Abstract
Cytotoxic activity of most chemotherapeutic agents is based on their ability to induce DNA damage. Interstrand crosslinks are among the most detrimental forms of DNA damage as both DNA strands are affected. As translesion polymerases participate in their repair, they may be important for response to chemotherapeutic agents that induce such lesions, including commonly used cisplatin. Altered expression of translesion polymerase genes REV1 and REV3L may modify sensitivity to cisplatin. As osteosarcoma patients are commonly treated with cisplatin-based chemotherapy, our aim was to investigate if REV1 and REV3L polymorphisms influence survival of osteosarcoma patients treated with cisplatin-based chemotherapy. We determined the genotypes of common functional tag REV1 and REV3L polymorphisms in 66 osteosarcoma patients. Cox regression was used for survival analysis. Carriers of at least one polymorphic REV1 rs3087403 allele had significantly shorter EFS and overall survival (OS) (p=0.004; HR=3.79; 95%CI=1.53–9.35 and p<0.001; HR=4.44; 95%CI=1.92–10.27, respectively). Combination of REV1 rs3087403 and REV3L rs462779 polymorphisms was also significantly associated with shorter OS (ptrend<0.001) and shorter EFS (ptrend=0.003). The results of this first study on polymorphisms in translesion polymerase genes in osteosarcoma suggest they could help predict outcome of cisplatin-based chemotherapy in osteosarcoma patients.
Introduction
Cytotoxic activity of many chemotherapeutic agents is based on the ability to induce DNA damage in rapidly dividing tumor cells (Woods and Turchi, 2013). Different mechanisms of action of individual therapeutics lead to formation of different types of DNA damage. Several different DNA repair pathways are involved in response to chemotherapy-induced DNA damage and can affect both the cytotoxic activity in tumor cells or toxicity in normal cells (Woods and Turchi, 2013).
Cisplatin is one of the most commonly used antitumor agents. The bifunctional electrophilic platinum compounds covalently bind to DNA and form DNA adducts, including intra- and interstrand crosslinks (ICLs), thus inhibiting DNA replication (Marsh et al., 2009). Intrastrand crosslinks are more common, but ICLs are more detrimental, as they compromise both strands and completely prevent DNA replication and transcription. Successful ICL repair is thus essential for genome stability and cell survival (Enoiu et al., 2012; Sharma and Canman, 2012) and requires interplay of different DNA repair pathways (Ho and Scharer, 2010). Apart from nucleotide excision repair and homologous recombination repair, translesion (TLS) polymerases have a crucial role in this process.
TLS polymerases polymerase zeta (Pol ζ) and DNA polymerase REV1 (DNA directed) (REV1) are involved in both replication-independent and replication-dependent mechanisms of ICL repair, as they enable the bypass of ICLs and restoration of one of the DNA strands (Ho and Scharer, 2010). Pol ζ, whose catalytic subunit is encoded by the REV3L gene, is involved in the extension step of TLS synthesis, while REV1 coordinates TLS synthesis through interactions with Pol ζ and other TLS polymerases, but also has a deoxycytidyltransferase activity (Ho and Scharer, 2010).
Differences in expression of either REV1 or REV3L have been associated with modified sensitivity to cisplatin (Doles et al., 2010; Lin et al., 2006), but the role of single nucleotide polymorphisms (SNPs) in these genes has not been explained yet. Some studies have shown that these SNPs may modify cancer susceptibility (He et al., 2008; Pan et al., 2012; Sakiyama et al., 2005; Varadi et al., 2011; Zhang et al., 2013). The influence on response to cisplatin treatment was previously unknown, so in our recent studies we have used a haplotype-based approach to select common functional REV1 and REV3L tag SNPs in the coding region or 3′ untranslated region that may influence enzyme function on expression. We have shown that they influence cisplatin-based treatment outcome in malignant mesothelioma (Goricar et al., 2014a). However the role of genetic variability of TLS polymerases in treatment of other cancers remains to be elucidated.
Cisplatin-based chemotherapy is used also in treatment of osteosarcoma, a bone malignancy that mostly affects adolescents and has a very low incidence of approximately 3 patients per million people per year (Ritter and Bielack, 2010). Current chemotherapy protocols include cisplatin in combination with doxorubicin, high-dose methotrexate, and/or ifosfamide (Quartuccio et al., 2013). Such multiagent chemotherapy significantly improved prognosis of the disease (Ritter and Bielack, 2010), but survival rates have not improved since the start of chemotherapy use, and poor outcome, local relapse, or metastases remain a challenge in as much as half of osteosarcoma patients (Chou and Gorlick, 2006; Salinas-Souza et al., 2010).
Apart from clinical characteristics, interindividual differences in treatment outcome may be attributed to genetic variability of mechanisms involved in response to chemotherapeutic agents and some polymorphisms have already been associated with response to methotrexate or cisplatin (Biason et al., 2012; Caronia et al., 2009; Goricar et al., 2014b; Hao et al., 2012; Yang et al., 2012), but the role of genetic variability in TLS polymerases has so far not been investigated in osteosarcoma. The aim of the present study was to evaluate the influence of polymorphisms and haplotypes in TLS polymerase genes on the survival of osteosarcoma patients treated with cisplatin-based chemotherapy.
Materials and Methods
Patients
Slovenian osteosarcoma patients diagnosed between 1990 and 2008 were eligible for inclusion in our retrospective study. We included all osteosarcoma patients who were treated with cisplatin-based chemotherapy at the Department of Hematology and Oncology, University Children's Hospital, Ljubljana, Slovenia, or at the Institute of Oncology, Ljubljana, Slovenia, and had sufficient formalin fixed, paraffin embedded (FFPE) material for DNA extraction and data from medical records available. The study was approved by the Slovenian Ethics Committee for Research in Medicine and was carried out according to the Declaration of Helsinki.
Assessment of treatment outcome
Demographic, clinical, and treatment parameters were obtained from the medical records. The primary outcome of interest was survival of osteosarcoma patients. Event-free survival (EFS) was defined as time from the beginning of treatment to disease recurrence, development of metastases, or death. Overall survival (OS) was defined as time from the beginning of treatment to death. If disease recurrence, development of metastases, or death did not occur to the time of the analysis, we censored patients at the date of the last follow-up.
DNA extraction and genotyping
Hematoxylin and eosin-stained sections of each FFPE sample obtained at surgery were examined by an experienced pathologist to confirm the diagnosis and select areas representative of tumor or normal tissue. Genomic DNA was isolated from two to three cores (1 mm in diameter) of histologically confirmed normal tissue using QIAamp DNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions (Goricar et al., 2014). Genotyping of REV1 rs3087403 (p.Val138Met), rs3087386 (p.Phe257Ser), and rs3087399 (p.Asn373Ser) and REV3L rs462779 (p.Thr1224Ile), rs455732 (p.Val1430=), rs3204953 (p.Val3064Ile), rs465646 (c.*461C>T) tag SNPs (Goricar et al., 2015) was performed using KASPar assay. Genotyping was repeated in 20% samples to check for genotyping accuracy.
Statistical analyses
Median and interquartile range or frequencies were used to describe the distribution of continuous and categorical variables, respectively. Deviation from Hardy–Weinberg equilibrium (HWE) was calculated with the standard chi-square test. Cox proportional hazards model was used in survival analysis to calculate hazard ratio (HR) and the 95% confidence interval (CI). A dominant genetic model was used in all statistical analyses. All statistical analyses were carried out by IBM SPSS Statistics, version 19.0 (IBM Corporation, Armonk, NY, USA). Thesias software was used for haplotype analysis (Tregouet and Garelle, 2007) as previously described (Erculj et al., 2012): the most frequent haplotype was used as reference and only haplotypes with frequencies above 5% were included in the analyses. Benjamini-Hochberg false discovery rate (FDR) was used to account for multiple comparisons (Benjamini and Hochberg, 1995) and p values less than 0.010 were considered significant after correction.
Results
Patients' characteristics
A total of 66 osteosarcoma patients treated with cisplatin-based chemotherapy were included in our study. Thirty-five (53.0%) were male. Patients' median age was 17.5 (13.8–33.3) years. The majority of patients had osteoblastic osteosarcoma located in the extremities [59 (89.4%) and 57 (86.4%), respectively]. Median tumor size was 9.5 (7.5–11.5) cm. Regarding stage, 18 (27.3%) patients had stage 1 osteosarcoma, 36 (54.5%) stage 2, and 10 (15.2%) stage 3, while the information was missing for two patients. Most patients, 38 (57.6%), were treated with multiagent chemotherapy that included both cisplatin and methotrexate, while 28 (42.4%) were receiving only combination of cisplatin and doxorubicin. Only four (6.1%) patients were not surgically treated and among the rest, ten (15.1%) patients were receiving adjuvant chemotherapy.
All patients were genotyped for REV1 and REV3L SNPs. Their minor allele frequencies (MAFs) are presented in Table 1. All of the genotype frequencies were in HWE except REV3L rs455732 (p=0.014), which was excluded from further analyses.
Table 1.
Association of Investigated Polymorphisms with Event-Free and Overall Survival in Osteosarcoma Patients
| Event-free survival | Overall survival | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Polymorphism | MAF | GenotypeCT+TT | HR (95% CI)0.85 (0.34–2.15) | pa0.729 | HR (95% CI)0.49 (0.22–1.11) | pa0.089 |
| REV1 | rs3087403 | CC | Reference | Reference | |||
| p.Val138Met | 0.24 | CT+TT | 3.79 (1.53–9.35) | 0.004 | 4.44 (1.92–10.27) | <0.001 | |
| rs3087386 | GG | Reference | Reference | ||||
| p.Phe257Serb | 0.45 | GA+AA | 1.19 (0.42–3.39) | 0.744 | 1.96 (0.72–5.33) | 0.189 | |
| rs3087399 | TT | Reference | Reference | ||||
| p.Asn373Ser | 0.22 | TC+CC | 1.38 (0.59–3.26) | 0.458 | 0.87 (0.41–1.82) | 0.707 | |
| REV3L | rs462779 | TT | Reference | Reference | |||
| p.Thr1224Ile | 0.14 | TC+CC | 2.49 (0.98–6.36) | 0.056 | 2.60 (1.18–5.74) | 0.018 | |
| rs3204953 | GG | Reference | Reference | ||||
| p.Val3064Ile | 0.14 | GA+AA | 0.99 (0.40–2.48) | 0.998 | 1.56 (0.71–3.51) | 0.265 | |
| rs465646 | AA | Reference | Reference | ||||
| c.*461C>T | 0.14 | AG+GG | 1.33 (0.46–3.83) | 0.601 | 1.50 (0.66–3.44) | 0.335 | |
p less than 0.010 was considered statistically significant after correction for multiple comparisons; bdata missing for 1 patient.
CI, confidence interval; HR, hazard ratio; MAF, minor allele frequency; OR, odds ratio.
Survival analysis
The median follow-up time for osteosarcoma patients in our study was 143.0 (109.1–205.6) months. A total of 39 (59.1%) patients experienced an event and 33 (50.0%) had died by the time of the analysis. The median EFS was 27.2 (10.9–136.3) months, while the median OS was 56.5 (25.4–140.7) months. Several clinical characteristics were associated with survival. Older patients had shorter EFS and OS (p=0.003; HR=1.02; 95%CI=1.01–1.04; and p=0.001; HR=1.03; 95%CI=1.01–1.05, respectively). Patients receiving multiagent chemotherapy including cisplatin and methotrexate had longer EFS and OS (p<0.001; HR=0.27; 95%CI=0.14–0.52; and p=0.002; HR=0.33; 95%CI=0.16–0.66, respectively) compared to patients treated only with the cisplatin and doxorubicin combination. Treatment with methotrexate was significantly correlated with age, as pediatric patients were more often treated with methotrexate according to the COSS protocols. Higher stage was also associated with shorter EFS and OS (p<0.001; HR=3.04; 95%CI=1.78–5.19; and p=0.003; HR=2.31; 95%CI=1.34–3.98, respectively). The size of the tumor was only associated with shorter EFS (p=0.033; HR=1.17; 95%CI=1.01–1.24). Due to correlations between clinical variables, only stage and age were included in the final multivariable model for OS, while the model for EFS was adjusted for stage, age, and tumor size.
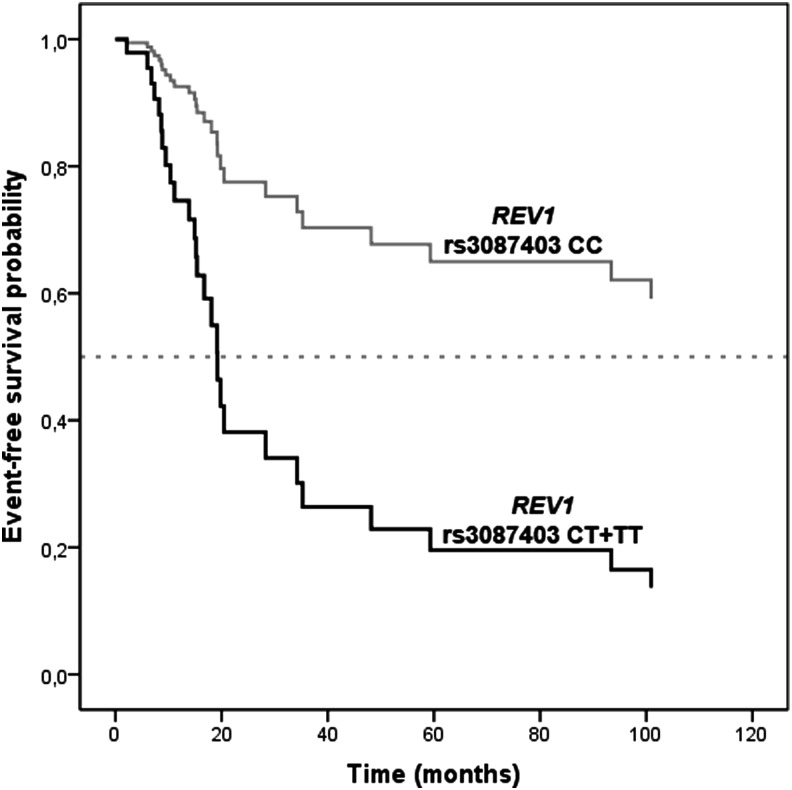
The association of investigated polymorphisms with EFS and OS after adjustment for clinical variables is presented in Table 1. Carriers of at least one polymorphic REV1 rs3087403 or REV3L rs462779 allele had significantly shorter OS (HR=4.44; 95%CI=1.92–10.27; p<0.001 and HR=2.60; 95%CI=1.18–5.74; p=0.018, respectively). Polymorphic REV1 rs3087403 was also associated with significantly shorter EFS (HR=3.79; 95%CI=1.53–9.35; p=0.004; Fig. 1).
FIG. 1.
The influence of REV1 rs3087403 on event-free survival of osteosarcoma patients.
Since REV1 and Pol ζ interact during DNA repair, we also evaluated the combined effect of REV1 rs3087403 and REV3L rs462779 SNPs on survival of osteosarcoma patients. The combination of both SNPs was also significantly associated with shorter OS (ptrend<0.001) and patients with at least one polymorphic allele for each SNP had even shorter OS (HR=7.52; 95%CI=2.14–26.43). The same combination of SNPs in these two genes also conferred to shorter EFS (ptrend=0.003) and individuals with at least one polymorphic allele for each SNP had much shorter EFS (HR=5.12; 95%CI=1.64–15.98). All these associations except the influence of REV3L rs462779 on OS remained significant after adjustment for multiple comparisons (p<0.010).
Haplotype analysis
As more than one SNP was investigated in most of the genes, we studied their combined effect using haplotype analysis. Four haplotypes with frequencies above 5% could explain approximately 88% of the REV1 variability (Table 2). Compared to the reference REV1 CAT haplotype, patients with the TGT haplotype had shorter OS, but the difference was not statistically significant (p=0.058). Four common haplotypes could explain around 93% of the REV3L variability. Carriers of the REV3L CGG haplotype had shorter OS and EFS (p=0.059 and p=0.020, respectively), but the influence did not remain significant after correction for multiple comparisons.
Table 2.
The Influence of Haplotypes on Survival in Osteosarcoma Patients
| Event-free survival | Overall survival | |||||
|---|---|---|---|---|---|---|
| Gene | Haplotype | Estimated frequency | HR (95 % CI) | pa | HR (95 % CI) | pa |
| REV1b | CAT | 0.45 | Reference | Reference | ||
| TGT | 0.18 | 1.58 (0.83–3.00) | 0.161 | 1.91 (0.98–3.71) | 0.058 | |
| CGC | 0.14 | 1.49 (0.77–2.91) | 0.239 | 1.20 (0.59–2.44) | 0.624 | |
| CGT | 0.11 | 0.73 (0.31–1.71) | 0.465 | 0.54 (0.21–1.38) | 0.197 | |
| REV3Lc | TGA | 0.65 | Reference | Reference | ||
| TAA | 0.14 | 1.11 (0.62–2.00) | 0.731 | 1.61 (0.85–3.05) | 0.142 | |
| CGG | 0.08 | 2.98 (0.96–9.26) | 0.059 | 3.27 (1.20–8.85) | 0.020 | |
| TGG | 0.06 | 0.67 (0.24–1.86) | 0.446 | 0.79 (0.21–2.96) | 0.715 | |
p less than 0.010 was considered statistically significant after correction for multiple comparisons; bThe SNPs are ordered from the 5′- to 3′-end as follows: rs3087403, rs3087386, and rs3087399; cThe SNPs are ordered from the 5′- to 3′-end as follows: rs462779, rs3204953, and rs465646.
CI, confidence interval; HR, hazard ratio.
Discussion
We present the results of the first study investigating SNPs in TLS polymerase genes and outcome of cisplatin-based treatment of osteosarcoma, showing that REV1 SNPs influenced both EFS and OS.
TLS polymerases have recently been described as important factors in DNA repair. But even though TLS polymerases might alter chemosensitivity to cisplatin due to their involvement in ICL repair (Makridakis and Reichardt, 2012), there is little information about the role of the genetic variability of REV1 and REV3L that encode two of the most important TLS polymerases. Only a handful of studies have previously studied REV1 and REV3L SNPs, and even those have focused mostly on cancer susceptibility (He et al., 2008; Pan et al., 2012; Sakiyama et al., 2005; Varadi et al., 2011; Zhang et al., 2013). Both genes are very polymorphic and to cover most of the genetic variability within REV1 and REV3L genes, we have previously used a tag SNP approach to select putative functional SNPs with MAF above 5% in the coding or 3′ untranslated region that could influence enzyme activity or expression (Goricar et al., 2014a).
In our recent study of cisplatin-treated malignant mesothelioma patients, we showed for the first time that REV1 and REV3L SNPs might contribute to interindividual differences in response to cisplatin (Goricar et al., 2014a). In the present study, polymorphic REV1 rs3087403 was associated with shorter survival of osteosarcoma patients. REV1 facilitates bypass of cisplatin-induced DNA damage (Lange et al., 2011) and decreased expression of REV1 increases genomic instability and sensitivity to cisplatin (Ho and Scharer, 2010; Lin et al., 2006). To the best of our knowledge, the non-synonymous REV1 rs3087403 (p.Val138Met) was not investigated in any previous study except in our study on malignant mesothelioma treatment, where it was associated with hematological toxicity. However, other REV1 SNPs have been associated with modified lung cancer and cervical cancer risk (He et al., 2008; Sakiyama et al., 2005). Osteosarcoma patients with at least one polymorphic REV3L rs462779 allele tended to have shorter OS. It was also shown that loss of Pol ζ increases genomic instability and that decreased REV3L expression increases sensitivity to cisplatin (Doles et al., 2010; Ho and Scharer, 2010). Studies have reported that REV3L rs462779 (p.Thr1224Ile) is associated with decreased breast cancer risk (Varadi et al., 2011), but increased colorectal cancer risk (Pan et al., 2012). Moreover, one study also observed that breast cancer patients with a polymorphic REV3L rs462779 allele had shorter time from surgery to an event (Varadi et al., 2011).
In our recent study on malignant mesothelioma, the REV3L rs462779 polymorphism conferred longer survival after cisplatin-based chemotherapy. The observed differences of the impact of REV1 and REV3L SNPs in different cancer types could partly be explained by differences in chemotherapy combinations and doses used for treatment of a particular malignancy. For example, compared to patients with malignant mesothelioma, patients with osteosarcoma had considerably different toxicity profiles as other chemotherapeutic agents also contribute to occurrence of adverse events. Moreover, other clinical factors might also contribute to differences in treatment response.
Because studies have shown that REV1 serves as a scaffold for other TLS polymerases (Tissier et al., 2004) and directly interacts with REV3L (Sharma et al., 2012), we also investigated the combined effect of both REV1 rs3087403 and REV3L rs462779 SNPs, associated with survival in single SNP analysis. Combination of REV1 rs3087403 and REV3L rs462779 polymorphic alleles was associated with further decrease in EFS and OS. In haplotype analysis, patients with the REV1 TGT haplotype tended to have shorter OS, while patients with the REV3L CGG haplotype tended to have shorter OS and EFS. These results were not adjusted for stage due to the small number of patients with late stage of disease. This could in part explain why the observed results that did not withstand the correction for multiple comparisons.
Although our study brings novel and interesting results, it is limited by the small sample size. This is mainly due to the fact that osteosarcoma is a very rare cancer, so large cohorts are difficult to obtain, especially in small populations. However, an advantage of our study was that all patients were from a ethnically homogenous population (Vidan-Jeras et al., 1998). Another limitation of our study is the fact that the patients were not all treated according to the same protocol. Even though cisplatin represents a cornerstone in osteosarcoma treatment, other chemotherapeutic agents contribute to treatment outcome, and validation studies should be limited to patients treated according to a single protocol.
Conclusions
The results of this first study investigating TLS polymerase genes in osteosarcoma identified TLS polymerase polymorphisms as a novel genetic factor associated with survival of osteosarcoma patients. Further studies are needed to validate our results and explore the potential role of TLS polymorphisms in personalization of osteosarcoma treatment.
Abbreviations Used
- CI
confidence interval
- EFS
event-free survival
- FDR
false discovery rate
- FFPE
formalin fixed, paraffin embedded
- HR
hazard ratio
- HWE
Hardy–Weinberg equilibrium
- ICL
interstrand crosslink
- MAF
minor allele frequency
- OS
overall survival
- Pol ζ
TLS polymerases polymerase zeta
- REV1
DNA polymerase REV1 (DNA directed)
- SNP
single nucleotide polymorphism
- TLS
translesion
Acknowledgments
The authors thank the staff of The Cancer Registry of Slovenia for providing information on all osteosarcoma patients in Slovenia. This work was financially supported by the Slovenian Research Agency (ARRS Grants No. P1-0170, P3-0343).
Author Disclosure Statement
This work was financially supported by the Slovenian Research Agency (ARRS Grants No. P1-0170, P3-0343). No competing financial interests exist.
References
- Benjamini Y, and Hochberg Y. (1995). Controlling the false dscovery rate—A practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57, 289–300 [Google Scholar]
- Biason P, Hattinger CM, Innocenti F, et al. (2012). Nucleotide excision repair gene variants and association with survival in osteosarcoma patients treated with neoadjuvant chemotherapy. Pharmacogenomics J 12, 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caronia D, Patino-Garcia A, Milne RL, et al. (2009). Common variations in ERCC2 are associated with response to cisplatin chemotherapy and clinical outcome in osteosarcoma patients. Pharmacogenomics J 9, 347–353 [DOI] [PubMed] [Google Scholar]
- Chou AJ, and Gorlick R. (2006). Chemotherapy resistance in osteosarcoma: Current challenges and future directions. Expert Rev Anticancer Therapy 6, 1075–1085 [DOI] [PubMed] [Google Scholar]
- Doles J, Oliver TG, Cameron ER, et al. (2010). Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci USA 107, 20786–20791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoiu M, Jiricny J, and Scharer OD. (2012). Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res 40, 8953–8964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erculj N, Kovac V, Hmeljak J, Franko A, Dodic-Fikfak M, and Dolzan V. (2012). The influence of gemcitabine pathway polymorphisms on treatment outcome in patients with malignant mesothelioma. Pharmacogenet Genomics 22, 58–68 [DOI] [PubMed] [Google Scholar]
- Goricar K, Kovac V, and Dolzan V. (2014a). Polymorphisms in translesion polymerase genes influence treatment outcome in malignant mesothelioma. Pharmacogenomics 15, 941–950 [DOI] [PubMed] [Google Scholar]
- Goricar K, Kovac V, Jazbec J, Zakotnik B, Lamovec J, and Dolzan V. (2014b). Influence of the folate pathway and transporter polymorphisms on methotrexate treatment outcome in osteosarcoma. Pharmacogenet Genomics 24, 514–521 [DOI] [PubMed] [Google Scholar]
- Goricar K, Kovac V, Jazbec J, Lamovec J, and Dolzan V. (2015). Homologous recombination repair polymorphisms and the risk for osteosarcoma. J Med Biochem 34, 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao T, Feng W, Zhang J, Sun YJ, and Wang G. (2012). Association of four ERCC1 and ERCC2 SNPs with survival of bone tumour patients. Asian Pac J Cancer Prev 13, 3821–3824 [DOI] [PubMed] [Google Scholar]
- He X, Ye F, Zhang J, Cheng Q, Shen J, and Chen H. (2008). REV1 genetic variants associated with the risk of cervical carcinoma. Eur J Epidemiol 23, 403–409 [DOI] [PubMed] [Google Scholar]
- Ho TV, and Scharer OD. (2010). Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen 51, 552–566 [DOI] [PubMed] [Google Scholar]
- Lange SS, Takata K, and Wood RD. (2011). DNA polymerases and cancer. Nat Rev Cancer 11, 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Okuda T, Trang J, and Howell SB. (2006). Human REV1 modulates the cytotoxicity and mutagenicity of cisplatin in human ovarian carcinoma cells. Mol Pharmacol 69, 1748–1754 [DOI] [PubMed] [Google Scholar]
- Makridakis NM, and Reichardt JK. (2012). Translesion DNA polymerases and cancer. Front Genet 3, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh S, McLeod H, Dolan E, et al. (2009). Platinum pathway. Pharmacogenet Genomics 19, 563–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Chi P, Lu X, and Xu Z. (2012). Genetic polymorphisms in translesion synthesis genes are associated with colorectal cancer risk and metastasis in Han Chinese. Gene 504, 151–155 [DOI] [PubMed] [Google Scholar]
- Quartuccio N, Treglia G, Salsano M, et al. (2013). The role of fluorine-18-fluorodeoxyglucose positron emission tomography in staging and restaging of patients with osteosarcoma. Radiol Oncol 47, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter J, and Bielack SS. (2010). Osteosarcoma. Ann Oncol 21, vii320–325 [DOI] [PubMed] [Google Scholar]
- Sakiyama T, Kohno T, Mimaki S, et al. (2005). Association of amino acid substitution polymorphisms in DNA repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int J Cancer 114, 730–737 [DOI] [PubMed] [Google Scholar]
- Salinas-Souza C, Petrilli AS, and de Toledo SR. (2010). Glutathione S-transferase polymorphisms in osteosarcoma patients. Pharmacogenet Genomics 20, 507–515 [DOI] [PubMed] [Google Scholar]
- Sharma S, and Canman CE. (2012). REV1 and DNA polymerase zeta in DNA interstrand crosslink repair. Environ Mol Mutagen 53, 725–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Hicks JK, Chute CL, et al. (2012). REV1 and polymerase zeta facilitate homologous recombination repair. Nucleic Acids Res 40, 682–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, and Cordonnier A. (2004). Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair (Amst) 3, 1503–1514 [DOI] [PubMed] [Google Scholar]
- Tregouet DA, and Garelle V. (2007). A new JAVA interface implementation of THESIAS: Testing haplotype effects in association studies. Bioinformatics 23, 1038–1039 [DOI] [PubMed] [Google Scholar]
- Varadi V, Bevier M, Grzybowska E, et al. (2011). Genetic variation in genes encoding for polymerase zeta subunits associates with breast cancer risk, tumour characteristics and survival. Breast Cancer Res Treat 129, 235–245 [DOI] [PubMed] [Google Scholar]
- Vidan-Jeras B, Jurca B, Dolzan V, Jeras M, Breskvar K, and Bohinjec M. (1998). Slovenian Caucasian normal. In: Gjertson D, Terasaki P. (Eds.), HLA 1998. American Society for Histocompatibility and Immunogenetics, Lenexa, Kansas, pp. 180–181 [Google Scholar]
- Woods D, and Turchi JJ. (2013). Chemotherapy induced DNA damage response: convergence of drugs and pathways. Cancer Biol Therapy 14, 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LM, Li XH, and Bao CF. (2012). Glutathione S-transferase P1 and DNA polymorphisms influence response to chemotherapy and prognosis of bone tumors. Asian Pac J Cancer Prev 13, 5883–5886 [DOI] [PubMed] [Google Scholar]
- Zhang S, Chen H, Zhao X, et al. (2013). REV3L 3'UTR 460 T>C polymorphism in microRNA target sites contributes to lung cancer susceptibility. Oncogene 32, 242–250 [DOI] [PubMed] [Google Scholar]