glucokinase | Gck | 0.24 | Insulin signaling |
| Glutamic pyruvic transaminase 1, soluble | Gpt1 | 0.25 | Biosynthetic enzyme |
| Carbonic anhydrase 3 | Ca3 | 0.25 | Nitrogen metabolism |
Pregnant:non-pregnant
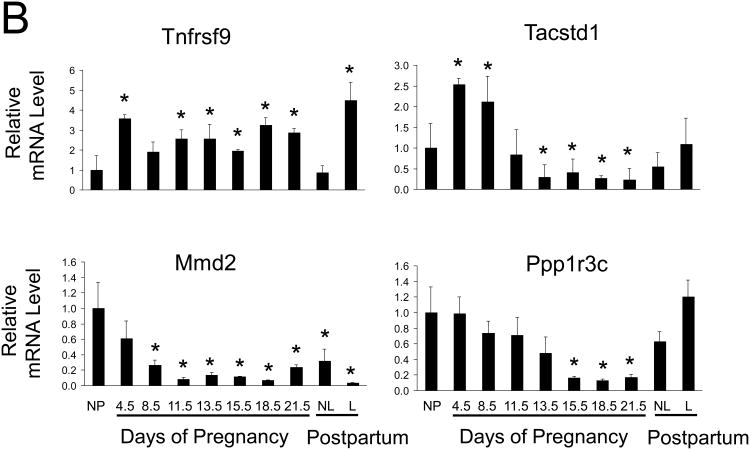
Fig. 5. Hepatic gene expression during pregnancy and lactation in the rat.
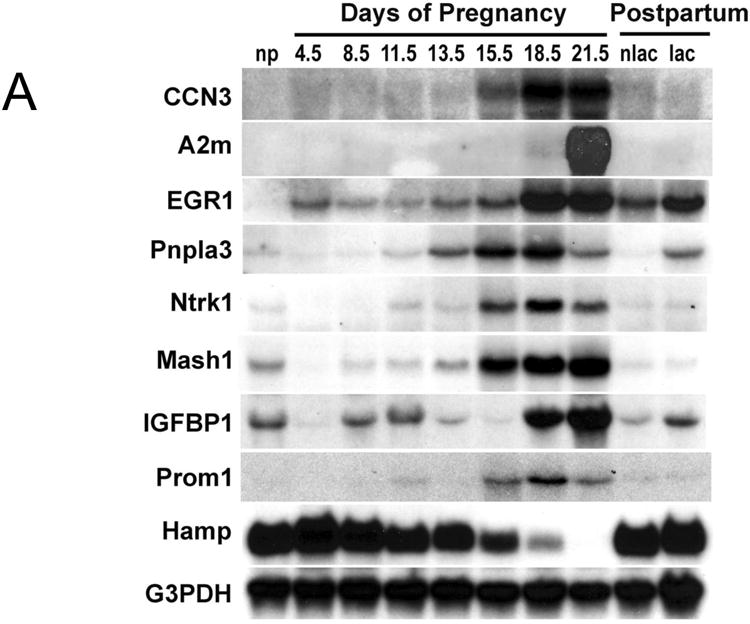
Total RNA was prepared from liver tissue of non-pregnant (np), pregnant (gestational d4.5, d8.5, d11.5, d13.5, d15.5, d18.5, and d21.5), and postpartum d10 non-lactating (nlac) and lactating (lac) rats. (A) The expression of indicated genes was analyzed by northern blotting. G3PDH served as control for loading and RNA integrity. (B) Maternal hepatic mRNA levels of indicated genes were assessed by quantitative RT-PCR and are expressed as means of fold changes compared to nonpregnant controls ± SD (n = 3 for each group). *, P < 0.05.
Genes upregulated in the liver during pregnancy exhibited distinct expression patterns (Fig. 5). Hepatic mRNA expression of nephroblastoma overexpressed gene (Nov/Ccn3), transcription factor mammalian achaete-scute homolog-1 (Ascl1), patatin-like phospholipase domain-containing protein 3 (Pnpla3), neurotrophic tyrosine kinase receptor type 1 (Ntrk1), transcription factor early growth response-1 (Egr1), and a cell surface protein prominin 1 (Prom1) increased as pregnancy advanced, especially during the second half of pregnancy in parallel with increases in maternal liver growth. Hepatic alpha-2 macroglobulin (A2m) transcript levels were prominently elevated at the end of pregnancy, coincident with the arrest of maternal hepatocyte proliferation (Fig. 4). mRNA for TNF receptor super family member 9 (Tnfrsf9) exhibited continued increases in hepatic expression throughout the course of pregnancy. Insulin-like growth factor binding protein 1 (Igfbp1) gene expression was highly dynamic. Igfbp1 mRNA decreased at gestation d4.5, rebounded to pre-pregnancy levels at gestation d8.5 and d11.5, decreased again at gestation days d13.5, and d15.5, and showed a dramatic upregulation prior to parturition (gestation d18.5 and d21.5).
Hepatic downregulation of several regulatory genes were verified by northern blotting or qRT-PCR analyses (Fig. 5). Hepcidin antimicrobial peptide (Hamp), protein phosphatase 1 regulatory subunit 3C (Ppp1r3c), and monocyte to macrophage differentiation-associated 2 (Mmd2) mRNAs declined as pregnancy progressed, inversely related to maternal liver growth. Tumor-associated calcium signal transducer 1 (Tacstd1) transcripts levels were elevated during the first half of gestation and declined during the second half of gestation.
Following parturition, expression levels for most genes investigated, including Ccn3, A2m, Ascl1, Ntrk1, Prom1, Hamp, Tacstd1, and Ppp1r3c, returned to pre-pregnancy levels in livers from both non-lactating and lactating rats (Fig. 5). The data suggest that this subset of genes is regulated in the liver by pregnancy, irrespective of postpartum events. However, the remainder of the tested genes was modulated by both pregnancy and lactation in hepatic expression. Lactation stimulated hepatic expression of Egr1, Pnpla3, Igfbp1, and Tnfrsf9 mRNA and inhibited Mmd2 transcript levels (Fig. 5).
Collectively, we demonstrate that the maternal liver adapts to pregnancy by altering its gene expression profile. Genes exhibiting robust pregnancy-dependent changes in maternal hepatic expression have been identified, providing us with a molecular basis for further investigation of the regulation of liver physiology during pregnancy. Pregnancy and lactation exert distinct effects on hepatic gene expression.
Discussion
The current study identified a robust growth response of the maternal liver to pregnancy, which was characterized by marked increases in maternal hepatic DNA content, hepatocyte proliferation, and increases in liver size. Lactation sustained pregnancy-dependent liver growth. Pregnancy represents a relatively unexplored model system to investigate the regulation of hepatocyte proliferation and liver remodeling. Reduction of liver mass by partial hepatectomy, a most commonly used model in the study of liver regeneration (23), induces intra- and/or extra-hepatic production of a variety of cytokines and growth factors, which coordinately stimulate liver cell proliferation and tissue remodeling (23, 24). Production of interleukin-6 and tumor necrosis factor alpha and activation of urokinase, Notch, β-catenin, signal transducer and activator of transcription protein 3, nuclear factor-kappa B, c-fos, c-jun, hepatocyte growth factor receptor, and epidermal growth factor receptor are among the preparative events for the entry of hepatocytes into the cell cycle. Concomitant with or subsequent to those early responses are the production of direct mitogens, including hepatocyte growth factor and transforming growth factor alpha, and comitogens, such as tumor necrosis factor and norepinephrine. These factors render the hepatocytes to enter into and progress through the cell cycle. As a consequence, the lost liver mass is restored by liver re-growth. Whether or not the factors involved in liver regeneration participate in modulating pregnancy-induced liver growth is under our investigation. Additionally, estrogen enhances the mitogenic effect of growth factors and therefore is classified as a comitogen for hepatocytes (25, 26). In vivo studies showed that estrogen induces transient hepatocyte proliferation and liver growth (27-29). It is evident that estrogen receptor is involved in the regulation of liver regeneration (29-32). Therefore, estrogen receptor signaling should be included in the future mechanistic study on pregnancy-induced liver growth.
Pregnancy induced maternal hepatocyte proliferation without affecting hepatocyte endoreduplication, an important feature of liver growth and physiology. Advanced polyploidy in mammalian cells is indicative of terminal differentiation and senescence (22, 33, 34), leading to a progressive loss of cell pluripotency and decreased replication capacity (35). The biological significance of hepatic polyploidy remains unclear (36). It has been suggested that the polyploid genome may provide protection against the dominant expression of mutated oncogenes for an organ heavily engaged in drug detoxification (37). Another suggestion is that hepatocyte endoreduplication is a normal process that occurs in response to oxidative stress and helps to maintain the detoxification capacity without having to proliferate (21). Remarkably, liver polyploidization is differentially regulated in two models of liver growth:i) following loss of liver mass; ii) liver tumor development. Ploidy of liver cells increases during liver re-growth induced by partial hepatectomy and results in an attenuation of proliferative capacity (38), whereas, hepatocellular tumor growth shifts to a nonpolyploidizing growth pattern manifested by expansion of the diploid hepatocyte population (39, 40). Our study revealed a distinct liver growth pattern, which exhibits massive hepatocyte proliferation without an alteration of polypoidization. This might be a unique feature of pregnancy-dependent liver growth.
Concomitant to its size adjustment to pregnancy, the maternal liver exhibits changes in its gene expression profile. Among the genes showing robust gestation-dependent increases in maternal hepatic expression were Ccn3, Ascl1, Ntrk1, Egr1, Tnfrsf9, Prom1, A2m, and Pnpla3. In contrast, Mmd2, Hamp, and Ppplr3c displayed marked gestation-dependent decreases in maternal hepatic expression. More complex pregnancy-dependent expression patterns were evident for Igfbp1 and Tacstd1. Some of these genes have been implicated in processes that may contribute to the regulation of pregnancy-dependent liver adaptations and more specifically to the control of hepatocyte proliferation and hepatic remodeling (see Supplemental Discussion).
In summary, as pregnancy advances, maternal hepatocytes exhibit an increased rate of proliferation without an increase in endoreduplication. The outcome is growth of the maternal liver proportional to increases in maternal body weight. It is presumed that these maternal hepatic adaptations are directed to meeting the needs of the mother and promoting growth of the placentas and fetuses. The maternal liver also changes its gene expression profile in response to pregnancy. Some of the differentially expressed genes may represent components of regulatory pathways controlling hepatic adaptations. Pregnancy-dependent growth of the maternal liver represents a physiologically-relevant model system for investigating the regulation of hepatocyte proliferation and hepatic remodeling.
Supplementary Material
Table S1. Genes exhibiting increased hepatic expression in pregnant rats
Table S2. Genes exhibiting decreased hepatic expression in pregnant rats
Acknowledgments
The authors thank Joyce Slusser for her assistance with the flow cytometry analysis and Clark Bloomer and the University of Kansas Medical Center Microarray Facility for assistance with the DNA microarray analysis.
This work was supported by grants from the National Institutes of Health (HD020676, HD048861) and the Hall Family Foundation
Abbrevations
- A2M
alpha-2 macroglobulin
- Ascl1
mammalian achaete-scute homolog-1
- Egr1
early growth response-1
- Hamp
hepcidin antimicrobial peptide
- Igfbp1
insulin-like growth factor binding protein 1
- Mmd2
monocyte to macrophage differentiation-associated 2
- NOV
nephroblastoma overexpressed gene
- Ntrk1
neurotrophic tyrosine kinase receptor type 1
- Pnpla3
patatin-like phospholipase domain-containing protein 3
- Ppp1r3c
protein phosphatase 1 regulatory subunit 3
- Tacstd1
tumor-associated calcium signal transducer 1
- Tnfrsf9
TNF receptor super family member 9
References
- 1.Nielsen JH, Svensson C, Galsgaard ED, Moldrup A, Billestrup N. Beta cell proliferation and growth factors. J Mol Med. 1999;77(1):62–6. doi: 10.1007/s001090050302. [DOI] [PubMed] [Google Scholar]
- 2.Bustamante JJ, Dai G, Soares MJ. Pregnancy and lactation modulate maternal splenic growth and development of the erythroid lineage in the rat and mouse. Reproduction, fertility, and development. 2008;20(2):303–10. doi: 10.1071/rd07106. [DOI] [PubMed] [Google Scholar]
- 3.Shingo T, Gregg C, Enwere E, Fujikawa H, Hassam R, Geary C, et al. Pregnancy-stimulated neurogenesis in the adult female forebrain mediated by prolactin. Science. 2003;299(5603):117–20. doi: 10.1126/science.1076647. [DOI] [PubMed] [Google Scholar]
- 4.Audus KL, Soares MJ, Hunt JS. Characteristics of the fetal/maternal interface with potential usefulness in the development of future immunological and pharmacological strategies. J Pharmacol Exp Ther. 2002;301(2):402–9. doi: 10.1124/jpet.301.2.402. [DOI] [PubMed] [Google Scholar]
- 5.Rahman TM, Wendon J. Severe hepatic dysfunction in pregnancy. Qjm. 2002;95(6):343–57. doi: 10.1093/qjmed/95.6.343. [DOI] [PubMed] [Google Scholar]
- 6.Smith JL, Lear SR, Forte TM, Ko W, Massimi M, Erickson SK. Effect of pregnancy and lactation on lipoprotein and cholesterol metabolism in the rat. Journal of lipid research. 1998;39(11):2237–49. [PubMed] [Google Scholar]
- 7.Smith RW. The effects of pregnancy and lactation on the activities in rat liver of some enzymes associated with glucose metabolism. Biochimica et biophysica acta. 1975;411(1):22–9. doi: 10.1016/0304-4165(75)90281-0. [DOI] [PubMed] [Google Scholar]
- 8.Smith RW, Walsh A. Composition of liver lipids of the rat during pregnancy and lactation. Lipids. 1975;10(10):643–5. doi: 10.1007/BF02532731. [DOI] [PubMed] [Google Scholar]
- 9.Smith RW, Walsh A. The composition of the liver lipids of the ewe during pregnancy and lactation. Res Vet Sci. 1975;19(2):230–2. [PubMed] [Google Scholar]
- 10.Mesbah MM, Baldwin RL. Effects of diet, pregnancy, and lactation on enzyme activities and gluconeogenesis in ruminant liver. J Dairy Sci. 1983;66(4):783–8. doi: 10.3168/jds.S0022-0302(83)81858-X. [DOI] [PubMed] [Google Scholar]
- 11.Anderson GD. Pregnancy-induced changes in pharmacokinetics: a mechanistic-based approach. Clin Pharmacokinet. 2005;44(10):989–1008. doi: 10.2165/00003088-200544100-00001. [DOI] [PubMed] [Google Scholar]
- 12.Bernus I, Hooper WD, Dickinson RG, Eadie MJ. Effects of pregnancy on various pathways of human antiepileptic drug metabolism. Clin Neuropharmacol. 1997;20(1):13–21. doi: 10.1097/00002826-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Tracy TS, Venkataramanan R, Glover DD, Caritis SN. Temporal changes in drug metabolism (CYP1A2, CYP2D6 and CYP3A Activity) during pregnancy. Am J Obstet Gynecol. 2005;192(2):633–9. doi: 10.1016/j.ajog.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Dickmann LJ, Tay S, Senn TD, Zhang H, Visone A, Unadkat JD, et al. Changes in maternal liver Cyp2c and Cyp2d expression and activity during rat pregnancy. Biochemical pharmacology. 2008;75(8):1677–87. doi: 10.1016/j.bcp.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waxman DJ, Holloway MG. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol. 2009;76(2):215–28. doi: 10.1124/mol.109.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He XJ, Ejiri N, Nakayama H, Doi K. Effects of pregnancy on CYPs protein expression in rat liver. Exp Mol Pathol. 2005;78(1):64–70. doi: 10.1016/j.yexmp.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Sweeney TR, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. Decreased nuclear hormone receptor expression in the livers of mice in late pregnancy. Am J Physiol Endocrinol Metab. 2006;290(6):E1313–20. doi: 10.1152/ajpendo.00071.2005. [DOI] [PubMed] [Google Scholar]
- 18.Kennaway EL, Kennaway NM. The Ascorbic Acid Content of the Liver in Pregnant Mice. Cancer research. 1944;4(11):704–06. [Google Scholar]
- 19.Kim SK, Abdelmegeed MA, Novak RF. The mitogen-activated protein kinase kinase (mek) inhibitor PD98059 elevates primary cultured rat hepatocyte glutathione levels independent of inhibiting mek. Drug Metab Dispos. 2006;34(4):683–9. doi: 10.1124/dmd.105.007666. [DOI] [PubMed] [Google Scholar]
- 20.Orwig KE, Ishimura R, Muller H, Liu B, Soares MJ. Identification and characterization of a mouse homolog for decidual/trophoblast prolactin-related protein. Endocrinology. 1997;138(12):5511–7. doi: 10.1210/endo.138.12.5628. [DOI] [PubMed] [Google Scholar]
- 21.Lu P, Prost S, Caldwell H, Tugwood JD, Betton GR, Harrison DJ. Microarray analysis of gene expression of mouse hepatocytes of different ploidy. Mamm Genome. 2007;18(9):617–26. doi: 10.1007/s00335-007-9048-y. [DOI] [PubMed] [Google Scholar]
- 22.Gupta S. Hepatic polyploidy and liver growth control. Seminars in cancer biology. 2000;10(3):161–71. doi: 10.1006/scbi.2000.0317. [DOI] [PubMed] [Google Scholar]
- 23.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213(2):286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology (Baltimore, Md) 2006;43(2 Suppl 1):S45–53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 25.Ni N, Yager JD. The co-mitogenic effects of various estrogens for TGF-alpha-induced DNA synthesis in cultured female rat hepatocytes. Cancer letters. 1994;84(2):133–40. doi: 10.1016/0304-3835(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 26.Ni N, Yager JD. Comitogenic effects of estrogens on DNA synthesis induced by various growth factors in cultured female rat hepatocytes. Hepatology (Baltimore, Md) 1994;19(1):183–92. [PubMed] [Google Scholar]
- 27.Payraudeau V, Sarsat JP, Sobczak J, Brechot C, Albaladejo V. Cyclin A2 and c-myc mRNA expression in ethinyl estradiol induced liver proliferation. Molecular and cellular endocrinology. 1998;143(1-2):107–16. doi: 10.1016/s0303-7207(98)00136-1. [DOI] [PubMed] [Google Scholar]
- 28.Yager JD, Zurlo J, Sewall CH, Lucier GW, He H. Growth stimulation followed by growth inhibition in livers of female rats treated with ethinyl estradiol. Carcinogenesis. 1994;15(10):2117–23. doi: 10.1093/carcin/15.10.2117. [DOI] [PubMed] [Google Scholar]
- 29.Fisher B, Gunduz N, Saffer EA, Zheng S. Relation of estrogen and its receptor to rat liver growth and regeneration. Cancer research. 1984;44(6):2410–5. [PubMed] [Google Scholar]
- 30.Kawai T, Yokoyama Y, Kawai S, Yokoyama S, Oda K, Nagasaka T, et al. Does estrogen contribute to the hepatic regeneration following portal branch ligation in rats? Am J Physiol Gastrointest Liver Physiol. 2007;292(2):G582–9. doi: 10.1152/ajpgi.00374.2006. [DOI] [PubMed] [Google Scholar]
- 31.Kitagawa T, Yokoyama Y, Kokuryo T, Kawai T, Watanabe K, Kawai K, et al. Estrogen promotes hepatic regeneration via activating serotonin signal. Shock. 2009;31(6):615–20. doi: 10.1097/SHK.0b013e31818ec195. [DOI] [PubMed] [Google Scholar]
- 32.Francavilla A, Polimeno L, Dileo A, Barone M, Ove P, Coetzee M, et al. The effect of estrogen and tamoxifen on hepatocyte proliferation in vivo and in vitro. Hepatology (Baltimore, Md) 1989;9(4):614–20. doi: 10.1002/hep.1840090417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brodsky WY, Uryvaeva IV. Cell polyploidy: its relation to tissue growth and function. International review of cytology. 1977;50:275–332. doi: 10.1016/s0074-7696(08)60100-x. [DOI] [PubMed] [Google Scholar]
- 34.Sigal SH, Gupta S, Gebhard DF, Jr, Holst P, Neufeld D, Reid LM. Evidence for a terminal differentiation process in the rat liver. Differentiation; research in biological diversity. 1995;59(1):35–42. doi: 10.1046/j.1432-0436.1995.5910035.x. [DOI] [PubMed] [Google Scholar]
- 35.Gorla GR, Malhi H, Gupta S. Polyploidy associated with oxidative injury attenuates proliferative potential of cells. Journal of cell science. 2001;114(Pt 16):2943–51. doi: 10.1242/jcs.114.16.2943. [DOI] [PubMed] [Google Scholar]
- 36.Guidotti JE, Bregerie O, Robert A, Debey P, Brechot C, Desdouets C. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. The Journal of biological chemistry. 2003;278(21):19095–101. doi: 10.1074/jbc.M300982200. [DOI] [PubMed] [Google Scholar]
- 37.Schwarze PE, Pettersen EO, Shoaib MC, Seglen PO. Emergence of a population of small, diploid hepatocytes during hepatocarcinogenesis. Carcinogenesis. 1984;5(10):1267–75. doi: 10.1093/carcin/5.10.1267. [DOI] [PubMed] [Google Scholar]
- 38.Sigal SH, Rajvanshi P, Gorla GR, Sokhi RP, Saxena R, Gebhard DR, Jr, et al. Partial hepatectomy-induced polyploidy attenuates hepatocyte replication and activates cell aging events. The American journal of physiology. 1999;276(5 Pt 1):G1260–72. doi: 10.1152/ajpgi.1999.276.5.G1260. [DOI] [PubMed] [Google Scholar]
- 39.Saeter G, Lee CZ, Schwarze PE, Ous S, Chen DS, Sung JL, et al. Changes in ploidy distributions in human liver carcinogenesis. Journal of the National Cancer Institute. 1988;80(18):1480–5. doi: 10.1093/jnci/80.18.1480. [DOI] [PubMed] [Google Scholar]
- 40.Seglen PO. DNA ploidy and autophagic protein degradation as determinants of hepatocellular growth and survival. Cell biology and toxicology. 1997;13(4-5):301–15. doi: 10.1023/a:1007487425047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes exhibiting increased hepatic expression in pregnant rats
Table S2. Genes exhibiting decreased hepatic expression in pregnant rats