Abstract
Perilipin proteins were discovered in the adipocyte, where they regulate lipid storage and lipolysis. Animal knockout models provided initial evidence of the critical role of perilipin 1, the most abundant of the adipocyte proteins, in energy and glucose metabolism. During a decade of study, genetic variation in perilipin 1 has been consistently but not invariably associated with body weight and obesity-related complications. Related phenotypes such as postprandial lipid metabolism and aerobic fitness are also modulated by perilipin 1 genotype, consistent with earlier metabolic studies. Investigations of gene-diet interactions, together with gene expression studies, have yielded increased understanding, but important questions about causal variants and mechanisms remain. The newest work examines perilipin 4, an adipocyte regulator of triglyceride synthesis and packaging. The novel discovery that a perilipin 4 variant creates a binding site for regulation of the perilipin gene (PLIN) by microRNA suggests intriguing new possibilities for additional mechanistic investigations of other perilipin proteins.
Keywords: epidemiology, genetics, nutrients, obesity
INTRODUCTION
In 2008, 1.46 billion people worldwide were estimated to be overweight, of which 502 million were obese and 347 million affected by diabetes, a major complication of obesity.1,2 Despite the undeniable impact of environment on obesity, some individuals appear to be obesity resistant, and the high heritability of obesity (up to 77%) reflects the strong impact of genetics.3 Dozens of genetic contributors to obesity have been identified, often through the agnostic approach provided by genome-wide association studies.4–6 In contrast, other genetic contributors have arisen through recognition of their key functional roles in energy and lipid homeostasis. Perilipin 1 is among these functionally well-characterized contributors.
Perilipin 1 was identified over 20 years ago as the most abundant protein surrounding the lipid droplet in the adipocyte.7,8 Previous nomenclature for the PAT9 (perilipin, adipocyte differentiation-related protein [ADRP], tail-interacting protein of 47 kDa [TIP47]) family of proteins was recently revised to clearly describe the locations of these proteins.10 Five sequentially numbered protein and gene names (perilipin 1 [PLIN1] through perilipin 5 [PLIN5]) are now used. Perilipin 1 (gene name: PLIN1) is the most well-characterized member of the perilipin family, and, except for perilipin 4 (PLIN4),11 the only perilipin protein for which genetic variability has been associated with human disease.
The role of perilipin 1 in energy metabolism and dysregulation centers on its ability to control access to the adipocyte triglyceride stores that supply most tissues with fuel. Depending on the energy state of the organism, perilipin 1 either limits lipase access to stored triglycerides (in the fed state) or facilitates hormonally stimulated lipolysis (in the fasted state).12 Upon catecholamine stimulation, protein kinase A phosphorylates perilipin 1, facilitating hormone-sensitive lipase translocation to the lipid droplet surface, where lipolysis occurs.13 Adipose triglyceride lipase (ATGL) was recognized during the last decade as an additional participant in lipolysis.14 In the basal state, hypophosphorylated perilipin 1 appears to modulate ATGL function by sequestering its coactivator, comparative gene identification-58 (CGI-58, also called Abhd5), which is released in response to perilipin phosphorylation.15,16 The understanding of relationships between the two new components (ATGL and CGI-58) and perilipin 1 is still incomplete. As a regulator of lipid storage and lipolysis, perilipin 1 is positioned to modify not only obesity risk but also the complications of obesity. Obesity is often accompanied by dysregulated lipolysis, in which excess release of fatty acids from the adipose tissue to the plasma contribute to insulin resistance and type 2 diabetes.17–19 These phenotypes have been explored extensively using animal models.
RESULTS FROM ANIMAL MODELS
Knockout of the perilipin 1 gene (plin1) in mice was instrumental in the development of functional understanding of perilipin 1. Compared with wild-type mice, plin1-null mice exhibited similar food consumption but resistance to obesity induced by a high-fat diet, as well as increased basal lipolysis and reduced isoproteronol-stimulated lipolysis.20 Along with a lean phenotype, knockout mice demonstrated greater glucose intolerance and increased insulin resistance, which were attributed to unregulated lipolysis.
Interestingly, overexpression of perilipin 1 in mice was recently shown to be protective against obesity, adipocyte hypertrophy, and glucose intolerance in mice exposed to a high-fat diet. In that model, both basal lipolysis and stimulated lipolysis were decreased.21 The metabolic benefits associated with overexpression were similar in mice expressing either murine or human perilipin and were accompanied by increased expression of the genes encoding the oxidative enzymes carnitine palmitoyltransferase 1 and 3-ketoacyl-CoA thiolase B, particularly in brown adipose tissue. The implications of these results for metabolic disease in humans are unclear.
PERILIPIN EXPRESSION AND CONTENT IN HUMAN OBESITY
Earlier human studies investigated links between adipocyte perilipin 1 content and obesity phenotypes but did not establish consistent results. Kern et al.22 found adipose perilipin 1 mRNA and protein levels to be greater in nondiabetic obese individuals than in lean individuals and to be positively correlated with percent body fat but unrelated to insulin resistance or inflammatory markers, including tumor necrosis factor alpha (TNF-α). Conversely, another study by Wang et al.23 reported lower perilipin 1 expression in severely obese individuals (mean body mass index [BMI], 53) than in nonobese individuals (mean BMI, 25). In the second study, perilipin 1 protein content was lower in obese individuals, but the mass of perilipin protein per fat cell did not differ between obese and nonobese subjects. In a third study, Mottagui-Tabar et al.24 confirmed a lower perilipin 1 protein content in the adipocytes of obese women (mean BMI, 40), which was associated with a greater rate of lipolysis.
The reasons for the inconsistencies among these three studies (e.g., higher versus lower perilipin content with obesity) are unclear and may be related to differences in the severity of obesity and the degree of metabolic dysregulation of the subjects, as well as to characteristics of the nonobese comparison groups. For example, the obese subjects studied by Wang et al.23 were bariatric surgery patients (mean BMI, 53) and were compared with overweight individuals (mean BMI, 25). In the study by Kern et al.,22 the mean BMI of obese subjects was 35 in men and 34 in women. Individuals in the Kern et al.22 study were normotriglyceridemic (on average) and not yet diabetic and had wide-ranging BMIs (21–65). In the study by Mottagui-Tabar et al.,24 the mean BMI of obese individuals was 40, and they were compared with normal-weight individuals (mean BMI, 23). Mottagui-Tabar et al.24 also noted substantial interindividual variability in adipocyte perilipin content in their population. Despite the apparently conflicting results, the studies support relationships between metabolic phenotypes and perilipin content, perhaps suggesting a bidirectional relationship in which metabolic phenotypes modulate perilipin content, and vice versa. The study by Mottagui-Tabar et al.24 was also the first of many to investigate genetic variability of PLIN1 in humans and to provide evidence for a potential impact of the PLIN1 genotype on disease risk in humans.
GENETIC VARIABILITY AND LIPOLYSIS: INTRODUCING GENETICS INTO THE STUDY OF PLIN1
The human chromosomal location of PLIN1 (15q26) was determined in the late 1990s,25 and several years later Mottagui-Tabar et al.24 genotyped the seven PLIN1 single-nucleotide polymorphisms (SNPs) then available in the National Center for Biotechnology Information (NCBI) Single-Nucleotide Polymorphism database (dbSNP). Two of these (rs1052700 and rs894160) were frequent enough for analysis in a population of 117 obese Swedish women. This group was the first to identify a relationship between a PLIN1 SNP in intron 6 (rs894160) and metabolic outcomes. Specifically, the group reported that adipocyte perilipin content was lower and norepinephrine-induced lipolysis was greater in homozygous minor participants (AA) than in homozygous major participants with GG genotype, and that heterozygotes (AG individuals) exhibited intermediate phenotypes. Further, when adipocyte perilipin 1 content was measured in a subset of 26 women and dichotomized into high and low, individuals with low perilipin 1 content had higher BMI, higher rates of basal lipolysis, and higher levels of serum nonesterified fatty acids. The investigators concluded that perilipin 1 content was directly linked to adipocyte lipolytic capacity. Interestingly, this study did not report any associations between PLIN1 genotype and BMI or lipid traits for either SNP in the small cohort. The results of this study, however, combined with strong evidence from animal and in vitro models, were an important impetus for many subsequent efforts in human populations.
PLIN1 GENOTYPE AND OBESITY PHENOTYPES
Qi et al.26 were among the earliest to systematically investigate PLIN1 genotype associations for obesity and related phenotypes, for which they analyzed large, multi-ethnic populations. They focused on a set of six SNPs (rs2289487, rs1561726, rs2304794, rs894160, rs2304795, rs1052700), which was to form the basis for many subsequent genetic association studies (Figure 1). Beginning with a population of 1,589 Spanish individuals, they detected gender-specific associations between two SNPs (rs2289487 and rs894160) and anthropometric and metabolic traits.26 In women only (n = 801), carriers of the minor alleles for rs2289487 and rs894160 exhibited a lower risk of obesity and related traits, including plasma glucose and triglycerides. None of the other four SNPs were associated with metabolic outcomes. The rs894160 SNP was the same variant for which Mottagui-Tabar et al.24 had detected differences in rates of lipolysis. These authors were also the first to investigate PLIN1 genetic structure, reporting that the two SNPs for which they detected similar protection against obesity were in strong linkage disequilibrium (LD).
Figure 1. Common PLIN1 single-nucleotide polymorphisms (SNPs).
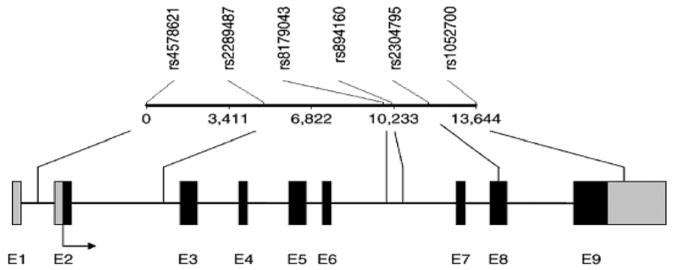
Common PLIN1 SNPs are indicated above and are not drawn to scale. Exons are designated by the letter E and numbered as black rectangles. Gray rectangles indicate the promoter and 3′ untranslated regions, with the arrow indicating the direction of transcription. Unique identifications for SNPs and their positions are indicated at the top, and the positions of SNPs relative to exons are indicated with lines. Adapted from Qi et al.47
A subsequent study by Qi et al.27 in a population of 734 white subjects in the United States (373 men and 361 women) confirmed the importance of the PLIN1 locus for obesity traits in women, but in this population, associations were detected for two other SNPs (rs2304795, rs1052700), both individually and as a haplotype, which were different from the ones that were protective in the Spanish women.27 Further, minor alleles for this pair of SNPs were associated with increased risk of obesity rather than protection from obesity.
Extension of these analyses to a third large population (n = 4,131; Chinese, Malays, and Asian Indians in Singapore) again confirmed the relationship between PLIN1 and obesity.28 In the Singaporeans, the strongest associations were for rs1052700, which was also associated with altered risk in the US women, but in the opposite direction. In contrast to the findings in US whites, in Malays and Indians rs1052700 was associated with protection from obesity rather than risk of obesity. Further, homozygous carriers of the minor alleles for rs894160 or rs2304795 were also at increased risk of obesity, although rs894160 had been protective in whites. Results from the Singapore study have been subsequently confirmed in Japanese males, in whom carrying two copies of the minor allele for rs894160 was associated with increased BMI.29 Ethnically based differences in LD may provide partial explanation for the apparent discrepancies: in Asians, rs1052700 and rs894160 (rather than rs2289487 and rs894160 in whites) were in LD. The results also collectively suggest that the SNPs evaluated are unlikely to be causal variants but are likely to be markers for an unidentified functional variant or variants.
Despite the persuasive evidence provided by Qi et al.,26–28 some studies failed to detect associations between PLIN1 and obesity-related phenotypes,30–35 and additional studies with negative findings undoubtedly remain unpublished. Since diet so strongly modifies the risk of obesity, examination of dietary modulations of PLIN1 associations represents a logical next investigative step, which has generated fruitful results. The first study of dietary modulation of PLIN1 analyzed the four SNPs that had been associated with baseline obesity in other populations (rs2289487, rs894160, rs2304795, rs1052700).36 Baseline analysis of 150 severely obese Spaniards confirmed results reported earlier in whites for a protective effect against obesity for the minor allele of rs894160. Further, when a subset of the Spanish cohort (n = 48) was exposed to an energy-restricted intervention, carriers of that protective allele were more resistant to weight loss compared with major allele homozygotes. Body weight in individuals carrying the minor allele for rs894160 may be generally less likely to change in response to environmental shifts.
In addition, the response of carriers of the minor allele for rs894160 to a low-energy diet may also be related to the macronutrient composition of the diet, which consisted of approximately 40% of total energy from fat, 20% from protein, and 40% from carbohydrate (CHO). The relatively high proportion of energy from fat and the low proportion from CHO (a 1:1 ratio) suggest the possibility that dietary composition might affect PLIN1-related resistance to weight loss, which was explicitly tested in a subsequent study. In an observational study of 920 Puerto Ricans living in the United States, dietary CHO interacted with PLIN1 genotype for the same SNP (rs894160), in that carriers of the minor allele were protected from increased adiposity in the context of high complex CHO intake, but at risk of increased adiposity when complex CHO intake was low.37
Results from two later weight-loss interventions, which were not designed to examine the role of macro-nutrients, might also be interpreted from this perspective. Ruiz et al.38 conducted an energy-restricted intervention (n = 78 obese Spanish women) in which carriers of the minor allele for rs894160 lost less central fat, and this loss of fat was positively correlated with lipid oxidation rates. Their observations of resistance to adiposity loss in carriers of the minor allele for rs894160 were consistent with the results from Corella et al.36 and with previously reported associations with lower lipid oxidation and weight-loss resistance.39 In the Ruiz et al.38 study, energy from CHO was 55% and energy from lipids 30%, representing a 1.8:1 CHO:fat ratio that was greater than that of the Corella et al.36 intervention, though it still contained moderate fat. In contrast, in another intervention of 188 obese Dutch individuals, women carriers of the minor allele for rs894160 demonstrated greater weight loss and fat loss.40 Of note, 42% of total energy was obtained from protein, 40% from CHO, and only 13% from fat, representing a CHO:fat ratio of 3:1. The greater ratio of CHO to fat (compared with ratios in the Corella et al.36 and Ruiz et al.38 studies) may have facilitated weight loss in carriers of the minor allele for rs894160, similar to the protective advantage observed by Smith et al.37 in Puerto Rican carriers of minor alleles in whom high CHO intake was associated with lower adiposity. Collectively, these studies support the hypothesis that a low-fat, high-CHO diet may be protective against obesity for individuals with the minor allele for rs894160.
Only one weight-loss intervention has included children and adolescents; the study cohort consisted of 234 Brazilians aged 7–14 years.41 In this study, PLIN1 genotype was not associated with baseline differences in anthropometric traits, but carriers of the minor allele for PLIN1 rs1052700 demonstrated greater weight loss than noncarriers. Although the individuals were reported to be of mixed ancestry that was not quantified, the strong LD between rs2289487 and rs894160 was similar to that described earlier in European populations. This study is the only intervention so far to report an association between improved weight loss and rs1052700, although the minor allele for this SNP has been associated with increased obesity risk in Singaporean Asians28 and US white women.27 The discrepancy between results from observational studies and this intervention is reminiscent of that described above for rs894160, in which an SNP observed to be protective at baseline was associated with resistance to weight loss in an interventional setting. Uncontrolled factors in observational studies may be interacting with PLIN1 genotype through mechanisms yet to be identified.
Two more interventions expand the biological understanding of PLIN1 through investigation of additional obesity-related phenotypes and confirm the relevance of two previously explored SNPs. In one energy-restricted intervention (n = 177 Koreans), Jang et al.42 demonstrated that carriers of the minor allele for either rs894160 or rs1052700, which are in strong LD in Asians, showed greater waist and fat reduction as well as a greater change in free fatty acids following weight loss. These two observations may be physiologically linked, since free fatty acids may reflect increased lipolysis accompanying weight loss, and rs894160 is the same SNP for which Mottagui-Tabar et al.24 reported greater rates of lipolysis in obese women. The second trial evaluated weight gain in the context of treatment with rosiglitazone, an insulin-sensitizing agent and peroxisome proliferator-activated receptor gamma (PPARγ) agonist.43 Treatment-associated weight gain in the diabetic treatment group (n = 160 Koreans; 84 men and 76 women) was lower in homozygous carriers of the minor allele for rs894160 compared with other genotypes. Homozygosity for the minor allele for this SNP had previously been reported to be associated with increased obesity risk or a tendency toward increased risk in Asians (Singaporean Malays and Indians).28 This study is also of interest in light of demonstrated PPARγ regulation of PLIN1 expression, suggesting that PLIN1 genotype may mediate responses to PPARγ stimulation.44,45
PLIN1 GENOTYPE AND GLUCOSE-RELATED PHENOTYPES
The well-recognized link between obesity and impaired glucose metabolism has led to extension of PLIN1 analyses to glucose and insulin phenotypes. In the PLIN1 knockout mouse, these relationships were uncoupled to some extent, as the deletion of PLIN1 created a lean mouse with increased insulin resistance that was attributed to unregulated lipolysis.20 In some human observational studies, PLIN1 genotype appears to be independently associated with obesity risk and plasma glucose concentrations. For example, in the cohort of Spanish women in whom linked PLIN1 SNPs rs2289487 and rs894160 were protective against adiposity, an association between these SNPs and lower plasma glucose was also found, and adjustment by BMI did not eliminate statistical significance of the association.26 In another population of US white women with (n = 185) and without (n = 120) polycystic ovarian syndrome,46 the minor allele for PLIN1 rs1052700 (previously associated with mixed risk for obesity traits in whites) was associated with glucose intolerance and higher fasting glucose. Unfortunately, rs894160 was not available in that study.
Glucose metabolism was also evaluated in the obese Brazilian children, in whom PLIN1 rs1052700 had conferred a weight-loss advantage during an energy-restricted intervention.41 The minor allele of the same SNP was associated with lower plasma insulin and HOMA-IR (homeostasis model assessment of insulin resistance) post-intervention in that study, but whether these results were driven by the weight loss is unclear. In the same group of children, PLIN1 rs894160 minor allele was associated with greater values for both baseline insulin concentration and insulin area under the curve (AUC) following an oral glucose tolerance test. These results may be unexpected, since rs894160 was protective against obesity in several groups; for glucose metabolism, however, the minor allele appears to increase risk.
These conflicting data may reflect the physiology demonstrated in animal models, in which PLIN1 knockout created a lean but insulin-resistant phenotype. Another possibility is that PLIN1 relationships with glucose metabolism may be “overcome” by the obesity phenotype, which interferes with the detection of genetic risk. This hypothesis is suggested by data from a nested case-control design consisting of 431 cases of incident type 2 diabetes and 791 controls in a group of mostly white US women.47 In that study, the two well-known and linked PLIN1 variants (rs2289487 and rs894160) and a third less-studied promoter SNP (rs8179043) were associated with risk of diabetes only in individuals without central obesity. The authors concluded that the centrally obese women, already metabolically stressed, were less sensitive to the effects of a single gene. In effect, obesity may be overwhelming the effects associated with genotype, a hypothesis that has been suggested for other metabolically important genes.48
Conflicting data may also be examined from a nutritional perspective, since dietary macronutrients may be modulating relationships between PLIN1 and glucose metabolism phenotypes. As was shown for obesity, gene-diet interactions have uncovered relationships that were not detectable as associations. For example, although PLIN1 genotype was not associated with plasma insulin independently of nutritional factors in Singaporean Asian women (Malays, Indians, and Chinese), Corella et al.49 detected interactions between PLIN1 rs894160 and PLIN1 rs1052700 and the macronutrients saturated fat and CHO, which were strongest when considered as a ratio. For homozygous-minor carriers of either SNP, the SFA:CHO ratio was associated with increased plasma insulin and HOMA-IR. Confirmation of gene-nutrient interactions in independent populations are important and, unfortunately, rare, but this relationship was replicated in women comprising a population of US whites for PLIN1 rs894160.50 In both Asian and white populations, inclusion of a measure of adiposity (BMI or waist) did not attenuate the significance of the interaction, suggesting that obesity was not the primary mediator of the SNP-related insulin resistance.
SUMMARY OF PLIN1 AND OBESITY-RELATED TRAITS
Several overall themes emerge from the wide range of observational and intervention studies investigating PLIN1 and outlined in Table 1. A number of studies support a protective role for a pair of strongly linked SNPs (rs894160 and rs2289487), as well as an association with increased obesity for a third SNP (rs1052700), in whites. LD differences appear to account for some ethnic-specific differences, as Tai and Ordovás51 recognized in an earlier review and as outlined in Table 2, but are less likely to explain differences within an ethnic group. Interaction with nutritional factors may represent another explanation for discrepancies and, although not yet investigated systematically, may underlie national differences, such as Spanish women in whom rs894160 and rs2289487 decreased obesity risk versus US women in whom rs1052700 and rs2304795 increased obesity risk.26,27 Whether dietary CHO and fat are actual regulators of PLIN1 function is unclear, although the effect of dietary fat on gene expression has been more commonly explored and will be discussed in a later section. Another recurrent theme is the strong but not universal gender specificity to PLIN1 that was reviewed earlier.52 Although a single study reported that PLIN1 transcription is activated by estrogen receptor-related receptor,53 the role of gender in PLIN1 may reflect wider relationships between gender, obesity, and the metabolic consequences of obesity.54 Finally, despite the identification of a key set of SNPs associated with several metabolic phenotypes, the causal SNP or SNPs have not been identified, and the mechanisms by which diet or other environmental factors alter PLIN1 function are still unknown.
Table 1.
Commonly studied PLIN1 single-nucleotide polymorphisms (SNPs) and obesity-related phenotypes in humans.
| SNP | Population | Main findings | References |
|---|---|---|---|
| rs2289487 | Spanish (n = 1,589) | Lower obesity, glucose, and related traits in women | Qi et al. (2004)26 |
| rs2289487 | US whites, nested case-control (n = 431 cases, n = 791 controls) | Higher diabetes risk | Qi et al. (2008)47 |
| rs2289487 | US whites (n = 271) Spanish men (n = 88) |
Lower postprandial response | Perez-Martinez et al. (2008)55 |
| rs894160 | Swedish women (n = 117) | Lower adipocyte perilipin, higher lipolysis | Mottagui-Tabar et al. (2003)24 |
| rs894160 | Spanish (n = 1,589) | Lower obesity, glucose, and related traits in women | Qi et al. (2004)26 |
| rs894160 | Singaporean Chinese, Indians, and Malays (n = 4,131) | Higher obesity risk in women | Qi et al. (2005)28 |
| rs894160 | Japanese men (n = 148) | Higher BMI | Sone et al. (2010)29 |
| rs894160 | Spanish women (n = 48) | Resistance to weight loss (energy-restricted intervention) | Corella et al. (2005)36 |
| rs894160 | US Puerto Ricans (n = 920) | Lower obesity traits with high complex carbohydrate intake | Smith et al. (2008)37 |
| rs894160 | Spanish (n = 78) | Resistance to weight loss (energy-restricted intervention), lower lipid oxidation | Ruiz et al. (2011)38 |
| rs894160 | Dutch (n = 188) | Higher weight loss and fat loss (weight-loss intervention) | Soenen et al. (2009)40 |
| rs894160 | Koreans (n = 277) | Higher weight loss, greater FFA change following weight loss | Jang et al. (2006)42 |
| rs894160 | Koreans (n = 160) | Lower weight gain with rosiglitazone (antiglycemic) | Kang et al. (2006)43 |
| rs894160 | US whites, nested case-control (n = 431 cases, n = 791 controls) | Higher diabetes risk | Qi et al. (2008)47 |
| rs894160 | Singaporean Chinese, Malays, and Indians, (n = 1,909 men, n = 2,198 women) | Interaction between saturated-fat-to-carbohydrate ratio and insulin resistance in women | Corella et al. (2006)49 |
| rs894160 | US whites (n = 462 men, n = 508 women) | Interaction between saturated-fat-to-carbohydrate ratio and insulin resistance in women | Smith et al. (2012)50 |
| rs894160 | US whites (n = 271) Spanish men (n = 88) |
Lower postprandial response | Perez-Martinez et al. (2008)55 |
| rs2304795 | US whites (n = 734) | Higher obesity risk in women | Qi et al. (2004)27 |
| rs2304795 | Singaporean Chinese, Indians, and Malays (n = 4,131) | Higher obesity risk in women | Qi et al. (2005)28 |
| rs2304795 | US whites (n = 56 women, n = 46 men) | Lower gains in aerobic capacity; altered glucose and lipid metabolism (response to endurance exercise intervention) | Jenkins et al. (2010)56 |
| rs1052700 | US whites (n = 734) | Higher obesity risk in women | Qi et al. (2004)27 |
| rs1052700 | Singaporean Chinese, Indians, and Malays (n = 4,131) | Lower obesity risk in women | Qi et al. (2005)28 |
| rs1052700 | Brazilian children, aged 7–14 (n = 234) | Higher weight loss (weight-loss intervention), lower insulin | Deram et al. (2008)41 |
| rs1052700 | Koreans (n = 277) | Higher weight loss, greater FFA change following weight loss | Jang et al. (2006)42 |
| rs1052700 | US women with (n = 185) and without (n = 120) polycystic ovarian syndrome | Glucose intolerance | Kawai et al. (2009)46 |
| rs1052700 | Singaporean Chinese, Malays, and Indians, (n = 1,909 men, n = 2,198 women) | Interaction with saturated-fat-to-carbohydrate ratio for insulin resistance | Corella et al. (2006)49 |
| rs1052700 | US whites (n = 56 women, n = 46 men) | Lower gains in aerobic capacity; altered glucose and lipid metabolism (response to endurance exercise intervention) | Jenkins et al. (2010)56 |
Abbreviations: BMI, body mass index; FFA, free fatty acids.
Table 2.
Linkage disequilibrium for common PLIN1 single-nucleotide polymorphisms (SNPs) by ethnicity.
PLIN1 GENOTYPE AND OTHER PHENOTYPES: LIPIDS, POSTPRANDIAL METABOLISM, AND AEROBIC FITNESS
Most PLIN1 studies have concentrated on phenotypes that are strongly related to adipocyte function, including obesity, and the effects of dysregulated lipolysis on glucose metabolism. A smaller number of studies report associations between PLIN1 genotype and baseline lipid concentrations, and a single study has evaluated post-prandial lipemia.55 In several studies discussed previously, the alleles that are associated with higher anthropometric measures or higher glucose are also associated with higher triglyceride or cholesterol (total or low-density lipoprotein cholesterol, or lower high-density lipoprotein cholesterol [HDL-C], as would be expected).26,41,46 In others, obesity and lipid trait differences are associated with different variants.27,41 Whether these results represent biological relationships is unclear.
Only one publication has reported PLIN1 genotype-related differences in postprandial lipid metabolism, although the results are strengthened by the similar responses observed in two white populations.55 In US whites (n = 271 men and women) and Spanish men (n = 88), lipid responses to an oral fat load were measured at intervals. In both populations, the presence of the minor alleles of PLIN1 rs2289487 and PLIN1 rs894160 were associated with lower postprandial concentrations of plasma triglycerides than those found in homozygous-major subjects. As observed in other white populations, the variants were in strong LD in both groups. Differences in BMI were not detected in either population, and baseline triglycerides were lower in carriers of the minor alleles only in the US population, which may have been detectable due to the larger US sample size. Physiological hypotheses to account for these results are plausible, based on the role of adipose as the major site of fatty acid storage in the postprandial period and the reported lower perilipin content in carriers of the minor alleles for PLIN1 rs894160.24 The authors propose that opposing effects on hormone-sensitive lipase (a lipolytic target of perilipin 1 during the fasted state) and lipoprotein lipase (which facilitates fatty acid uptake by adipose in the fed state) during the postprandial period may underlie these appealing results.
Only one study so far has examined relationships between endurance exercise and PLIN1 genotype for a series of phenotypes, including aerobic fitness, along with the more commonly studied metabolic phenotypes.56 Sedentary study participants (56 women and 46 men) completed a 6-month training program consisting of varied endurance activities performed three times a week at 70% VO2max for 40 minutes. Jenkins et al.56 evaluated a PLIN1 haplotype of rs2304795 and rs1052700 (reported previously to be related to obesity in US white women27). At baseline, minor allele subjects exhibited higher BMI and higher body fat compared with major allele subjects, and this difference persisted after training. Although aerobic capacity did not differ by genotype at baseline, minor allele carriers achieved smaller increases in aerobic capacity. Glucose and insulin responses to the oral glucose tolerance test were better in major allele subjects, but only male minor allele carriers showed improvement in insulin response post-intervention. However, only women who were minor allele carriers experienced improvements in HDL (increase) and triglyceride (decrease) levels in response to training.
The results from the exercise intervention are largely consistent with earlier studies of obesity and related traits. To summarize, the presence of the minor alleles for rs2304795 and rs1052700 predisposed individuals to obesity and also to the lower level of fitness that might be expected to accompany adiposity. Similarly, these individuals appear resistant to the improvements in body weight and fitness that exercise provides. Despite the apparent physiological disadvantages of this genotype in the context of an obesogenic environment, the metabolic complications of obesity (dysregulated glucose in men and lipids in women) showed improvement in response to exercise only in carriers of minor alleles. Whether the gender-associated differences can be interpreted in light of the small sample sizes is unclear, but additional investigation of the modulation of PLIN1-associated risk through exercise is warranted.
IN VITRO AND ANIMAL MODELS OF NUTRITIONAL MODULATION OF PERILIPIN 1
The human observational and intervention studies discussed above provide substantial evidence that environmental factors, especially nutrition, modify metabolic differences related to PLIN1. Functional data to support mechanisms through which diet modifies perilipin 1 function have usually examined dietary fat or specific fatty acids. In particular, high-fat feeding (60% of total energy from fat for 8 weeks) in mice resulted in reduced perilipin 1 content in adipocytes and reduced phosphorylation of its target, hormone-sensitive lipase, but also increased ATGL content and increased expression of its coactivator, CGI-58.57 Visceral and subcutaneous adipocytes from these high-fat fed mice demonstrated increased basal lipolysis and reduced epinephrine-stimulated lipolysis, both of which are consistent with the reduced perilipin 1 content. Palmitate uptake and oxidation by visceral adipocytes from high-fat fed mice was decreased under basal and insulin-stimulated conditions compared with that by control adipocytes. These results may lend support to studies described earlier in which PLIN1 variants interacted with dietary macronutrients (CHO, SFA:CHO ratio) to modulate obesity and insulin resistance.49,50 However, a complication in the interpretation of “high-fat feeding” studies is that, in animal models, high-fat feeding generally produces obese animals. Although these models are useful in elucidating the biology of perilipin 1 and related proteins, they cannot be used to differentiate the effects of a specific nutrient from those of excess energy.
A series of additional studies have examined the effects of a particular fatty acid, conjugated linoleic acid (CLA), on expression of a series of genes and proteins related to adipocyte metabolism. House et al.58 reported that body-fat reduction with trans-10, cis-12 CLA feeding was linked to decreased adipose expression of PPARγ and PLIN1 and to increased adipose expression of TNF-α. TNF-α had been previously shown to increase lipolysis in adipocytes, and this effect is believed to be mediated through downregulation of perilipin 1.59 Two other groups have investigated the specific effects of CLA. Chung et al.60 confirmed that trans-10, cis-12 CLA reduced lipolysis and induced delipidation. They also expanded the characterization of CLA-induced changes by describing the displacement of perilipin 1 on the lipid droplet surface and its replacement by perilipin 2 (adipose differentiation-related protein). In a complex dietary design, a third group examined the response of perilipin 1 and related lipases to intake of CLA in conjunction with other sources of fatty acids. The feeding of CLA in combination with soy oil but not coconut oil was associated with reduced body fat, increased lipolysis, and increased expression of activated perilipin 1. Unexpectedly, the increased expression of perilipin 1 was not accompanied by increased concentrations of hormone-sensitive lipase and ATGL.61 In summary, it is unclear whether this single fatty acid, CLA, has substantial implications for human obesity. However, this set of studies supports the possibility that nutrient ratios (e.g., saturated fatty acids to polyunsaturated fatty acids [PUFA]) or combinations of nutrients may regulate PLIN1.
In addition to fatty acids, several other dietary compounds derived from plants have been recently investigated for their roles in either perilipin 1 expression in preadipocytes or perilipin content in adipocytes. The garlic-derived organosulfur compound 1,2-vinyldithiin (1,2-DT) was demonstrated to reduce gene expression of PPARγ and expression of perilipin 1 protein in preadipocytes derived from subcutaneous adipose in nonobese women.62 The authors suggest that the antiadipogenic effects of 1,2-DT are evidence of its potential as an antiobesity agent. In contrast, the compound phloretin, a dietary flavonoid found in apples and strawberries, was shown to increase the expression of PLIN1 (and other PPARγ targets) via PPARγ activation in 3T3-L1 adipocytes.63 The authors propose that the enhanced adipocyte differentiation induced by phloretin resembles that of the insulin-sensitizing agents, the thiazolidinediones, and that the compound will provide similar insulin-sensitizing benefits. A third study examined the effects of Aralia mandshurica and Engelhardtia crysolepsis extracts, which provided triterpene saponines (aralosides) and the flavonoid dehydroquercetin.64 In that study, obese women (n = 32) in an energy-restricted intervention who were given these plant extracts as supplements exhibited greater fat loss, greater reduction of adipocyte perilipin content, and greater hormone-sensitive lipase activity compared with placebo-treated women. Mechanisms to account for the effects of the plant extracts on either weight loss or perilipin content are unclear. Of interest in these studies is that the dietary compounds being evaluated for potential protective qualities modify perilipin 1 in two opposite directions, reduced and increased expression or content, with one action addressing adiposity and the other insulin resistance. In light of the tight linkages between these conditions in people, the health implications of these opposing effects are unclear.
PERILIPIN 4: NOVEL GENETIC FINDINGS
All of the studies discussed thus far have focused efforts on the relationship between PLIN1 genotype and metabolic risk. In a single study, Richardson et al.11 demonstrated that genetic variability of another perilipin, perilipin 4 (encoded by PLIN4), also modulates obesity-related traits. Their approach combined epidemiological, in silico, and in vitro methods to reveal potential regulatory mechanisms involving microRNA (miRNA). Perilipin 4 (previously referred to S3–12) is not as well characterized as perilipin 1, but it appears to participate in triglyceride synthesis. In the basal state, perilipin 4 is located throughout the adipocyte cytoplasm, but when stimulated by insulin and oleic acid, lipid droplets coated with perilipin 4 form and relocate to the periphery of the adipocyte.65
In the study by Richardson et al.,11 meta-analysis of seven PLIN4 SNPs in two white populations (n = 2,611 and n = 1,395) detected associations and interactions for two SNPs. The minor allele of PLIN4 rs8887 was associated with increased adiposity and was also shown to interact with omega-3 PUFA, such that higher omega-3 PUFA was associated with lower anthropometric traits. A second SNP (PLIN4 rs884164) interacted with omega-6 PUFA (higher omega-6 PUFA was associated with higher triglyceride and lower HDL-C) and with omega-3 PUFA (higher omega-3 intake was associated with increased anthropometric measures). Evaluation of these relationships in other populations is warranted.
What is particularly interesting about the findings for PLIN4 rs8887 is that the SNP is located in the 3′-UTR region of PLIN4 mRNA, suggesting the possibility that it may be regulated by miRNA. Software designed to predict miRNA binding sites revealed that the rs8887 variant (minor allele A) creates a binding site for miRNA 522. Carrying the investigation one step further, Richardson et al.11 confirmed that miRNA 522 was expressed in adipose cells and that the 3′-UTR containing the minor allele was associated with reduced PLIN4 expression. Explanations for the ways in which decreased PLIN4 expression leads to increased adiposity can only be hypothesized, since one might expect that decreased availability of perilipin 4 protein for the packaging of triglyceride might have the opposite effect (e.g., decreased triglyceride storage). However, the ways in which a different perilipin protein might “compensate” for reduced expression of a given perilipin are largely unknown and could provide alternative hypotheses. In summary, the discovery that a genetic variant of perilipin 4 creates a binding site for a known miRNA is an important contribution that suggests new avenues for investigating other perilipin loci.
CONCLUSION
Twenty years after the discovery of “perilipin,” the perilipin proteins and the genes encoding them remain an active focus of research on obesity and adipocytes. Perilipin 1, in particular, is recognized as a reliable marker of adipogenesis, and its function has been well characterized. While not entirely consistent, observational and interventional genetic association studies have confirmed the importance of a selected set of PLIN1 variants. Results from these SNPs have been informed by gene-diet interaction studies and reinforced through evidence that the same variants modulate responses to the metabolic challenges of fat load and endurance exercise. The discovery that PLIN1 is a PPARγ target, along with the identification of two new protein participants in lipolysis (ATGL and its activator CGI-58), raises the possibility that additional sites of genetic variability may potentially interact with PLIN1 variants to alter perilipin 1 expression or function. Despite this continued progress towards greater understanding, it is still unknown which PLIN1 variant or variants mediate changes in lipolysis to alter metabolic outcomes. One of the major challenges is the identification of “functional” variants at frequencies high enough to be investigated in populations. As resequencing becomes readily available and statistical methods are developed to analyze and interpret rare variants identified through sequencing, perhaps substantial strides towards a more complete understanding of this interesting locus will be made. New functional approaches have begun to be applied to a second perilipin protein, perilipin 4. The findings that a variant in PLIN4 creates an miRNA binding site and that post-transcriptional downregulation by a specific miRNA appears to result in obesity represent important breakthroughs in the understanding of how genetic variants ultimately alter phenotypes.
Acknowledgments
Funding. Supported by the National Institutes of Health, National Institute on Aging, grant no. 5P01AG023394-02, the National Institutes of Health, National Heart, Blood, and Lung Institute, grant no. HL54776, the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, grant no. DK075030, and contracts 53-K06-5-10 and 58–1950-9–001 from the US Department of Agriculture Research Service.
Footnotes
Declaration of interest. The authors have no relevant interests to declare.
Contributor Information
Caren E Smith, Jean Mayer US Department of Agriculture Human Nutrition Research Center on Aging at Tufts University, Boston, Massachusetts, USA.
José M Ordovás, Jean Mayer US Department of Agriculture Human Nutrition Research Center on Aging at Tufts University, Boston, Massachusetts, USA, and Tufts University School of Nutrition, Boston, Massachusetts, USA.
References
- 1.Finucane MM, Stevens GA, Cowan MJ, et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9·1 million participants. Lancet. 2011;377:557–567. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danaei G, Finucane MM, Lu Y, et al. Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating Group (Blood Glucose). National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2.7 million participants. Lancet. 2011;378:31–40. doi: 10.1016/S0140-6736(11)60679-X. [DOI] [PubMed] [Google Scholar]
- 3.Wardle J, Carnell S, Haworth CM, et al. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am J Clin Nutr. 2008;87:398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- 4.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 5.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egan JJ, Greenberg AS, Chang MK, et al. Control of endogenous phosphorylation of the major cAMP-dependent protein kinase substrate in adipocytes by insulin and β-adrenergic stimulation. J Biol Chem. 1990;265:18769–18775. [PubMed] [Google Scholar]
- 8.Greenberg AS, Egan JJ, Wek SA, et al. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- 9.Miura S, Gan JW, Brzostowski J, et al. Functional conservation for lipid storage droplet association among erilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- 10.Kimmel AR, Brasaemle DL, McAndrews-Hill M, et al. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. 2010;51:468–471. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson K, Louie-Gao Q, Arnett DK, et al. The PLIN4 variant rs8887 modulates obesity related phenotypes in humans through creation of a novel miR-522 seed site. PLoS ONE. 2011;6:e17944. doi: 10.1371/journal.pone.0017944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brasaemle DL, Rubin B, Harten I, et al. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J Biol Chem. 2000;275:38486–38493. doi: 10.1074/jbc.M007322200. [DOI] [PubMed] [Google Scholar]
- 13.Miyoshi H, Souza SC, Zhang HH, et al. Perilipin promotes hormone-sensitive lipase-mediated adipocyte lipolysis via phosphorylation-dependent and -independent mechanisms. J Biol Chem. 2006;281:15837–15844. doi: 10.1074/jbc.M601097200. [DOI] [PubMed] [Google Scholar]
- 14.Zimmermann R, Strauss JG, Haemmerle G, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 15.Subramanian V, Rothenberg A, Gomez C, et al. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279:42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- 16.Granneman JG, Moore HP. Location, location: protein trafficking and lipolysis in adipocytes. Trends Endocrinol Metab. 2008;19:3–9. doi: 10.1016/j.tem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Boden G, Shulman GI. Free fatty acids in obesity and type 2 diabetes: defining their role in the development of insulin resistance and beta-cell dysfunction. Eur J Clin Invest. 2002;32(Suppl 3):14–23. doi: 10.1046/j.1365-2362.32.s3.3.x. [DOI] [PubMed] [Google Scholar]
- 18.Koutsari C, Jensen MD. Thematic review series: patient-oriented research. Free fatty acid metabolism in human obesity. J Lipid Res. 2006;47:1643–1650. doi: 10.1194/jlr.R600011-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Haus JM, Solomon TP, Marchetti CM, et al. Free fatty acid-induced hepatic insulin resistance is attenuated following lifestyle intervention in obese individuals with impaired glucose tolerance. J Clin Endocrinol Metab. 2010;95:323–327. doi: 10.1210/jc.2009-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tansey JT, Sztalryd C, Gruia-Gray J, et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci U S A. 2001;98:6494–6499. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi H, Souza SC, Endo M, et al. Perilipin overexpression in mice protects against diet-induced obesity. J Lipid Res. 2010;51:975–982. doi: 10.1194/jlr.M002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kern PA, Di Gregorio G, Lu T, et al. Perilipin expression in human adipose tissue is elevated with obesity. J Clin Endocrinol Metab. 2004;89:1352–1358. doi: 10.1210/jc.2003-031388. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Sullivan S, Trujillo M, et al. Perilipin expression in human adipose tissues: effects of severe obesity, gender, and depot. Obes Res. 2003;11:930–936. doi: 10.1038/oby.2003.128. [DOI] [PubMed] [Google Scholar]
- 24.Mottagui-Tabar S, Ryden M, Lofgren P, et al. Evidence for an important role of perilipin in the regulation of human adipocyte lipolysis. Diabetologia. 2003;46:789–797. doi: 10.1007/s00125-003-1112-x. [DOI] [PubMed] [Google Scholar]
- 25.Nishiu J, Tanaka T, Nakamura Y. Isolation and chromosomal mapping of the human homolog of perilipin (PLIN), a rat adipose tissue-specific gene, by differential display method. Genomics. 1998;48:254–257. doi: 10.1006/geno.1997.5179. [DOI] [PubMed] [Google Scholar]
- 26.Qi L, Corella D, Sorlí JV, et al. Genetic variation at the perilipin (PLIN) locus is associated with obesity-related phenotypes in white women. Clin Genet. 2004;66:299–310. doi: 10.1111/j.1399-0004.2004.00309.x. [DOI] [PubMed] [Google Scholar]
- 27.Qi L, Shen H, Larson I, et al. Gender-specific association of a perilipin gene haplotype with obesity risk in a white population. Obes Res. 2004;12:1758–1765. doi: 10.1038/oby.2004.218. [DOI] [PubMed] [Google Scholar]
- 28.Qi L, Tai ES, Tan CE, et al. Intragenic linkage disequilibrium structure of the human perilipin gene (PLIN) and haplotype association with increased obesity risk in a multiethnic Asian population. J Mol Med. 2005;83:448–456. doi: 10.1007/s00109-004-0630-4. [DOI] [PubMed] [Google Scholar]
- 29.Sone Y, Yamaguchi K, Fujiwara A, et al. Association of lifestyle factors, polymorphisms in adiponectin, perilipin and hormone sensitive lipase, and clinical markers in Japanese males. J Nutr Sci Vitaminol (Tokyo) 2010;56:123–131. doi: 10.3177/jnsv.56.123. [DOI] [PubMed] [Google Scholar]
- 30.Yan W, Chen S, Huang J, et al. Polymorphisms in PLIN and hypertension combined with obesity and lipid profiles in Han Chinese. Obes Res. 2004;12:1733–1737. doi: 10.1038/oby.2004.214. [DOI] [PubMed] [Google Scholar]
- 31.Meirhaeghe A, Thomas S, Ancot F, et al. Study of the impact of perilipin polymorphisms in a French population. J Negat Results Biomed. 2006;5:10. doi: 10.1186/1477-5751-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergmann A, Li J, Reimann M, et al. Polymorphisms in perilipin gene (PLIN) are not associated with obesity and weight variation in people with high risk of type 2 diabetes. Exp Clin Endocrinol Diabetes. 2008;116(Suppl 1):S56–S58. doi: 10.1055/s-2008-1081493. [DOI] [PubMed] [Google Scholar]
- 33.Hu DS, Xie J, Yu DH, et al. Perilipin gene 1237 T>C polymorphism is not associated with obesity risk in northern Chinese Han adults. Biomed Environ Sci. 2009;22:442–447. doi: 10.1016/S0895-3988(10)60023-2. [DOI] [PubMed] [Google Scholar]
- 34.Peeters A, Beckers S, Verrijken A, et al. Possible role for ENPP1 polymorphism in obesity but not for INSIG2 and PLIN variants. Endocrine. 2009;36:103–109. doi: 10.1007/s12020-009-9194-y. [DOI] [PubMed] [Google Scholar]
- 35.Angeli CB, Kimura L, Auricchio MT, et al. Multilocus analyses of seven candidate genes suggest interacting pathways for obesity-related traits in Brazilian populations. Obesity (Silver Spring) 2011;19:1244–1251. doi: 10.1038/oby.2010.325. [DOI] [PubMed] [Google Scholar]
- 36.Corella D, Qi L, Sorli JV, et al. Obese subjects carrying the 11482G>A polymorphism at the perilipin locus are resistant to weight loss after dietary energy restriction. J Clin Endocrinol Metab. 2005;90:5121–5126. doi: 10.1210/jc.2005-0576. [DOI] [PubMed] [Google Scholar]
- 37.Smith CE, Tucker KL, Yiannakouris N, et al. Perilipin polymorphism interacts with dietary carbohydrates to modulate anthropometric traits in Hispanics of Caribbean origin. J Nutr. 2008;138:1852–1858. doi: 10.1093/jn/138.10.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz JR, Larrarte E, Margareto J, et al. Preliminary findings on the role of PLIN1 polymorphisms on body composition and energy metabolism response to energy restriction in obese women. Br J Nutr. 2011;106:486–490. doi: 10.1017/S0007114511000432. [DOI] [PubMed] [Google Scholar]
- 39.Labayen I, Diez N, Gonzalez A, et al. Effects of protein versus carbohydrate-rich diets on fuel utilisation in obese women during weight loss. Forum Nutr. 2003;56:168–170. [PubMed] [Google Scholar]
- 40.Soenen S, Mariman EC, Vogels N, et al. Relationship between perilipin gene polymorphisms and body weight and body composition during weight loss and weight maintenance. Physiol Behav. 2009;96:723–728. doi: 10.1016/j.physbeh.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 41.Deram S, Nicolau CY, Perez-Martinez P, et al. Effects of perilipin (PLIN) gene variation on metabolic syndrome risk and weight loss in obese children and adolescents. J Clin Endocrinol Metab. 2008;93:4933–4940. doi: 10.1210/jc.2008-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jang Y, Kim OY, Lee JH, et al. Genetic variation at the perilipin locus is associated with changes in serum free fatty acids and abdominal fat following mild weight loss. Int J Obes (Lond) 2006;30:1601–1608. doi: 10.1038/sj.ijo.0803312. [DOI] [PubMed] [Google Scholar]
- 43.Kang ES, Cha BS, Kim HJ, et al. The 11482G>A polymorphism in the perilipin gene is associated with weight gain with rosiglitazone treatment in type 2 diabetes. Diabetes Care. 2006;29:1320–1324. doi: 10.2337/dc05-2466. [DOI] [PubMed] [Google Scholar]
- 44.Dalen KT, Schoonjans K, Ulven SM, et al. Adipose tissue expression of the lipid droplet-associating proteins S3-12 and perilipin is controlled by peroxisome proliferator-activated receptor-[gamma] Diabetes. 2004;53:1243–1252. doi: 10.2337/diabetes.53.5.1243. [DOI] [PubMed] [Google Scholar]
- 45.Arimura N, Horiba T, Imagawa M, et al. The peroxisome proliferator-activated receptor gamma regulates expression of the perilipin gene in adipocytes. J Biol Chem. 2004;279:10070–10076. doi: 10.1074/jbc.M308522200. [DOI] [PubMed] [Google Scholar]
- 46.Kawai T, Ng MC, Hayes MG, et al. Variation in the perilipin gene (PLIN) affects glucose and lipid metabolism in non-Hispanic white women with and without polycystic ovary syndrome. Diabetes Res Clin Pract. 2009;86:186–192. doi: 10.1016/j.diabres.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qi L, Zhang C, Greenberg A, et al. Common variations in perilipin gene, central obesity, and risk of type 2 diabetes in US women. Obesity (Silver Spring) 2008;16:1061–1065. doi: 10.1038/oby.2008.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruiz-Narvaez EA, Sacks FM, Campos H. Abdominal obesity and hyperglycemia mask the effect of a common APOC3 haplotype on the risk of myocardial infarction. Am J Clin Nutr. 2008;87:1932–1938. doi: 10.1093/ajcn/87.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corella D, Qi L, Tai ES, et al. Perilipin gene variation determines higher susceptibility to insulin resistance in Asian women when consuming a high-saturated fat, low-carbohydrate diet. Diabetes Care. 2006;29:1313–1319. doi: 10.2337/dc06-0045. [DOI] [PubMed] [Google Scholar]
- 50.Smith CE, Arnett DK, Corella D, et al. Perilipin polymorphism interacts with saturated fat and carbohydrates to modulate insulin resistance. Nutr Metab Cardiovasc Dis. 2012;22:449–455. doi: 10.1016/j.numecd.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tai ES, Ordovas JM. The role of perilipin in human obesity and insulin resistance. Curr Opin Lipidol. 2007;18:152–156. doi: 10.1097/MOL.0b013e328086aeab. [DOI] [PubMed] [Google Scholar]
- 52.Ordovas JM. Gender, a significant factor in the cross talk between genes, environment, and health. Gend Med. 2007;4(Suppl B):S111–S122. doi: 10.1016/s1550-8579(07)80052-0. [DOI] [PubMed] [Google Scholar]
- 53.Akter MH, Yamaguchi T, Hirose F, et al. Perilipin, a critical regulator of fat storage and breakdown, is a target gene of estrogen receptor-related receptor alpha. Biochem Biophys Res Commun. 2008;368:563–568. doi: 10.1016/j.bbrc.2008.01.102. [DOI] [PubMed] [Google Scholar]
- 54.Power ML, Schulkin J. Sex differences in fat storage, fat metabolism, and the health risks from obesity: possible evolutionary origins. Br J Nutr. 2008;99:931–940. doi: 10.1017/S0007114507853347. [DOI] [PubMed] [Google Scholar]
- 55.Perez-Martinez P, Yiannakouris N, Lopez-Miranda J, et al. Postprandial triacyl-glycerol metabolism is modified by the presence of genetic variation at the perilipin (PLIN) locus in 2 white populations. Am J Clin Nutr. 2008;87:744–752. doi: 10.1093/ajcn/87.3.744. [DOI] [PubMed] [Google Scholar]
- 56.Jenkins NT, McKenzie JA, Damcott CM, et al. Endurance exercise training effects on body fatness, VO2max, HDL-C subfractions, and glucose tolerance are influenced by a PLIN haplotype in older Caucasians. J Appl Physiol. 2010;108:498–506. doi: 10.1152/japplphysiol.01018.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaidhu MP, Anthony NM, Patel P, et al. Dysregulation of lipolysis and lipid metabolism in visceral and subcutaneous adipocytes by high-fat diet: role of ATGL, HSL, and AMPK. Am J Physiol Cell Physiol. 2010;298:C961–C971. doi: 10.1152/ajpcell.00547.2009. [DOI] [PubMed] [Google Scholar]
- 58.House RL, Cassady JP, Eisen EJ, et al. Functional genomic characterization of delipidation elicited by trans-10, cis-12-conjugated linoleic acid (t10c12-CLA) in a polygenic obese line of mice. Physiol Genomics. 2005;21:351–361. doi: 10.1152/physiolgenomics.00244.2004. [DOI] [PubMed] [Google Scholar]
- 59.Green A, Rumberger JM, Stuart CA, et al. Stimulation of lipolysis by tumor necrosis factor-alpha in 3T3-L1 adipocytes is glucose dependent: implications for long-term regulation of lipolysis. Diabetes. 2004;53:74–81. doi: 10.2337/diabetes.53.1.74. [DOI] [PubMed] [Google Scholar]
- 60.Chung S, Brown JM, Sandberg MB, et al. Trans-10, cis-12 CLA increases adipocyte lipolysis and alters lipid droplet-associated proteins: role of mTOR and ERK signaling. J Lipid Res. 2005;46:885–895. doi: 10.1194/jlr.M400476-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ippagunta S, Hadenfeldt TJ, Miner JL, et al. Dietary conjugated linoleic acid induces lipolysis in adipose tissue of coconut oil-fed mice but not soy oil-fed mice. Lipids. 2001;46:821–830. doi: 10.1007/s11745-011-3574-9. [DOI] [PubMed] [Google Scholar]
- 62.Keophiphath M, Priem F, Jacquemond-Collet I, et al. 1,2-vinyldithiin from garlic inhibits differentiation and inflammation of human preadipocytes. J Nutr. 2009;139:2055–2060. doi: 10.3945/jn.109.105452. [DOI] [PubMed] [Google Scholar]
- 63.Hassan M, El Yazidi C, Malezet-Desmoulins C, et al. Gene expression profiling of 3T3-L1 adipocytes exposed to phloretin. J Nutr Biochem. 2010;21:645–652. doi: 10.1016/j.jnutbio.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 64.Abidov MT, del Rio MJ, Ramazanov TZ, et al. Effects of Aralia mandshurica and Engelhardtia chrysolepis extracts on some parameters of lipid metabolism in women with nondiabetic obesity. Bull Exp Biol Med. 2006;141:343–346. doi: 10.1007/s10517-006-0167-3. [DOI] [PubMed] [Google Scholar]
- 65.Wolins NE, Skinner JR, Schoenfish MJ, et al. Adipocyte protein S3-12 coats nascent lipid droplets. J Biol Chem. 2003;278:37713–37721. doi: 10.1074/jbc.M304025200. [DOI] [PubMed] [Google Scholar]
- 66.Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 67.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]


