Abstract
The ethanolamine utilization (eut) locus of Enterococcus faecalis, containing at least 19 genes distributed over four polycistronic messenger RNAs, appears to be regulated by a single adenosyl cobalamine (AdoCbl)–responsive riboswitch. We report that the AdoCbl-binding riboswitch is part of a small, transacting RNA, EutX, which additionally contains a dual-hairpin substrate for the RNA binding–response regulator, EutV. In the absence of AdoCbl, EutX uses this structure to sequester EutV. EutV is known to regulate the eut messenger RNAs by binding dual-hairpin structures that overlap terminators and thus prevent transcription termination. In the presence of AdoCbl, EutV cannot bind to EutX and, instead, causes transcriptional read through of multiple eut genes. This work introduces riboswitch-mediated control of protein sequestration as a posttranscriptional mechanism to coordinately regulate gene expression.
Riboswitches are an important class of regulatory RNA; they typically regulate expression in cis of downstream open reading frames in response to binding of metabolic ligands, usually by affecting the transcription or translation (1–3). Small regulatory RNAs (sRNAs) are another class, and they commonly regulate gene expression by interacting with target mRNAs to affect translation or stability (3). Herein, we report the discovery of an sRNA, EutX, that contains a riboswitch aptamer and a EutV-binding site named P3/P4 (fig. S1A). The presence of ethanolamine (EA) causes the sensor histidine kinase EutW to phosphorylate the response regulator, EutV, which thereby converts it into an active RNA binding protein (4, 5). Thus, in the absence of EA, EutV is unphosphorylated and inactive. Adenosyl cobalamine (AdoCbl) is a cofactor required in the first enzymatic step of EA catabolism (6). Under conditions in which EA is present, but AdoCbl is not, our model states that EutX uses P3/P4 to bind and sequester active EutV~P, which prevents its antitermination activity (fig. S1A). When AdoCbl is also present, it binds to the AdoCbl riboswitch and induces termination before generating the P3/P4 loops, which prevents sequestration. Unhindered, EutV is then able to promote antitermination at the four eut polycistronic RNAs to drive gene expression. By this mechanism, the Eut system demands the presence of both the catabolic substrate (EA) and the key cofactor (AdoCbl) (fig. S1A).
To gain a more complete understanding of the effect of AdoCbl and EA on eut gene expression, Enterococcus faecalis cells were cultured to exponential growth phase in modified minimal medium in the presence or absence of EA and AdoCbl. Total RNA was then extracted and subjected to RNA-sequencing (RNA-seq) analysis (Fig. 1 and table S1). This revealed increased expression across the eut locus for cells that had been cultured with EA and AdoCbl compared with those cultured only in minimal medium. Metabolism of EA is believed to occur within a protein-bound organelle called a microcompartment [reviewed by (7)]. Electron microscopy revealed microcompartments only for cells cultured in EA and AdoCbl (fig. S1B). These aggregate data demonstrate that eut gene expression and microcompartment formation can indeed be induced in the presence of both EA and AdoCbl.
Fig. 1. EA and AdoCbl induce eut gene expression.

By RNA-seq, all the eut genes were expressed in the presence of EA and AdoCbl, whereas little or no expression was observed in minimal medium.
Of the four constitutive promoters in the eut locus, three are followed by a EutV RNA binding site (P1/P2, P5/P6, and P7/P8). The distal hairpin in each case overlaps the 5′ sequence of an adjacent, intrinsic terminator. Phosphorylated EutV dimerizes, which increases the affinity for the dual hairpin, which prevents the formation of the terminator structure and thereby induces gene expression (fig. S1A) (4, 5, 8, 9). A fourth promoter was detected just upstream of the AdoCbl riboswitch (8). Moreover, we discovered that another two-hairpin motif (P3/P4) is located downstream of the AdoCbl riboswitch (fig. S1A). To more closely examine the transcripts in this region, we analyzed total RNA by Northern blotting. An antisense RNA probe that spanned 301 nucleotides (nt) of the eutT-eutG intergenic region (Fig. 2A) revealed an RNA species of ~300 nt in length that was not present in medium containing AdoCbl, detectable in minimal medium, and notably enhanced (about fivefold) upon addition of EA without AdoCbl (Fig. 2B). A second RNA of ~150 nt was produced upon addition of AdoCbl. To resolve which parts of the eutT-eutG intergenic region corresponded to these different RNA species, the membranes were hybridized with antisense probes corresponding to the left and right halves of the 301-nt antisense probe (Fig. 2A). This analysis revealed that the ~300-nucleotide and ~150-nucleotide RNAs both included the AdoCbl riboswitch. Quantitative reverse transcription polymerase chain reaction (qRT-PCR), using primers within and outside of EutX, detected full-length EutX primarily in medium containing only EA (fig. S2). Together, these data demonstrate that an sRNA, coined EutX, is produced from the eutT-eutG intergenic region.
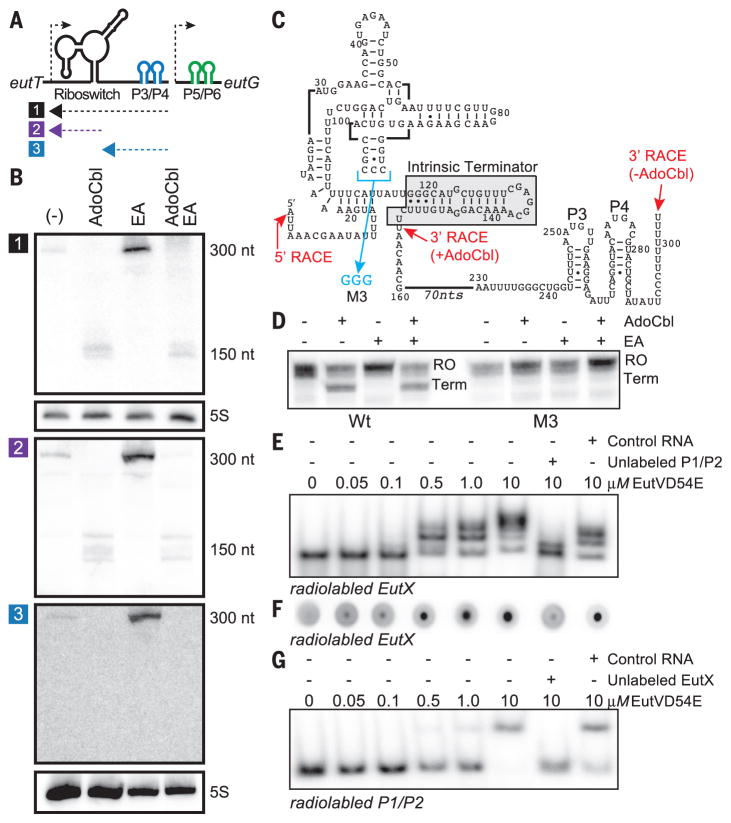
Fig. 2. AdoCbl binding to the riboswitch causes premature termination within a sRNA, preventing generation of the P3/P4 hairpins that bind EutV.
(A) Location of 32P-radiolabeled RNA anti-sense probes used to analyze the eutT-eutG intergenic region. (B) Total RNA was isolated from cells grown in the indicated medium conditions, resolved by denaturing polyacrylamide gel electrophoresis, transferred to a membrane, and hybridized with different antisense probes (1, 2 and 3). (C) The mapped 5′ and 3′ termini and the M3 mutation are indicated on a secondary structure depiction of EutX. A putative intrinsic terminator is highlighted by a gray-shaded box. (D) In vitro transcription termination assays with wild-type EutX template display a premature termination product only in the presence of AdoCbl. Termination is lost in the M3 mutant template. (E) EMSA of binding of EutV to EutX. A 117-nt fragment of EutX (117EutX) and EutVD54E protein [constitutively active for RNA binding (5)] were used. Controls included addition of 30 μM of an unlabeled competitor P1/P2 RNA and an unrelated RNA. (F) DRaCALA to assess EutV binding to the 117EutX RNA by using the same reaction conditions and control RNAs as in (E). (G) EMSA to investigate whether EutX could compete for EutV binding to P1/P2.
To map the difference between the 300-nucleotide and 150-nucleotide RNAs, we used 3′ rapid amplification of cDNA ends (3′ RACE). By using cells grown in EA without AdoCbl, 3′ RACE located the 3′ terminus within an oligouridylate tract that occurs downstream of P3/P4 and just before the promoter of eutG, a length consistent with the Northern blot analysis (301 nts) (Fig. 2C). Using RNA from cells grown with just AdoCbl revealed a 3′ terminus at the end of the riboswitch aptamer where another stretch of U’s exists. The latter sequence appears to be part of an intrinsic terminator hairpin, which is formed from sequences at the 3′ end of the AdoCbl riboswitch aptamer (Fig. 2C), consistent with previous in vitro studies (8). Indeed, addition of low-micromolar AdoCbl promoted transcription termination at this site in vitro (Fig. 2D and fig. S3). AdoCbl riboswitches have a small loop containing three cytosines that forms a pseudoknot with downstream guanosines and is necessary for ligand binding (10, 11). The eut AdoCbl riboswitch is predicted to contain this characteristic L5 loop (Fig. 2C) (4). Consistent with a role in sensing AdoCbl, when we mutated these residues, it disrupted termination in response to AdoCbl (Fig. 2D). Also, structural probing revealed that AdoCbl-induced changes were limited to the riboswitch portion of EutX (fig. S4). In addition, a construct in which lacZ is fused downstream of the terminator resulted in moderately decreased expression upon addition of AdoCbl (fig. S5).
To further characterize the function of EutX, the ability to bind EutV was confirmed by electrophoretic mobility assays (EMSAs) and differential radial capillary action of ligand assays (DRaCALA) (12). Upon addition of micromolar concentrations, purified EutV bound to a portion of EutX that included P3/P4, as shown in Fig. 2, E and F. On the basis of comparative data of EutV binding to the P1/P2 hairpins, we speculate that the binding affinity to EutX may be moderately better (Fig. 2G) (5). Unlabeled 30 μM competitor RNA (EutX or P1/P2) could fully compete for binding to EutV in the context of radiolabeled EutX or P1/P2 (Fig. 2, E to G). However, an unrelated RNA of similar length was unable to compete for EutV, which demonstrated that binding of EutV to EutX is specific.
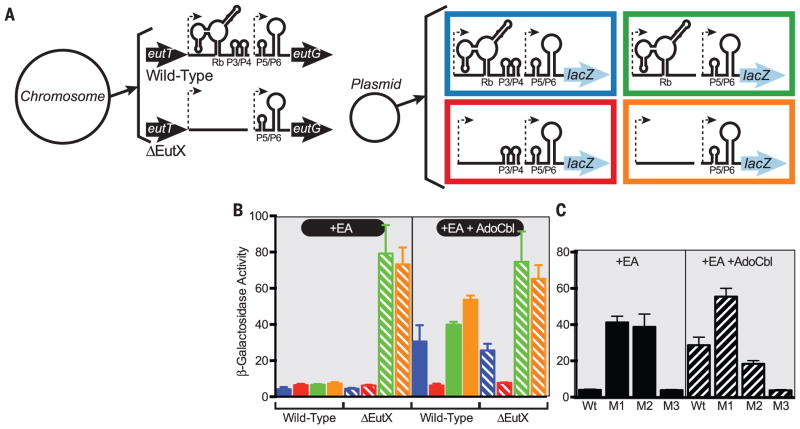
On the basis of the observation that the riboswitch exerts transcription attenuation–mediated control over a EutV binding site, we hypothesized that the EutX sRNA affects eut expression in trans. We therefore generated a eutG-lacZ fusion that contained the entire intergenic region between eutT and eutG (Fig. 3, A and B, blue construct), as well as fusions that either deleted P3/P4 (green), the riboswitch (red), or both (orange). These plasmids were transformed into a strain lacking the endogenous eutX or a wild-type strain. Expression of wild-type eutG-lacZ (blue) was induced by the presence of EA and AdoCbl when cells were grown in a modified minimal medium (Fig. 3B). No expression was observed for this construct in minimal medium, medium containing only AdoCbl, or in a background lacking EutV and EutW (fig. S6). The following data indicate that EutX acts in trans. First, the constructs that lacked P3/P4 were normally regulated in a wild-type strain that still contains P3/P4 in the chromosome (Fig. 3B), which suggested that P3/P4 is not needed in cis. Second, in the strain that lacked endogenous EutX, these same constructs were induced both in +EA and +EA+AdoCbl conditions (Fig. 3B), which suggested that the endogenous EutX prevents eut mRNA expression by sequestering EutV. Third, the construct that lacked the riboswitch aptamer, but still contained P3/P4, remained uninduced even in the presence of EA and AdoCbl, consistent with constitutive sequestration of EutV by P3/P4 (Fig. 3B). Because the regulation of the lacZ constructs could be altered by the presence or absence of EutX in the chromosome, these data demonstrate that the sRNA can regulate in trans and are consistent with the sequestration model (fig. S1A). Remarkably, in addition to inducing eutG-lacZ expression with only EA, the eutX strain has the additional property of supporting the formation of visually normal microcompartments in this medium, unlike the wild-type strain (fig. S7). Together, these data show that, in the absence of EutX, EA is sufficient to induce functional expression of the entire eut locus.
Fig. 3. Both the riboswitch and the P3/P4 hairpins are required for inducible expression in vivo.
(A) A diagram of the experimental design. A plasmid containing a eutG-lacZ fusion was manipulated to contain a deletion of the riboswitch, the P3/P4 hairpin, or both. The plasmid was introduced into E. faecalis strains containing either wild-type EutX or a deletion. (B) β-Galactosidase assays of strains shown in (A) when grown in modified minimal medium containing EA or both EA and AdoCbl. (C) β-Galactosidase assays of strains missing EutX but carrying eutG-lacZ plasmids that contain either the M1 or M2 mutations. The M1 mutations are A249U and G252U and the M2 mutations are A273U and G276U site-directed changes. The data are presented as the average of three or more independent experiments, and the error bars represent the standard deviation.
To further test the sequestration model, we introduced point mutations into the eutG-lacZ construct instead of deletions. An adenosine (A) at the first position and a guanine (G) at the fourth position of the six-nucleotide loops were previously shown to be required for EutV binding of dual-hairpin substrates (5). We tested one mutant, in which these critical residues of the P3 loop were mutated (M1) and a second, in which the same changes were made to the P4 loop (M2). Similar to deletion of P3/P4, these mutant constructs were both induced by EA alone in the eutX strain, in contrast to wild-type eutG-lacZ (Fig. 3D). Therefore, these data confirm that P3/P4 mutations that prevent EutV binding affect regulation. We also examined the M3 mutation predicted to abrogate AdoCbl binding and cause constitutive inclusion of P3/P4 in EutX (Fig. 2, C and D). As expected, this resulted in uninducible expression (Fig. 3C).
In conclusion, we present evidence of a ribo-switch acting in trans, the second described (13). In the prior example, a classical sRNA that acts by base-pairing to a target mRNA can be prematurely terminated by a riboswitch. However, EutX acts in a signal transduction pathway that is subject to riboswitch-mediated control within the confines of an sRNA. A few other sRNAs, like EutX, have been shown to affect gene expression by sequestering RNA-binding proteins. The best-studied examples are CsrB and CsrC, which titrate CsrA, a translational inhibitor protein, away from its mRNA targets [reviewed by (14)]. EutX demonstrates a mechanism for how protein sequestration can be placed under signal-responsive regulatory control by a riboswitch.
Supplementary Material
Acknowledgments
This work was supported by the NIH grants R01AI076406 (D.A.G.), R56AI110432 (D.A.G. and W.C.W.) and R01GM099790 (A.V.H.), a NSF grant MCB-1051440 (W.C.W.) and a Welch Foundation grant AU-1773 (A.V.H.). For transmission electron microscopy imaging, we thank S. Kolodziej and P. Navarro; J. R. Mellin and P. Cossart are acknowledged for valuable discussions.
Footnotes
The data reported are presented in the main paper and the supplementary materials.
REFERENCES AND NOTES
- 1.Roth A, Breaker RR. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serganov A, Nudler E. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waters LS, Storz G. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox KA, et al. Proc Natl Acad Sci USA. 2009;106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramesh A, et al. PLOS Genet. 2012;8:e1002666. doi: 10.1371/journal.pgen.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scarlett FA, Turner JM. J Gen Microbiol. 1976;95:173–176. doi: 10.1099/00221287-95-1-173. [DOI] [PubMed] [Google Scholar]
- 7.Garsin DA. Nat Rev Microbiol. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker KA, Perego M. J Bacteriol. 2011;193:2575–2586. doi: 10.1128/JB.00217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Papa MF, Perego M. J Bacteriol. 2008;190:7147–7156. doi: 10.1128/JB.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson JE, Jr, Reyes FE, Polaski JT, Batey RT. Nature. 2012;492:133–137. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peselis A, Serganov A. Nat Struct Mol Biol. 2012;19:1182–1184. doi: 10.1038/nsmb.2405. [DOI] [PubMed] [Google Scholar]
- 12.Roelofs KG, Wang J, Sintim HO, Lee VT. Proc Natl Acad Sci USA. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loh E, et al. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 14.Babitzke P, Romeo T. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.