Abstract
In response to wounding or infection, insects produce a battery of antimicrobial peptides (AMPs) and other defense molecules to kill the invading pathogens. To study their structures, functions, and transcriptional regulation, we synthesized Manduca sexta moricin, a 42-residue peptide (GKIPVKAIKQAGKVIGKGLRAINIAGTTHDVVSFFRPKKKKH, 4539 Da). The compound exhibited potent antimicrobial activities against a broad spectrum of Gram-positive and Gram-negative bacteria with a minimum inhibitory concentration of 1.4 µm. The mRNA levels of M. sexta moricin increased substantially in fat body and hemocytes after the larvae were challenged with bacterial cells. We determined the solution structure of this AMP by two-dimensional 1H–1H-nuclear magnetic resonance spectroscopy. The tertiary structure is composed of an eight-turn α-helix spanning almost the entire peptide. Insights of relationships between the structure and function are also presented.
Keywords: antimicrobial peptide, insect immunity, NMR spectroscopy, tobacco hornworm
INTRODUCTION
Antimicrobial peptides (AMPs) are a critical component of the natural defense system in various organisms ranging from prokaryotes to plants, arthropods, and vertebrates [1]. Most of them are less than 10 kDa, hydrophobic, membrane active, and have an overall net positive charge. Since the isolation of cecropin from bacteria-challenged diapausing pupae of Hyalophora cecropia [2], over 200 AMPs have been identified in arthropods. Upon microbial infection, expression of Drosophila AMP genes is up-regulated by transcriptional factors from the Rel family (i.e. Dif, Relish, and Dorsal) through the Toll and Imd pathways [3]. Identification of orthologous genes in the genomes of Anopheles gambiae, Apis mellifera, and Tribolium castaneum suggests the existence of similar signaling pathways for induced synthesis of immune-responsive proteins in other insect species [4–6]. Fat body (a tissue analogous to human liver) synthesizes AMPs and secretes these heat-stable molecules into the plasma to kill a broad spectrum of microorganisms. Insect AMPs can be categorized into three major groups: peptides with α-helical conformation (e.g. cecropin, sarcotoxin, and moricin), peptides with β-sheet or α-helix/β-sheet mixed structures stabilized by disulfide bond(s) (e.g. defensin, thanatin, and drosomycin), and peptides overrepresented with Pro or Gly residues (e.g. lebocin, apidaecin, attacin, diptericin, and gloverin). In the tobacco hornworm Manduca sexta, transcription of AMP and other immunity-related genes is up-regulated in response to bacterial infection [7,8].
Exclusively found in lepidopteran insects so far, moricin-like AMPs are active against both Gram-positive and Gram-negative bacteria [9]. They affect structural integrity of bacterial plasma membrane. Moricin synthesis is induced by microbial cell wall components [10]. NMR structural studies show that residues 5–36 of Bombyx mori and Spodoptera litura moricins form a long, single α-helix [11,12]. Their amino-terminal segment forms an amphipathic helix important for the bactericidal activity, whereas their carboxyl-terminus contains a cluster of 5–6 positively charged residues accounting for the broad specificity.
To determine the role of AMPs in humoral responses of the tobacco hornworm, we chemically synthesized M. sexta moricin, tested its biological activity, and solved its tertiary structure by NMR spectroscopy. A comparison with cecropin provided insights of the structure–function relationships for AMPs with α-helical conformation. The inducibility, tissue specificity, and developmental profile of the moricin gene expression were also examined.
MATERIALS AND METHODS
Peptide Synthesis, Bacterial Strains, Insect Rearing, and RNA Preparation
M. sexta moricin (~10 mg) was prepared by automated protocols of stepwise solid-phase synthesis using Fmoc-amino acid derivatives (Bio-Synthesis Inc, Lewisville, TX, USA). Following deprotection and cleavage, the peptide was purified by RP-HPLC and analyzed by MS. The peptide concentration was determined along with bovine serum albumin standards by a modified Bradford method using a commercial kit (Pierce). Pathogenic strains of Klebsiella pneumoniae, Salmonella typhimurium, Escherichia coli O157:H7, S. typhimurium DT104, Listeria monocytogenes, Staphylococcus aureus, and two strains of methicillin-resistant S. aureus were provided by Dr Guolong Zhang in the Department of Animal Science at Oklahoma State University. M. sexta eggs were purchased from Carolina Biological Supplies and larvae were reared on an artificial diet [13]. Hemocytes and fat body total RNA samples were prepared from the naïve and induced, day 3, fifth instar larvae according to Wang et al. [14]. Fat body, nerve tissue (thoracic ganglia), salivary gland, midgut, Malpighian tubule, trachea, muscle, and integument were dissected from bled insects (day 3, fifth instar, uninjected larvae) for RNA isolation. Fat body tissues were dissected from unchallenged larvae (4th, 5th and wandering) and adults at different days for expression analysis.
Antibacterial Activity Assay
The synthetic peptide was separately tested against four different strains of Gram-negative and Gram-positive bacteria including E. coli, S. typhimurium, K. pneumoniae, S. typhimurium DT104, L. monocytogenes, S. aureus, and the resistant strains of S. aureus. The minimum inhibitory concentrations (MICs) were determined in a broth microdilution assay [15]. Briefly, overnight bacterial cultures were subcultured into 4 ml of Trypticase Soy Broth for 3–5 h until the bacteria reached mid-log phase. After centrifugation at 1000 g at 4 °C and washing with 10 mm sodium phosphate (pH 7.4), the cells were suspended in Muller-Hinton Broth (5 × 105 cfu/ml). Aliquots of the diluted cultures (90 µl) were mixed with 10 µl of the synthetic peptide at 500, 250, 125, 62.5, 31.2, 15.6, 7.8, and 3.9 µg/ml. All bacteria were cultured at 37 °C overnight in a 96-well cell culture plate, and the lowest concentration of peptide that caused no visible growth was recorded. The experiment was performed at least 3 times for each strain to obtain MICs of M. sexta moricin against the bacterial strains.
Expression Analysis by RT-PCR
The RNA sample (2–4 µg), oligo(dT) (0.5 µg), and dNTPs (1 µl, 10 mm each) were mixed with diethylpyrocarbonate-treated H2O in a final volume of 12 µl, denatured at 65 °C for 5 min, and quickly chilled on ice for 3 min. M-MLV reverse transcriptase (1 µl, 200 U/µl, Invitrogen), 5 × buffer (4 µl), 0.1 m dithiothreitol (2 µl), and RNase OUT (1 µl, 40 U/µl, Invitrogen) were added to the denatured RNA sample (12 µl) for cDNA synthesis at 37 °C for 50 min. The M. sexta ribosomal protein S3 (rpS3) mRNA was used as an internal control to normalize the cDNA samples in a preliminary PCR using primers 501 (5′-GCCGTTCTTGCCCTGTT-3′) and 504 (5′-CGCGAGTTGACTTCGGT-3′). Moricin cDNA fragment was amplified using forward (5′-TTGTTATTGTCGCGTTGTCGC-3′) and reverse (5′-GGCCTGAAGAAACTAACAACATCG-3′) primers under conditions empirically chosen to avoid saturation: 30 cycles of 94 °C, 30 s; 52 °C, 30 s; 72 °C, 20 s. The relative levels of moricin transcripts in the normalized samples were determined by 1.5% agarose gel electrophoresis.
Circular Dichroism (CD) Spectroscopy
CD analysis was performed on a Jasco-720 Spectropolarimeter (JASCO, Tokyo, Japan) installed with a standard analysis program. Spectra were recorded using a quartz cell of 0.1-cm path length with an acquisition time of 3 s/nm and 1-nm spectral bandwidth over a wavelength of range 190–250 nm. Each spectrum was an average of eight consecutive scans, subtracted with the signal from peptide-free references. Samples were prepared by dissolving the peptide (0.2 mg/ml) in 0%, 10%, 30%, 50%, and 100% TFE in H2O or 100% methanol. CD analyses were also performed in SDS micelles (0.5% and 2.0%).
Nuclear Magnetic Resonance (NMR) Spectroscopy
The NMR experiments were performed on an 11.75 T Varian NMR System (Varian Inc, Palo Alto, CA, USA) operating at 499.96 MHz for proton frequency using a 3-mm triple resonance inverse detection pulse-field gradient probe. One-dimensional (1D) and two-dimensional (2D) NMR data were collected as previously described [16]. Because NMR signals in methanol were much sharper and stronger than in TFE or SDS micelles, the synthetic moricin was dissolved in 400-µl methanol (CD3OH) in the final concentration of 2.0 mm for detailed NMR studies. 2D 1H–1H TOCSY NMR spectra at a spin-locked field (B1) strength of 7 kHz were recorded with isotropic mixing times of 50 and 100 ms; 2D 1H–1H NOESY NMR data were collected with mixing times of 100, 200, 300, and 400 ms. The data sets were acquired at different temperatures from 10 to 35 °C. The spectra acquired at 25 °C provided the optimal resolution of overlapping proton resonances. The invariant nature of the proton chemical shifts and line widths upon ten-fold dilution suggested that moricin was monomeric in solution at the concentration used for 2D NMR analysis. A total of 256 increments of 4K data points were collected for these experiments. All data sets were obtained in hypercomplex phase-sensitive mode. The hydroxyl proton resonance of the solvent was suppressed with the watergate sequence [17]. The residual methyl proton peak (3.30 ppm at 25 °C) of the solvent (CD3OH) was used for referencing the chemical shift assignments. Peak picking and assignment were performed using Sparky [18]. Side chain proton resonances were assigned by comparing cross peaks in the NOESY spectra with those in the TOCSY spectra acquired under similar experimental conditions.
Structural Calculations
Interproton distance constraints were obtained from NOESY spectra observed with different mixing times. Initially a mixing time of 300 ms was used for distance constraint measurements, and then the assigned NOE peaks were checked with the spectrum obtained with a 100-ms mixing time. For structural calculations, only NOE peaks present in the NOESY spectrum (tmixing = 100 ms) were used to rule out the peaks due to spin diffusion. NOE-derived distance restraints were classified into three ranges (1.8–2.5, 1.8–3.5 and 1.8–5.0 Å), corresponding to strong, medium, and weak NOE intensities. Upper distance limits for methyl and nonstereospecifically assigned methylene protons (by adding 0.5 Å) were corrected appropriately for center averaging; additional 0.5 Å was added for NOEs involving methyl protons. Structural calculations were performed using the simulated annealing procedure in CNS program [19] and the annealing module. Force constants were scaled throughout the procedure using the default parameters of CNS. Distance constraints were used to calculate an ensemble of 100 structures, 20 of which with the lowest energy were selected to represent the three-dimensional NMR structure of M. sexta moricin. The calculated structures were analyzed using MOLMOL [20] and PROCHECK-NMR [21]. Figures of the structures were generated using MOLMOL and PyMOL (http://pymol.sourceforge.net).
RESULTS
Sequence Features of M. sexta Moricin
M. sexta moricin was identified as a differentially expressed gene in response to bacterial injection in a subtractive suppression hybridization experiment [7]. The expressed sequence tag contained a complete open reading frame encoding a 67-residue sequence: MKLTSLFIFVIVAL SLLFSSTDAAPGKIPVKAIKQAGKVIGKGLRAINIAGTTHDVVSFFRPKKKKH with the underlined part representing a predicted signal peptide rich in hydrophobic residues. Based on the amino acid sequences of moricins isolated from B. mori and S. litura plasma [11,12], we suggest the mature M. sexta moricin actually start with GKIPV instead of APGKIPV. The discrepancy between predicted and observed amino terminus exists in moricins from the silkworm and tobacco cutworm.
The chemically synthesized M. sexta moricin (GKIPVKAIKQAGKVIGKGLRAINIAGTTHDVVSFFRPKKKKH) has a purity of >95% and an experimental mass of 4539.5 Da, nearly identical to the theoretical value of the peptide (4539 Da). Containing one Asp, two Arg, and nine Lys residues, this peptide is highly cationic with a calculated isoelectric point of 11.4. As there is no Trp, Tyr, or Cys residue in the sequence, it does not absorb UV at 280 nm. M. sexta and S. litura moricins are different in nine positions (Q10–K10, K13V14–A13A14, G26–S26, T28–A28, V32–Y32, R36–K36, K39–H39, and H42–K42), whereas M. sexta and B. mori moricins are even less similar (G1–A1, V5–I5, Q10A11–T10V11, V14I15–A14V15, G26–S26, T28H29–A28N29, V32S33–F32N33, F35R36–L35K36, K40–R40). Most of the changes involve substitutions of residues with similar chemical properties.
Antibacterial Activity and Spectrum
We examined the antimicrobial activity of M. sexta moricin against four Gram-negative and four Gram-positive bacterial strains and found that the synthetic peptide killed all these bacteria at low micromolar range (Table 1). Except for S. aureus BAA-39, which required 12.5 µg/ml of the peptide for complete growth inhibition, the moricin was effective at 6.25 µg/ml or 1.4 µM against all the strains tested, including multidrug-resistant S. typhimurium DT104 and methicillin-resistant S. aureus ATCC 43 300.
Table 1.
MICs of Manduca sexta moricina
| Bacterial species | ATCC number | Moricin (µg/ml) |
|---|---|---|
| Gram-negative | ||
| E. coli 0157H7 | 25 922 | 6.25 |
| S. typhimurium | 14 028 | 6.25 |
| K. pneumoniae | 13 883 | 6.25 |
| S. typhimurium DT104 | 700 408 | 6.25 |
| Gram-positive | ||
| L. monocytogenes | 19 115 | 6.25 |
| S. aureus | 25 923 | 6.25 |
| S. aureus (MRSA) b | 43 300 | 6.25 |
| S. aureus (MRSA) b | BAA-39 | 12.5 |
MIC is defined as the lowest peptide concentration that gives no visible growth after overnight incubation in 100% Muller-Hinton broth. Conditions for the broth microdilution assay are specified by Clinical Laboratory Standard Institute. The experiments were repeated at least 3 times for each bacterial strain.
Methicillin-resistant S. aureus.
Tissue Specificity, Immune Inducibility, and Developmental Regulation of Moricin Expression
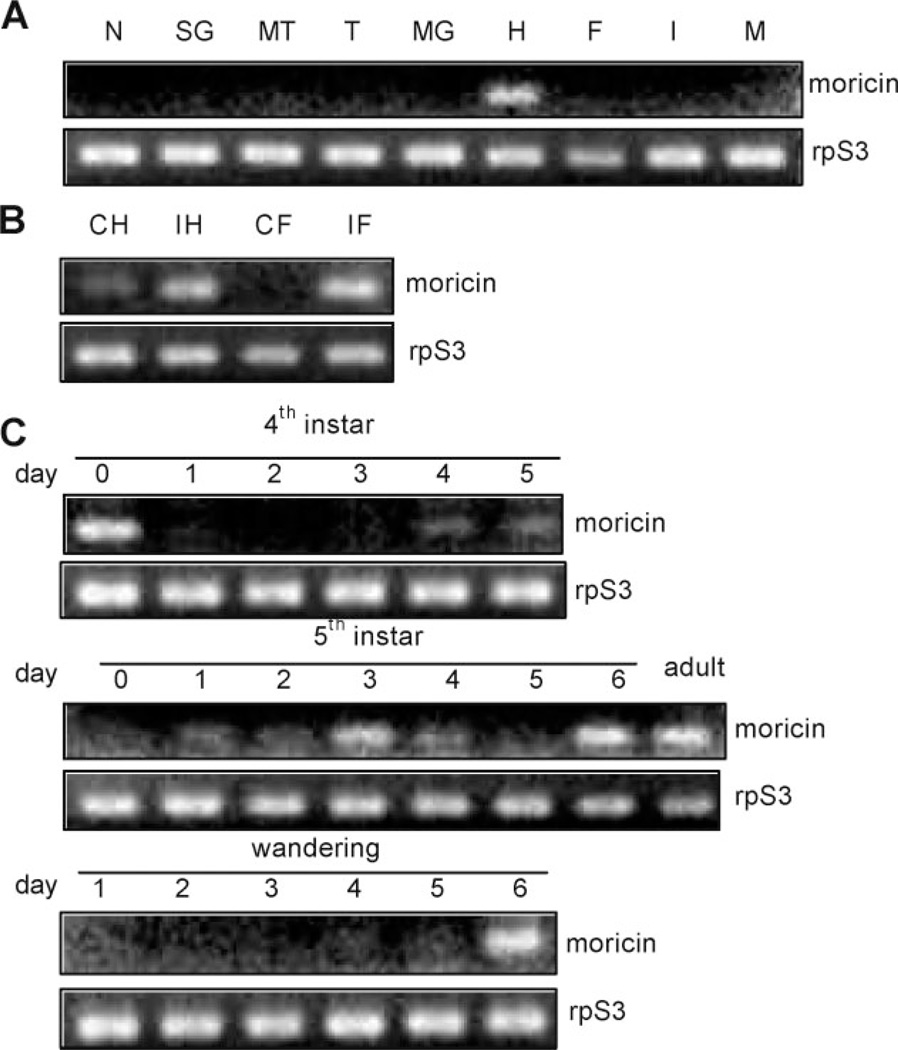
M. sexta moricin transcripts were present at low levels in nerve tissue, salivary gland, Malpighian tubule, trachea, midgut, fat body, integument, or muscle from day 3, fifth instar insects (Figure 1). A moderate level of moricin mRNA was observed in the hemocytes from the naïve larvae. After an immune challenge, the transcripts became more abundant in hemocytes and fat body consistent with the observation that moricin expression was up-regulated by injecting B. mori spätzle-1 into the M. sexta larvae [22]. The transcription of moricin gene may also be controlled by developmental signals: we detected moderate levels of its mRNA in fat body from the 4th instar larvae (day 0), 5th instar larvae (days 3 and 6), wandering larvae (day 6), and adults in the absence of infection.
Figure 1.
Profiling of M. sexta moricin transcript levels by reverse transcription-PCR. (A) Expression in different tissues from day 3, 5th instar naïve larvae. N, nerve tissue; SG, salivary gland; MT, Malpighian tubule; T, trachea; MG, midgut; H, hemocytes, F, fat body; I, integument; M, muscle. (B) Induced transcription in hemocytes (H) and fat body (F) from naïve (C) and bacteria-injected (I) larvae. (C) mRNA level changes in fat body from insects at different developmental stages.
CD Spectroscopy and Secondary Structural Prediction
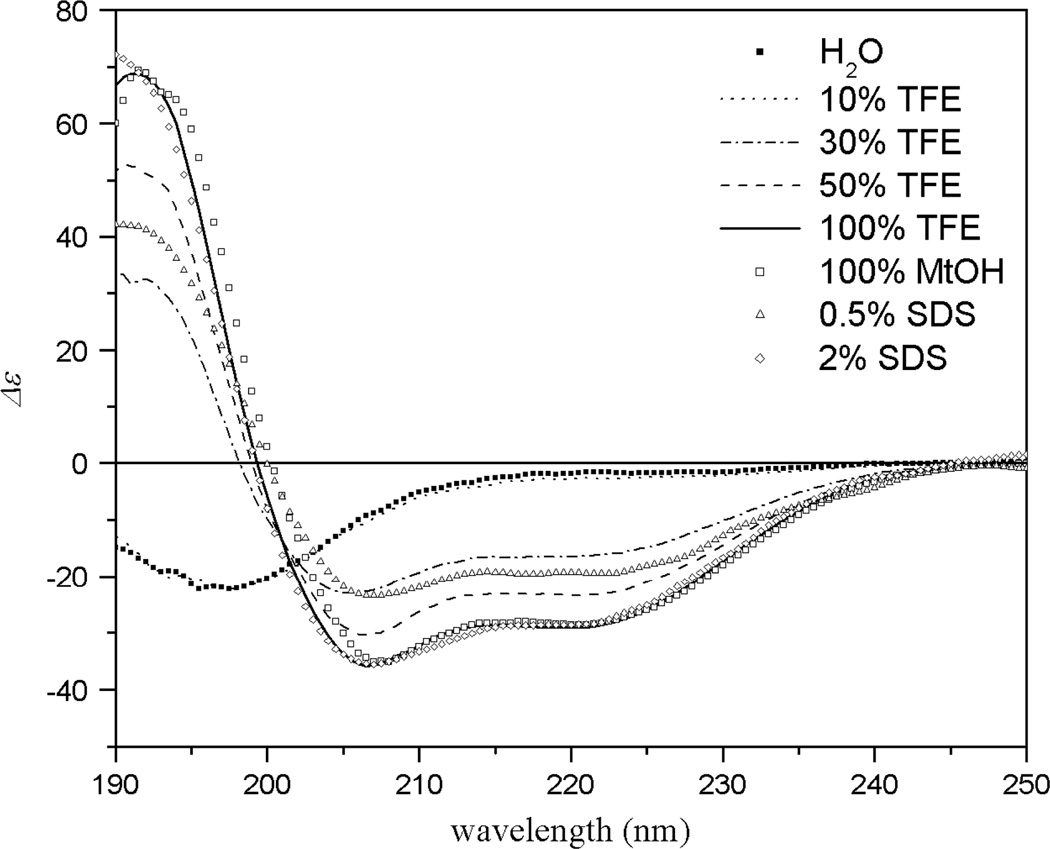
The CD spectrum indicated that M. sexta moricin was in the conformation of random coil in water (Figure 2). In the presence of 30% TFE in water, however, the spectrum exhibited two minima at 208 and 222 nm, indicative of α-helical conformation. The helical content increased as the TFE concentration arose to 50 and 100%. The spectrum of moricin in 100% methanol highly resembled that in 100% TFE and 2% SDS respectively, suggesting that the peptide adopted a maximal α-helical structure in methanol. These results indicated that moricin was likely to adopt a predominantly α-helical conformation when interacting with bacterial membrane. Similar conformational transition from random coil in aqueous solution to α-helix in hydrophobic environments (e.g. CH3OH, TFE–H2O, and HFIP-H2O) was also observed in cecropin-like AMPs including cecropins and mellitin [23]. Since the α-helical content of moricin peaked in 100% methanol and strong, sharp NMR signals were observed in this solvent system, therefore methanol-d3 solvent was selected for NMR studies to determine 3D solution structure.
Figure 2.
CD spectra of M. sexta moricin in different concentration of TFE, SDS, and CH3OH.
NMR Resonance Assignment and Secondary Structure
The spin systems and sequential proton assignments were completed using 2D 1H–1H TOCSY NMR spectra for intraresidue 1H–1H correlations and NOESY spectra for inter-residue 1H–1H correlations as described by Wüthrich [24]. In these assignments, Hα(i) − Hδ(i + 1:Pro) (dαδ) or Hα(i) − Hα(i + 1:Pro) (dαα) NOEs instead of dαN were used for Pro4 and Pro37. Both proline in moricin were assigned to be in trans conformation, through strong dαδ NOEs. The absence of dαα NOEs indicated that any cis form if existed was below detection. Short-mixing time (50 ms) TOCSY spectra combined with intraresidue NH–Hβ and Hα—Hβ NOEs were used to obtain information on 3JHαHβ coupling constant estimated qualitatively and to assign stereo-specific β-methylene protons [25,26]. The 1H chemical shifts for M. sexta moricin were deposited in BioMagResBank (15 323). The sequential and medium distance NOE connectivity and Cα-proton chemical shift index (CSI) [27] are illustrated in Figure 3. A number of nonsequential dαN(i, i + 3) and dαβ (i, i + 3) NOEs characteristic of α-helical conformation were observed for the moricin from Val5 to Arg36. A continuous stretch of dNN(i, i + 1) also extended from Val5 to Arg36. The helicity of M. sexta moricin was further supported by the up-field shift of Cα proton in CSI values of residues 5–35. Thus, M. sexta moricin is very similar in the secondary structure to B. mori and S. litura moricins.
Figure 3.
Schematic diagram of sequential and medium distance NOE connectivity and CαH chemical shift index for M. sexta moricin. Thickness of a bar reflects the strength of NOE connectivity.
Tertiary Structure of M. sexta Moricin
A simulated annealing molecular dynamics analysis was performed with distance constraints derived from the NOEs and found to converge to a single family of conformers regardless of the starting structure. We determined the three-dimensional structure of M. sexta moricin by hybrid distance geometry-simulated annealing calculations using 283 NOE-derived restraints (including 87 intraresidue, 102 sequential and 94 medium-range ones). From the 100 calculated structures, we selected 20 with the lowest energy as the final ensemble. These representative structures were in good agreement with the experimental data, with no distance violations larger than 0.3 Å. The statistical data (Table 2) for these 20 energy-minimized structures demonstrated good covalent geometry and stereochemistry. Ramachandran plot [28] confirmed the high quality of these 20 structures: 89.6% of φ and ψ angles are located in the most favored stereochemically allowed regions while 0.6% of φ and ψ angles are found in the disallowed region.
Table 2.
Structural statistics of the 20 lowest energy structures
| Total NOE restraints | 283 | |
| Intraresidue | 87 | |
| Sequential | 102 | |
| Medium (1 < |i − j| < 5) | 94 | |
| Long (|i − j| ≥ 5) | 0 | |
| Energies (kcal/mol) | ||
| Overall | 21.82 ± 0.70 | |
| Bonds | 0.83 ± 0.06 | |
| Angles | 12.27 ± 0.29 | |
| Impropers | 0.61 ± 0.10 | |
| Vdw | 3.48 ± 0.48 | |
| NOE | 4.63 ± 0.46 | |
| RMSD from experimental restraints | ||
| NOE distance restraints (Å) | 0.0148 ± 0.0007 | |
| RMSD from ideal covalent geometry | ||
| Bonds (Å) | 0.0011 ± 0.0001 | |
| Angles (°) | 0.251 ± 0.003 | |
| Impropers | 0.108 ± 0.008 | |
| Ramachandran analysis | ||
| Residues in most favored regions | 89.6% | |
| Residues in additionally allowed regions | 8.2% | |
| Residues in generously allowed regions | 1.6% | |
| Residues in disallowed regions | 0.6% | |
| RMSD relative to the mean structure (Å) | Backbone | All non-H |
| Residues 5–36 | 1.91 ± 0.53 | 2.97 ± 1.05 |
| Residues 5–22 | 0.66 ± 0.23 | 1.24 ± 0.22 |
| Residues 23–36 | 0.64 ± 0.20 | 1.37 ± 0.22 |
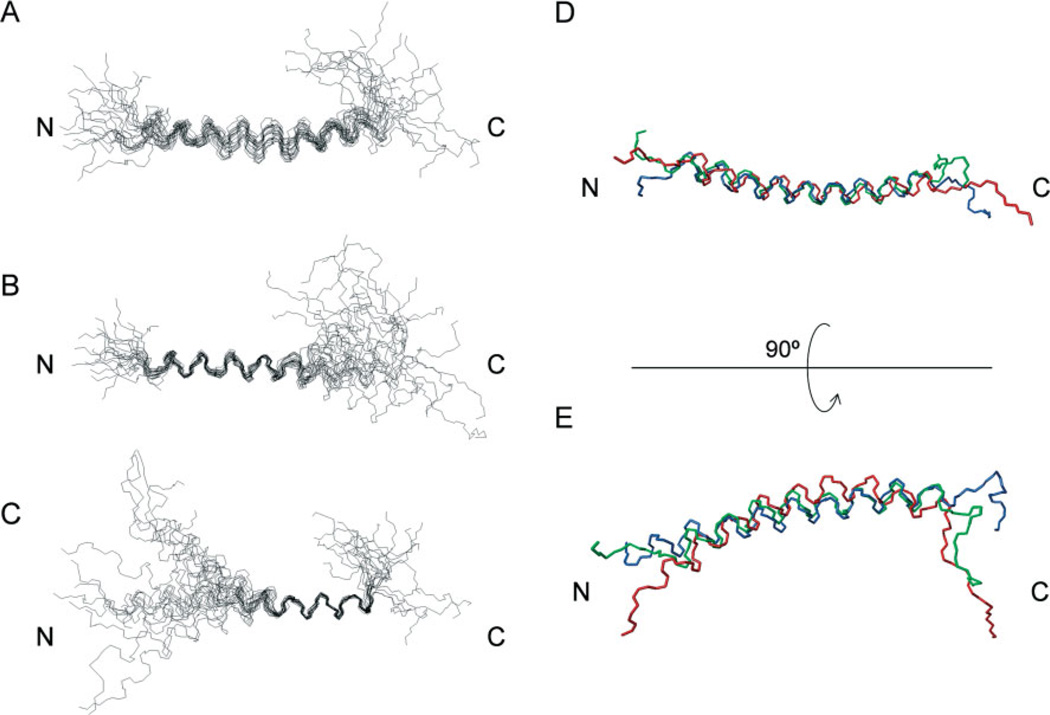
The structural coordinates, deposited in Protein Data Bank (PDB) with the entry code of 2JR8, indicate that M. sexta moricin contains a long α-helix spanning from V5 to R36 (Figure 4(A)). The amino-terminal G1KIP4 and carboxyl-terminal P37KKKKH42 were disordered. The root-mean-square deviation (RMSD) of the 20 structures from the average was 1.91 ± 0.53 Å for the backbone heavy atoms in the α-helical region. This relatively high RMSD was caused by random bending of the entire helix. The RMSDs for the backbone heavy atoms in the regions of 5–22 and 23–36 were 0.66 ± 0.23 and 0.64 ± 0.20 Å (Figure 4(B) and (C); Table 2), respectively. Superposition of the average backbone structures (Figure 4(D) and (E)) revealed that these three moricins are highly similar in their overall structure, with a curved α-helix spanning residues 5–36.
Figure 4.
The tertiary structure of M. sexta moricin in 100% CD3OH. (A)–(C) superimposition of the backbones of 20 lowest energy structures of M. sexta moricin best fitted to residues 5–36 (A), to residues 5–22 (B), and to residues 23–36 (C). (D) Superimposition of the average, lowest energy structures of B. mori (blue), S. litura (green) and M. sexta (red) moricins; (E) the same as (D) but rotated 90° around the horizontal axis.
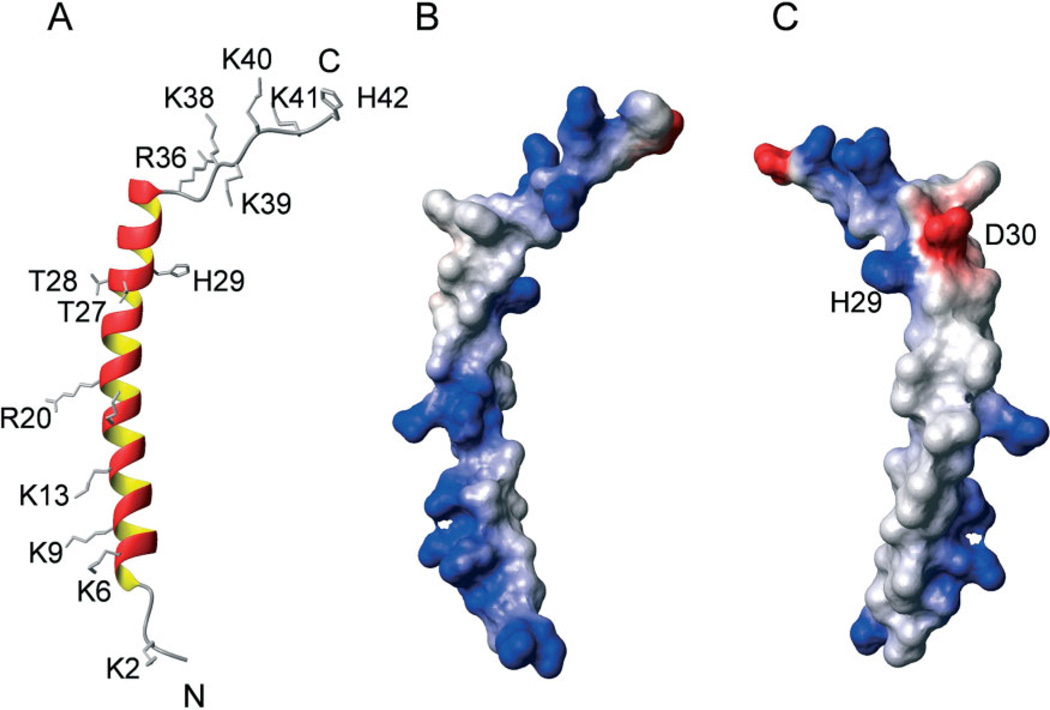
The schematic ribbon diagram and surface electrostatic surface of the energy-minimized average structure (Figure 5) indicate the amino-terminal segment of M. sexta moricin (V5 to I22) is amphipathic, with one side of the helix carrying 5 positive charges and the other side consisting of 12 hydrophobic residues. The carboxyl-terminal segment (N23 to K36) is slightly more hydrophilic: half of the residues carrying polar or charged side chains (N23, T27, T28, H29, D30, S32, K36) and the other half being hydrophobic. The long α-helix (V5 to K36) is followed by Pro37, connected to a stretch of highly basic amino acid residues. The charged surface potentials of M. sexta moricin exhibit a pattern remarkably similar to those of S. litura and B. mori moricins.
Figure 5.
Charge properties of the average, lowest energy structure of M. sexta moricin. (A) Ribbon diagram of the moricin structure with side chains of charged residues and two sequential threonine residues (T27 and T28) shown in sticks. (B) Surface electrostatic potentials of the average structure of M. sexta moricin with positive charges shown in blue and negative charges in red; (C) the same as (B) but rotated 180° around the vertical axis.
DISCUSSION
Insects and other multicellular organisms rely on a network of host defense mechanisms to survive and prosper in microbe-thriving environments [3]. One such mechanism involves the synthesis and operation of AMPs that are effective against a broad spectrum of pathogens including bacteria, fungi, viruses, and protozoa [1]. Analysis of the silkworm genome has revealed over 30 AMP genes belonging to the cecropin, moricin, gloverin, attacin, lebocin, and enbocin families [29]. Similar genes also exist in M. sexta [8]. While defensins are found in several orders of insects (Diptera, Hymenoptera, Coleoptera, Trichoptera, Hemiptera, and Odonata) and are highly effective against Grampositive bacteria, they have not been found in any lepidopteran species. Instead, a unique group of AMPs named moricins are identified in moths, which kill Gram-positive and Gram-negative bacteria at a low micromolar range [7,9,12]. Moricins drastically differ in tertiary structure from defensins that are stabilized by three disulfide bonds [30]. Rather, they are similar to cecropins, AMPs that are mainly active against Gram-negative bacteria.
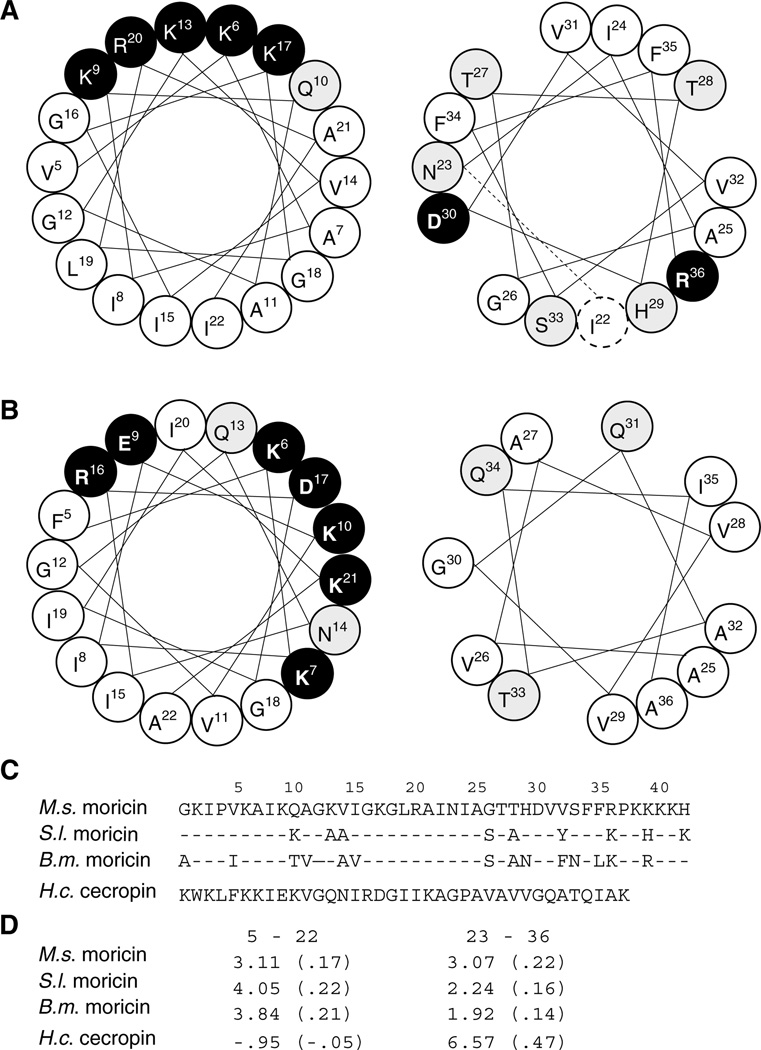
H. cecropia cecropin contains a long basic amphipathic α-helix (residues 5–22) at the amino terminus, a G23–P24 hinge, and a shorter hydrophobic α-helix at the carboxyl-terminus (residues 25–37) (Figure 6). Although cecropins and moricins have no significant similarity in their primary structures to suggest common ancestry, this sharing of structural properties strongly suggests the functional importance of the amino-terminal segments. In fact, Hara and Yamakawa [9] have demonstrated that B. mori moricin (residues 1–29) permeabilizes liposomal membrane.
Figure 6.
Helical wheel projections and side chain properties of the helices in moricins and cecropin. Residues 5–22 (left panel) and 23–36 (right panel) of M. sexta moricin (A) and H. cecropia cecropin (B). Hydrophobic, hydrophilic, and charged residues are shown in white, gray and black backgrounds, respectively. While cecropin contains two α-helices interrupted by G23–P24, the extended α-helix of moricin is divided into two parts (by a dashed line in right panel of (A) for comparing with the cecropin structure and for showing property differences in these two parts). (C) Amino acid sequences of M. sexta, B. mori, and S. litura moricins and H. cecropia cecropin. (D) Calculated overall and per residue (in parenthesis) hydrophobicity.
Not being interrupted by Gly-Pro as in cecropin, moricins take the shape of a long bent α–helix from residue 5–36 (Figures 4 and 5), whose carboxyl-terminal segment is slightly more hydrophilic than its counterpart in the cecropin (Figure 6(D)). Such a feature of moricins may limit the possibility for this segment to span lipid bilayer. While this variation could affect their mode of action, the absence of a positively charged tail in the cecropin is likely related to its specific killing of Gram-negative bacteria.
The solution structure of M. sexta moricin is suggestive of its action mechanism:
Initially, the positively charged regions such as G1KI3, K6–R20, and K38KKKH42 may be attracted by the anionic bacterial surface. Through electrostatic interactions with negatively charged lipopolysaccharides, teichoic acids, and peptidoglycans, the peptide molecules traverse capsular polysaccharides and attach to phospholipids of the cytoplasmic membrane. Due to structural flexibility of the terminal regions (Figure 4) and the dynamic nature of charge–charge interactions, the clustered basic residues at the carboxyl-terminus can be valuable for tighter binding, especially to Gram-positive bacteria such as S. aureus. In this human pathogen, products of the dlt operon reduce the net negative surface charges by transporting d-alanine from the cytosol to teichoic acids on the surface – esterification of deprotonized phosphate group of teichoic acids with D-alanine reduces the net negative charge [31]. We plan to use S. aureus to test whether or not removal of the positive tail affects MIC and rate of killing.
After the peptides attach to the plasma membrane P4, P37-flanked central α-helix may be partially embedded. The 5 positively charged and 12 hydrophobic residues of the amphipathic segment should stabilize the peptide–phospholipid complexes (Figure 6(A), left panel). In cecropin, there are two hydrophilic, five positively charged, and two negatively charged residues in the same region, which spread half of the helical wheel (Figure 6(B), left panel). Deeper embedding into the plasma membrane is anticipated for moricins in this part of the helix. While the second α-helix of cecropin is strongly hydrophobic (Figure 6(B), right panel; Figure 6(D)), the carboxyl-terminal region of moricin (residues 23–36) contains five hydrophilic and two charged residues (Figure 6(A), right panel). This shorter, less hydrophobic carboxyl-terminal α-helical segment is expected to less strongly associate with the membrane.
When the peptide concentration exceeds certain threshold, moricin molecules could cause membrane bending followed by pore formation, as depicted in the toroidal model [32]. To confirm this mode of action, we could test whether or not the pores formed by moricin molecules allow selective passage of certain ions. Based on the charge properties of M. sexta moricin, we hypothesize that the amino-terminal α-helical segment forms ion channels more favorable for Cl− or other negatively charged ions.
Acknowledgements
We thank Drs Michael Massiah and Guolong Zhang for the critical comments on the manuscript. Dr Guolong Zhang kindly provided technical guidance and laboratory space for the biological activity assays. We also thank KSU Targeted Excellence Programs for supporting NMR studies. Financial support for this work was provided by NIH (GM58634 and S10RR022392) and NSF (EPS0236913). This article was approved for publication by the Director of Oklahoma Agricultural Experimental Station and supported in part under project OKLO2450.
REFERENCES
- 1.Bulet P, Stöcklin R, Menin L. Antimicrobial peptides: from invertebrates to vertebrates. Immunol. Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- 2.Steiner H, Hultmark D, Engström A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 3.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Ann. Rev. Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 4.Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Müller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 5.Evans JD, Aronstein K, Chen YP, Hetru C, Imler JL, Jiang H, Kanost MR, Thompson GJ, Zou Z, Hultmark D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006;15:645–656. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou Z, Evans J, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. Comparative genome analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Y, Johnson T, Kanost MR. Identification of differentially expressed genes in the immune response of the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 2003;33:541–559. doi: 10.1016/s0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 8.Kanost MR, Jiang H, Yu X. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 9.Hara S, Yamakawa M. Moricin, a novel type of antibacterial peptide isolated from the silkworm, Bombyxmori. J. Biol. Chem. 1995;270:29923–29927. doi: 10.1074/jbc.270.50.29923. [DOI] [PubMed] [Google Scholar]
- 10.Furukawa S, Tanaka H, Nakazawa H, Ishibashi J, Shono T, Yamakawa M. Inducible gene expression of moricin, a unique antibacterial peptide from the silkworm Bombyx mori. Biochem. J. 1999;340:254–271. [PMC free article] [PubMed] [Google Scholar]
- 11.Hemmi H, Ishibashi J, Hara S, Yamakawa M. Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori. FEBS Lett. 2002;518:33–38. doi: 10.1016/s0014-5793(02)02637-6. [DOI] [PubMed] [Google Scholar]
- 12.Oizumi Y, Hemmi H, Minami M, Asaoka A, Yamakawa M. Isolation, gene expression and solution structure of a novel moricin analogue, antibacterial peptide from a lepidopteran insect, Spodoptera litura. Biochim. Biophys. Acta. 2005;1752:83–92. doi: 10.1016/j.bbapap.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Dunn P, Drake D. Fate of bacteria injected into naïve and immunized larvae of the tobacco hornworm, Manduca sexta. J. Invert. Pathol. 1983;41:77–85. [Google Scholar]
- 14.Wang Y, Zou Z, Jiang H. An expansion of the dual clipdomain serine proteinase family in Manduca sexta: gene organization, expression, and evolution of prophenoloxidase-activating proteinase-2, hemolymph proteinase 12, and other related proteinases. Genomics. 2006;87:399–409. doi: 10.1016/j.ygeno.2005.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. 6th edn. Wayne, PA: NCCLS; 2003. Approved Standard M7-A6. [Google Scholar]
- 16.Bommineni YR, Dai H, Gong YX, Soulages JL, Fernando SC, Desilva U, Prakash O, Zhang G. Fowlicidin-3 is an alpha-helical cationic host defense peptide with potent antibacterial and lipopolysaccharide-neutralizing activities. FEBS J. 2007;274:418–428. doi: 10.1111/j.1742-4658.2006.05589.x. [DOI] [PubMed] [Google Scholar]
- 17.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–666. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 18.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; 2002. [Google Scholar]
- 19.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Chen T, Rayaprolu S, Zou Z, Xia Q, Xiang Z, Jiang H. Proteolytic activation of pro-spätzle is required for the induced transcription of antimicrobial peptide genes in lepidopteran insects. Dev. Comp. Immunol. 2007;31:1002–1012. doi: 10.1016/j.dci.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shai Y. Mode of action of antibacterial peptides. In: Brey PT, Hultmark D, editors. Molecular Mechanisms of Immune Responses in Insects. London: Chapman and Hall; 1997. pp. 111–134. [Google Scholar]
- 24.Wüthrich K. NMR of Proteins and Nucleic Acids. New York, NY: John Wiley and Sons; 1986. [Google Scholar]
- 25.Cai M, Gong Y, Kao JL, Krishnamoorthi R. A practical method for stereospecific assignments of gamma- and delta-methylene hydrogens via estimation of vicinal 1H-1H coupling constants. J. Magn. Reson., Ser. B. 1995;107:172–178. doi: 10.1006/jmrb.1995.1074. [DOI] [PubMed] [Google Scholar]
- 26.Wagner G, Braun W, Havel TF, Schaumann T, Go N, Wüthrich K. Protein structures in solution by nuclear magnetic resonance and distance geometry. The polypeptide fold of the basic pancreatic trypsin inhibitor determined using two different algorithms, DISGEO and DISMAN. J. Mol. Biol. 1987;196:611–639. doi: 10.1016/0022-2836(87)90037-4. [DOI] [PubMed] [Google Scholar]
- 27.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 28.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 29.Cheng T, Zhao P, Liu C, Xu P, Gao Z, Xia Q, Xiang Z. Structures, regulatory regions, and inductive expression patterns of antimicrobial peptide genes in the silkworm Bombyx mori. Genomics. 2006;87:356–365. doi: 10.1016/j.ygeno.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Bulet P, Stöcklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept. Lett. 2005;12:3–11. doi: 10.2174/0929866053406011. [DOI] [PubMed] [Google Scholar]
- 31.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 32.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]